Open Access Article
Open Access ArticleThe anti-apoptotic proteins NAF-1 and iASPP interact to drive apoptosis in cancer cells†
Anat
Iosub-Amir‡
a,
Fang
Bai‡
 b,
Yang-Sung
Sohn
c,
Luhua
Song
d,
Sagi
Tamir
c,
Henri-Baptiste
Marjault
c,
Guy
Mayer
a,
Ola
Karmi
c,
Patricia A.
Jennings
e,
Ron
Mittler
b,
Yang-Sung
Sohn
c,
Luhua
Song
d,
Sagi
Tamir
c,
Henri-Baptiste
Marjault
c,
Guy
Mayer
a,
Ola
Karmi
c,
Patricia A.
Jennings
e,
Ron
Mittler
 f,
José N.
Onuchic
*b,
Assaf
Friedler
f,
José N.
Onuchic
*b,
Assaf
Friedler
 *a and
Rachel
Nechushtai
*c
*a and
Rachel
Nechushtai
*c
aInstitute of Chemistry, The Hebrew University of Jerusalem, Edmond J. Safra Campus at Givat Ram, Jerusalem 91904, Israel. E-mail: assaf.friedler@mail.huji.ac.il
bCenter for Theoretical Biological Physics, Department of Physics, Rice University, Houston, TX 77005, USA. E-mail: jonuchic@rice.edu
cThe Alexander Silberman Institute of Life Science, The Hebrew University of Jerusalem, Edmond J. Safra Campus at Givat Ram, Jerusalem 91904, Israel. E-mail: rachel@mail.huji.ac.il
dDepartment of Biological Sciences, University of North Texas, Denton, TX 76203, USA
eDepartment of Chemistry & Biochemistry, University of California at San Diego, La Jolla, CA 92093, USA
fDepartment of Surgery, University of Missouri School of Medicine, Christopher S. Bond Life Sciences Center, University of Missouri, 1201 Rollins St, Columbia, MO 65201, USA
First published on 20th November 2018
Abstract
Suppression of apoptosis is a key Hallmark of cancer cells, and reactivation of apoptosis is a major avenue for cancer therapy. We reveal an interaction between the two anti-apoptotic proteins iASPP and NAF-1, which are overexpressed in many types of cancer cells and tumors. iASPP is an inhibitory member of the ASPP protein family, whereas NAF-1 belongs to the NEET 2Fe–2S protein family. We show that the two proteins are stimulated to interact in cells during apoptosis. Using peptide array screening and computational methods we mapped the interaction interfaces of both proteins to residues 764–778 of iASPP that bind to a surface groove of NAF-1. A peptide corresponding to the iASPP 764–780 sequence stabilized the NAF-1 cluster, inhibited NAF-1 interaction with iASPP, and inhibited staurosporine-induced apoptosis activation in human breast cancer, as well as in PC-3 prostate cancer cells in which p53 is inactive. The iASPP 764–780 IC50 value for inhibition of cell death in breast cancer cells was 13 ± 1 μM. The level of cell death inhibition by iASPP 764–780 was altered in breast cancer cells expressing different levels and/or variants of NAF-1, indicating that the peptide activity is associated with NAF-1 function. We propose that the interaction between iASPP and NAF-1 is required for apoptosis activation in cancer cells. This interaction uncovers a new layer in the highly complex regulation of cell death in cancer cells and opens new avenues of exploration into the development of novel anticancer drugs that reactivate apoptosis in malignant tumors.
Introduction
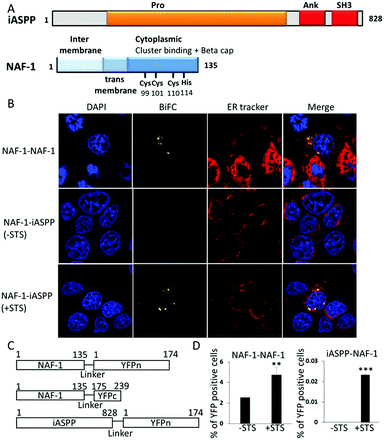
Protein–protein interactions (PPI) are at the core of numerous cell death pathways such as apoptosis, autophagy, and necrosis.1,2 Affecting specific PPI involved in these pathways can change the fate of cells. For example, inhibiting the interactions of anti-apoptotic proteins can result in promoting apoptotic cell death. Here we show that iASPP and NAF-1, two proteins that are important inhