Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Polymerase-amplified release of ATP (POLARA) for detecting single nucleotide variants in RNA and DNA†
Michael G.
Mohsen
,
Debin
Ji
and
Eric T.
Kool
 *
*
Department of Chemistry, Stanford University, Stanford, CA 94305, USA. E-mail: kool@stanford.edu
First published on 30th January 2019
Abstract
The identification of single nucleotide polymorphisms (SNP) is increasingly important for diagnosis and treatment of disease. Here we studied the potential use of ATP-releasing nucleotides (ARNs) for identifying SNPs in DNA and RNA targets. Synthesized as derivatives of the four canonical deoxynucleotides, ARNs can be used in the place of deoxynucleoside triphosphates to elongate a primer hybridized to a nucleic acid template, with the leaving group being ATP rather than pyrophosphate. The released ATP is then harnessed in conjunction with luciferase to generate chemiluminescence. Extension on a long target DNA or RNA generates many equivalents of ATP per target strand, providing isothermal amplification of signal. In principle, allele-specific primers could be used in conjunction with ARNs to generate differential luminescence signals with respect to distinct genetic polymorphisms. To test this, varied primer designs, modifications, enzymes and conditions were tested, resulting in an optimized strategy that discriminates between differing nucleic acid templates with single nucleotide resolution. This strategy was then applied to diagnostically relevant alleles resulting in discrimination between known polymorphisms. SNP detection was successfully performed on transcribed mRNA fragments from four different alleles derived from JAK2, BCR-ABL1, BRAF, and HBB. To investigate background interference, wild-type and mutant transcripts of these four alleles were tested and found to be easily distinguishable amid total cellular RNA isolated from human blood. Thus, ARNs have been employed with specialized allele-specific primers to detect diagnostically important SNPs in a novel method that is sensitive, rapid, and isothermal.
Introduction
At the nucleotide level, genetic variation among all human individuals is estimated to be as low as 0.1–0.5%.1,2 However, given a genome size of 3.2 billion base pairs, this implies that several million bases differ from person to person. The vast majority of these disparities occur as single base substitutions, known as single nucleotide polymorphisms (SNP).3 By definition, SNPs are known substitutions occurring at specific genomic locations and they are becoming increasingly important as genetic markers.3,4 Most SNPs are of little biological consequence, and of those that are biologically functional, the majority are benign and can account for variations as innocuous as eye color.3 There is, however, a smaller subset of SNPs that are considered risk-associated alleles, which are linked to diseases.3Examples of medically relevant SNPs include JAK2 V617F (1849 G → T), BCR-ABL1 T315I (944 C → T), BRAF V600E (1799 T → A), and HBB V6E (20 A → T). In the case of the JAK2 V617F polymorphism, a change in the amino acid sequence from valine to phenylalanine causes Janus kinase 2 to be conformationally compromised.5 Patients who have this mutation suffer from polycythemia vera (PV), a form of myeloproliferative disease.5 This SNP occurs in approximately 95% of patients who suffer from PV, so it is imperative for physicians to identify patients who have the V617F mutation.5 The T315I mutation in the BCR-ABL1 fusion gene is known to provide resistance to Gleevec (Imanitib), a small-molecule inhibitor of leukemia-initiating BCR-ABL tyrosine kinase.6 Imatinib is the therapeutic standard for patients diagnosed with chronic myeloid leukemia (CML), but approximately one-third of CML patients have the BCR-ABL1 T315I mutation and are consequently resistant to this treatment.6 The BRAF V600E mutation is linked to a host of cancers including melanoma, non-small cell lung cancer, colorectal cancer, and thyroid cancer.7 The mutation from valine to glutamic acid at this position results in enhanced BRAF kinase activity and increased phosphorylation of downstream targets.8 Vemurafenib (Zelboraf) is a BRAF kinase inhibitor administered to cancer patients who test positive for the V600E mutation.9 Finally, the V6E mutation in the HBB gene is established as the cause of 60–70% of sickle cell disease cases in the United States.10 For all of these alleles and many others as well, the ability to detect SNPs is essential because the genetic differences affect disease diagnosis, prognosis, and treatment.
A rapid and accurate method for detecting SNPs is important in informing prognoses and treatments for patients who possess these genetic variations. For example, BRAF-mutated melanoma is known to manifest more aggressively in patients than BRAF wild-type melanoma.11BRAF-mutant tumors are more likely to metastasize to the brain than BRAF wild-type tumors, and are also linked to decreased likelihood of survival in patients with stage IV cancer.12 It is thus crucial to quickly determine whether melanoma patients have BRAF-mutated tumors to select the appropriate treatment.
Recent advances in high-throughput sequencing have significantly reduced costs of whole-genome analysis,13,14 but these techniques remain prohibitively expensive, require specialized equipment, and provide much more data than needed for SNP analysis.15 The most common genotyping method for SNP analysis is allele-specific PCR, which is more practical for assaying SNPs in a laboratory setting.16–19 The basis of allele-specific PCR involves thermostable polymerase extension of a primer only when the primer's 3′ end is perfectly complementary to the template.20 Discrimination is achieved by extension when the SNP is complementary to the primer at the 3′-terminal nucleotide, and inhibited extension in the case of a mismatch. However, allele-specific PCR requires thermal cycling equipment and necessitates the use of gel electrophoresis or real-time PCR technology for data readout.21 Because PCR is a sensitive method with risks of contamination and artifacts, it is carried out at specialized facilities separate from clinics, requiring added time and increasing costs. The often-urgent nature of medical diagnostic decision-making places value on more rapid techniques that may be less specialized and sensitive to conditions and contamination, and some point-of-care applications (such as in economically limited environments) may find it difficult to obtain real-time PCR equipment.
There are several existing methods for SNP detection that are isothermal and so do not require a thermal cycler.22 Pyrosequencing involves a sequence-by-synthesis approach in which nucleotide triphosphates (dNTPs) are added one at a time, and the released PPi is used to generate chemiluminescence through a cascade of enzymatic reactions.23,24 Padlock probe-initiated rolling circle amplification (RCA) uses the target nucleic acid as a ligation template, and can discriminate between targets at single nucleotide resolution.25,26 The Invader® assay utilizes an invader probe complementary to the polymorphic site, and two allele-specific probes complementary to either polymorphism with an overhang at their 5′ ends.27,28 When hybridized to a target, the invader and matched allele-specific probe form a structure recognized and cleaved by a flap endonuclease enzyme, causing a fluorescence signal.27,28 However, these methods are either expensive or tedious, require specialized technical experience to perform, and may not be readily available in a clinical laboratory setting.
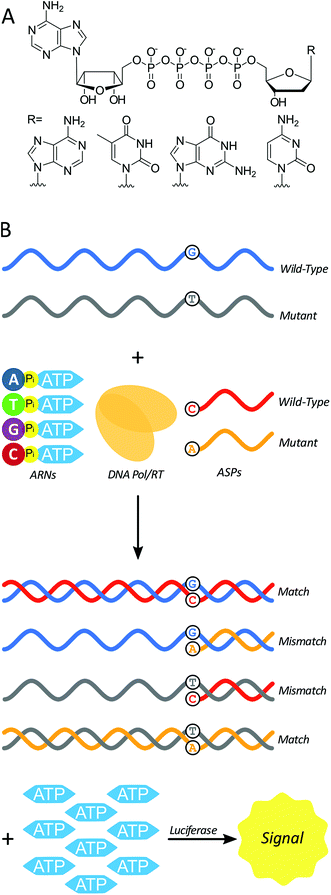
Here we test a new isothermal approach for amplified detection of SNPs. In this approach, luminescence signals are generated by release of ATP from chimeric nucleotides (ATP-releasing nucleotides (ARNs, Fig. 1A)) during polymerase extension of primers. For SNP discrimination, allele-specific oligonucleotide primers are used to discriminate between single-stranded nucleic acid targets differing by a single nucleotide. In the case of a match between an allele-specific probe (ASP) oligonucleotide and the target, extension of the probe to form double-stranded nucleic acid material also yields ATP as a by-product (Fig. 1B). In previous studies, polymerase with 3′ → 5′ exonuclease activity digested the primer starting from the 3′ end.29 As a result, allele-specific extension was not possible since the ASP's 3′ end contains the allele-specific residue. However, a polymerase lacking 3′ → 5′ exonuclease activity (exo-) should allow for primer extension without altering the ASP, and thus the possibility of single-nucleotide discrimination. The amount of ATP produced is stoichiometrically large, because there is one ATP molecule released for each ARN that is incorporated into the growing strand by polymerase. The produced ATP can then be quantified using a luciferase-based assay to generate a luminescence signal. In principle, this approach could be used to distinguish SNPs in DNAs (using a DNA polymerase) or in RNAs (using a reverse transcriptase). The proposed polymerase-amplified release of ATP (POLARA) method to detect SNPs is isothermal, time-efficient, simple in practice, and highly sensitive due to the large amount of signal generated from priming long genetic targets.
Results
In order to determine the feasibility of the POLARA method for SNP discrimination, we first constructed a model system using 50 nt synthetic single-stranded DNA targets. The sequences were excerpts from the four aforementioned genes (BRAF, JAK2, BCR-ABL1, HBB) containing the polymorphic site at the 18th nucleotide from the 5′ end. Corresponding 18mer synthetic allele-specific probe (ASP) oligonucleotides were designed to have the 3′-terminal base complementary to either the wild-type or mutant SNP. In total, 8 DNA targets and 8 ASPs were used, corresponding to the wild-type or mutant SNP for each allele. The components of a reaction included equimolar DNA target and ASP, the four ARNs, and Klenow exo- DNA polymerase.The initial results showed that in the case of a mismatch at the SNP site, the DNA polymerase does not extend the ASP, or extends it to a far lesser degree. Using a commercial luciferase-based assay to quantify the relative amounts of ATP in solution, luminescence signals ranged from 4.0-fold to 5.5-fold higher when ASPs and target DNA sequences were correctly matched, while mismatches generated signals that were only slightly above background luminescence (Fig. S1A†). As discussed in a previous study,29 background luminescence primarily arises from trace ATP quantities that could not be completely eliminated from ARNs during purification. After subtracting background luminescence from all luminescence signals, the average signal for matches between ASP and target was 29-fold greater than that of mismatches for JAK2 targets (Fig. S1B†). Clear discrimination between SNPs was achieved with 50mers corresponding to all four alleles. To test whether this approach would be suitable for long targets, two 500 nt single-stranded DNAs corresponding to wild-type and mutant BRAF sequences were generated by PCR and strand separation (see ESI for details†). Under similar reaction conditions that succeeded for short targets, wild-type and mutant 500 nt single-stranded BRAF DNA targets could also be distinguished using this approach, yielding a 10-fold increase in signal for matches over mismatches after subtracting background luminescence (Fig. S2A†). Though significantly longer, signals generated from 500 nt targets were approximately the same magnitude as those of 50 nt targets (Fig. S2B†). This could be a result of secondary structure in the long single-stranded DNAs, which could impede polymerase from extending the ASP fully. The match-to-mismatch signal ratio for longer targets decreased somewhat relative to that of 50 nt targets, possibly due to mispriming events on the longer, more complex sequences.
After success with DNA targets, long single-stranded RNA targets corresponding to fragments of the allele mRNAs (87–500 nt in length) were transcribed from appropriate DNAs containing T7 RNA polymerase promoters as a more realistic model system for cellular mRNAs. We generated both wild-type and SNP mutant RNA transcripts for each allele. The resulting synthetic RNAs are human mRNA mimics as they contain identical sequence information, albeit in truncated form. To detect SNPs in the mRNA mimics, the approach was modified from that of DNA targets to use reverse transcriptase (RT) lacking RNase H activity in place of DNA polymerase. Initial experiments with Maxima H Minus RT under similar conditions to the DNA targets resulted in satisfactory discrimination between SNPs for JAK2 and BRAF mRNA mimics (Fig. S4†). However, attaining selectivity for BCR-ABL1 and HBB targets proved unsuccessful. It was hypothesized that secondary structure of long RNAs could interfere with the ability of the ASP to hybridize to the target at the SNP site, limiting ASP extension and thus positive signal. Additionally, since RT only requires a few hybridized base pairs to initiate DNA synthesis, even transient nonspecific hybridization of the ASP 3′ end elsewhere on the target RNA could result in nondiscriminatory signals.
To address these issues, the reaction conditions and ASP designs were re-examined in order to increase specificity. Firstly, dCp4A was replaced with dCTP in order to improve signal to noise ratios as described in a previous study.29 By varying incubation time with working targets, it was found that optimal selectivity for matches over mismatches was observed after 30 minutes (Fig. S5†). Modulating the incubation temperature of the reaction yielded increases in signal as the temperature approached 55 °C, although selectivity suffered (Fig. S6†). This trade-off is likely due to the optimal temperature of Maxima H Minus RT (55 °C) being higher than the ASP melting temperature (Tm) (45 °C on average). Thus 37 °C was selected as the temperature for the reaction. Pre-incubating the reaction mixture at room temperature for 0–90 minutes to allow for slow hybridization had no discernible effect on signal or selectivity (Fig. S7†). Thermal annealing of the ASP on the target also gave no change in selectivity (Fig. S8†). To determine whether accidental 3′-end homology could be a cause of selectivity issues, five different ASPs were used where the first was fully complementary to the target at the polymorphic site, and the other four had the last 6 nucleotides at the 3′ end conserved, as this sequence was repeated a few times throughout the target, and the remaining 12 nucleotides randomly selected using a random number generator. The fully complementary ASP provided nondiscriminatory signal regardless of match or mismatch, while the scrambled ASPs did not generate any signal over background (Fig. S9†). This provided evidence against the hypothesis that non-specific hybridization was contributing substantially to signal.
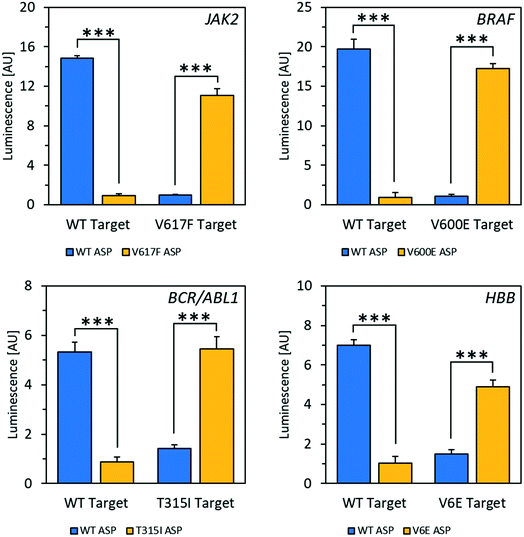
Using the mfold web server to predict folding of the target RNAs showed that there was likely an extensive degree of self-complementarity near the polymorphic site.30 In order to outcompete secondary structure, locked nucleic acid (LNA)31 nucleotides were introduced into the ASPs to increase affinity between probe and target.32 Several LNA-containing ASPs were designed with 1–2 LNA nucleotides at or near the 3′ end (Fig. S10†) as well as one with four LNA bases evenly spaced throughout (Fig. S11†). Placing the LNA nucleotides directly at the ASP's 3′ end, which is the most common design used in allele-specific PCR, resulted in decreased specificity (Fig. S10†).33 The disparity observed in this case may be because the modification negatively affects RT's ability to differentiate residues with specificity on RNA templates, while allele-specific PCR is typically performed with DNA polymerase on DNA templates. However, including an LNA residue at the second position from the 3′ end enhanced specificity to an acceptable level (Fig. S10†); with 2 LNA nucleotides at the second and third positions from the 3′ end, specificity was improved dramatically. Using the optimized reaction conditions and ASPs containing two LNA nucleotides, excellent selectivity for matches over mismatches was observed in mRNA fragments for all four alleles, with signal increases ranging from 4.3-fold to 18-fold (Fig. 2). Discrimination between wild-type and mutant targets could be reliably determined down to a target RNA concentration of 2.5 nM for JAK2 and 1 nM for BRAF (Fig. S12†). This latter concentration corresponds to 25 femtomoles of target at the test volume. We also tested a strategy using one of the alleles to increase signal for matches over mismatches via a double-extension experiment, wherein after 30 minutes of extension by RT, RNase H was used to digest target RNA hybridized to DNA. After this, a primer complementary to the 5′ end of the resulting cDNA was added along with DNA polymerase and incubated for another 30 minutes. For the JAK2 allele, this resulted in a 29-fold increase in signal for matches over mismatches, improved from 14-fold for a single extension (Fig. S14†). Therefore, this double extension appears to be a viable approach for improving signal over background.
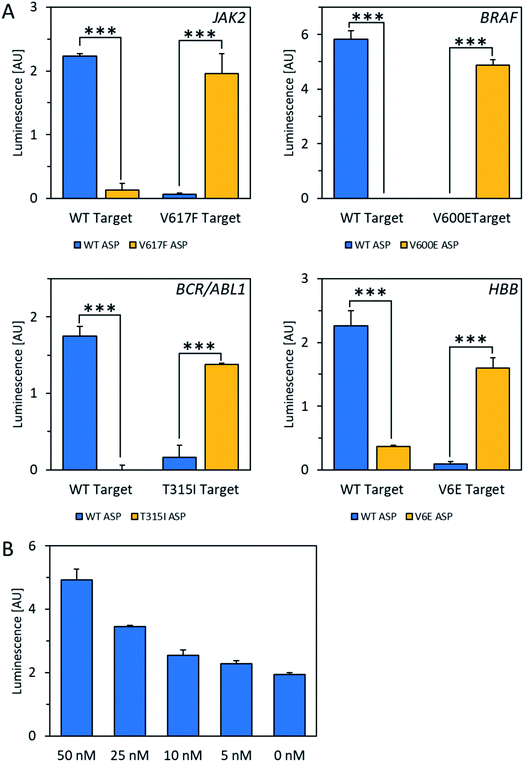
For the POLARA method to be viable as a diagnostic tool, it should have the capacity to detect the target RNA from a pool of total cellular RNA derived from blood, which could cause background interference. To test this issue, total cellular RNA was extracted from a human blood sample and spiked with wild-type and mutant RNA fragments from each of the four alleles. The data show that in all cases, wild-type and mutant RNAs could clearly be discriminated using the standard single extension technique at a concentration of 50 nM amid total cellular RNA (Fig. 3A). After subtracting background luminescence, signal increases for matches over mismatches ranged from 8.5-fold for HBB/beta-globin to 20- to 22-fold for BCR-ABL1 and JAK2. Mismatches for the BRAF allele were discriminated with even higher specificity, yielding no signal over background, while the matched primers yielded robust signals. Additionally, in dilution experiments the wild-type HBB mRNA fragment could be detected above background amid total cellular RNA down to a concentration of 5 nM of the test allele (Fig. 3B). Thus, we conclude that total cellular RNA from blood does not interfere either with allele discrimination or sensitivity over the concentrations tested.
Taken together, our data show that the POLARA approach offers clear single-nucleotide discriminating ability and succeeds in identifying four clinically important alleles even in the background of total cellular RNA. In comparison, AS-PCR performed by gel electrophoresis intrinsically provides a qualitative readout, where mismatch discrimination is assessed by the presence or absence of a target band.16,34 More recently, allele-specific real-time PCR (RT-PCR) makes use of fluorescent dyes that generate increasing signal as amplification proceeds.35 Amplification is observed in the case of both a match and a mismatch but occurs earlier for matches.36 This effect is described quantitatively by measuring the difference in cycle threshold (ΔCT) for matched vs. mismatched primers and targets.36 Although ΔCT is highly dependent on primer design, allele, and several other factors, a ΔCT of ca. 5–7 is typical for a single mismatch relative to a perfect match using a primer designed to bind to the polymorphic site at its 3′ end.36 A ΔCT of 5–7 corresponds to a maximal 32 to 128-fold signal increase assuming that the number of amplicons is doubled with every cycle of PCR. Although such fold changes associated with AS-PCR are greater than that of POLARA for three of the four alleles tested here, the signal increase observed with POLARA is obvious and easy to interpret since both alleles are tested side by side. In the case of the BRAF allele tested here, essentially complete specificity was realized, yielding no signal over background for mismatches, and highlighting POLARA's potential as an especially promising method for detecting the V600E mutation.
Conclusions
Although more studies will be needed to test authentic mRNA targets in a series of clinical samples, these early studies suggest the combination of POLARA and specialized allele-specific primers as a novel approach to genetic diagnosis of SNPs. In principle, this method is highly generalizable, as ASPs can easily be synthesized complementary to any SNP site. It is also low-cost and time-efficient; the entire reaction can be completed in 45–75 minutes and is carried out without thermal cycling. A luminometer is needed to measure luminescence produced, and such equipment is common in biomedical laboratories due to the popularity of luminescent reporter assays. Comparatively, AS-PCR is considerably slower, requiring several hours to complete, and necessitates the use of gel electrophoresis or real-time PCR instrumentation.21 The new method has the potential to significantly decrease the time and cost of characterizing SNPs in a clinical setting. Future studies will test this possibility.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank the U.S. National Institutes of Health (GM130704 and CA217809) for support.Notes and references
- D. N. Cooper, B. A. Smith, H. J. Cooke, S. Niemann and J. Schmidtke, Hum. Genet., 1985, 69, 201–205 CrossRef CAS PubMed.
- S. Levy, G. Sutton, P. C. Ng, L. Feuk, A. L. Halpern, B. P. Walenz, N. Axelrod, J. Huang, E. F. Kirkness, G. Denisov, Y. Lin, J. R. MacDonald, A. W. C. Pang, M. Shago, T. B. Stockwell, A. Tsiamouri, V. Bafna, V. Bansal, S. A. Kravitz, D. A. Busam, K. Y. Beeson, T. C. McIntosh, K. A. Remington, J. F. Abril, J. Gill, J. Borman, Y. H. Rogers, M. E. Frazier, S. W. Scherer, R. L. Strausberg and J. C. Venter, PLoS Biol., 2007 DOI:10.1371/journal.pbio.0050254.
- P. Y. Kwok and X. Chen, Curr. Issues Mol. Biol., 2003 DOI:10.1007/978-1-60327-411-1.
- A. Vignal, D. Milan, M. SanCristobal and A. Eggen, Curr. Opin. Hematol., 2002, 34, 275 CAS.
- R. L. Levine and D. G. Gilliland, Curr. Opin. Hematol., 2007, 14, 43–47 CrossRef CAS PubMed.
- D. Bixby and M. Talpaz, Leukemia, 2011, 25, 7–22 CrossRef CAS PubMed.
- H. Davies, G. R. Bignell, C. Cox, P. Stephens, S. Edkins, S. Clegg, J. Teague, H. Woffendin, M. J. Garnett, W. Bottomley, N. Davis, E. Dicks, R. Ewing, Y. Floyd, K. Gray, S. Hall, R. Hawes, J. Hughes, V. Kosmidou, A. Menzies, C. Mould, A. Parker, C. Stevens, S. Watt, S. Hooper, H. Jayatilake, B. A. Gusterson, C. Cooper, J. Shipley, D. Hargrave, K. Pritchard-Jones, N. Maitland, G. Chenevix-Trench, G. J. Riggins, D. D. Bigner, G. Palmieri, A. Cossu, A. Flanagan, A. Nicholson, J. W. C. Ho, S. Y. Leung, S. T. Yuen, B. L. Weber, H. F. Seigler, T. L. Darrow, H. Paterson, R. Wooster, R. Wooster, M. R. Stratton and P. A. Futreal, Nature, 2002, 417, 949–954 CrossRef CAS PubMed.
- P. T. C. Wan, M. J. Garnett, S. M. Roe, S. Lee, D. Niculescu-Duvaz, V. M. Good, C. G. Project, C. M. Jones, C. J. Marshall, C. J. Springer, D. Barford and R. Marais, Cell, 2004 DOI:10.1016/S0092-8674(04)00215-6.
- K. M. Ilieva, I. Correa, D. H. Josephs, P. Karagiannis, I. U. Egbuniwe, M. J. Cafferkey, J. F. Spicer, M. Harries, F. O. Nestle, K. E. Lacy and S. N. Karagiannis, Mol. Cancer Ther., 2014, 13, 2769–2783 CrossRef CAS.
- M. Bender, Sickle Cell Disease, 1993 Search PubMed.
- L. Cheng, A. Lopez-Beltran, F. Massari, G. T. Maclennan and R. Montironi, Mod. Pathol., 2018, 31, 24–38 CrossRef CAS PubMed.
- A. Ribas and K. T. Flaherty, Nat. Rev. Clin. Oncol, 2011 Search PubMed.
- J. Shendure and H. Ji, Nat. Biotechnol, 2008, 26, 1135–1145 CrossRef CAS PubMed.
- A. Rokas and P. Abbot, Trends Ecol. Evol., 2009, 24, 192–200 CrossRef PubMed.
- C. P. Van Tassell, T. P. L. Smith, L. K. Matukumalli, J. F. Taylor, R. D. Schnabel, C. T. Lawley, C. D. Haudenschild, S. S. Moore, W. C. Warren and T. S. Sonstegard, Nat. Methods, 2008, 5, 247–252 CrossRef CAS PubMed.
- C. R. Newton, A. Graham, L. E. Heptinstall, S. J. Powell, C. Summers, N. Kalsheker, J. C. Smith and A. F. Markham, Nucleic Acids Res., 1989 DOI:10.1093/nar/17.7.2503.
- R. S. Cha, H. Zarbl, P. Keohavong and W. G. Thilly, Genome Res., 1992, 2, 14–20 CrossRef CAS.
- K. R. Tindall and T. A. Kunkel, Biochemistry, 1988, 27, 6008–6013 CrossRef CAS.
- J. Liu, S. Huang, M. Sun, S. Liu, Y. Liu, W. Wang, X. Zhang, H. Wang and W. Hua, Plant Methods, 2012 DOI:10.1186/1746-4811-8-34.
- J. Petruska, M. F. Goodman, M. S. Boosalis, L. C. Sowers, C. Cheong and I. Tinoco, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 6252–6256 CrossRef CAS.
- A. H. Lang, H. Drexel, S. Geller-Rhomberg, N. Stark, T. Winder, K. Geiger and A. Muendlein, J. Mol. Diagn., 2011, 13, 23–28 CrossRef CAS PubMed.
- S. Kim and A. Misra, Annu. Rev. Biomed. Eng., 2007, 9, 289–320 CrossRef CAS PubMed.
- P. Nyrén, B. Pettersson and M. Uhlén, Anal. Biochem., 1993, 208, 171–175 CrossRef PubMed.
- T. Langaee and M. Ronaghi, Mutat. Res., Fundam. Mol. Mech. Mutagen., 2005, 573, 96–102 CrossRef CAS PubMed.
- J. Banér, M. Nilsson, M. Mendel-Hartvig and U. Landegren, Nucleic Acids Res., 1998, 26, 5073–5078 CrossRef.
- M. G. Mohsen and E. T. Kool, Acc. Chem. Res., 2016 DOI:10.1021/acs.accounts.6b00417.
- V. Lyamichev, A. L. Mast, J. G. Hall, J. R. Prudent, M. W. Kaiser, T. Takova, R. W. Kwiatkowski, T. J. Sander, M. De Arruda, D. A. Arco, B. P. Neri and M. A. D. Brow, Nat. Biotechnol., 1999, 17, 292–296 CrossRef CAS PubMed.
- M. Olivier, Mutat. Res., Fundam. Mol. Mech. Mutagen., 2005, 573, 103–110 CrossRef CAS PubMed.
- D. Ji, M. G. Mohsen, E. M. Harcourt and E. T. Kool, Angew. Chem., Int. Ed., 2016, 55, 2087–2091 CrossRef CAS PubMed.
- M. Zuker, Nucleic Acids Res., 2003 DOI:10.1093/nar/gkg595.
- A. A. Koshkin, P. Nielsen, M. Meldgaard, V. K. Rajwanshi, S. K. Singh and J. Wengel, J. Am. Chem. Soc., 1998, 120, 13252–13253 CrossRef CAS.
- Y. You, B. G. Moreira, M. A. Behlke and R. Owczarzy, Nucleic Acids Res., 2006 DOI:10.1093/nar/gkl175.
- D. Latorra, K. Campbell, A. Wolter and J. M. Hurley, Hum. Mutat., 2003, 22, 79–85 CrossRef CAS PubMed.
- S. Ayyadevara, J. J. Thaden and R. J. Shmookler Reis, Anal. Biochem., 2000, 284, 11–18 CrossRef CAS PubMed.
- C. A. Heid, J. Stevens, K. J. Livak and P. M. Williams, Genome Res., 1996 DOI:10.1006/meth.2001.1260.
- S. Lefever, F. Pattyn, J. Hellemans and J. Vandesompele, Clin. Chem., 2013, 59, 1470–1480 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8sc03901a |
| This journal is © The Royal Society of Chemistry 2019 |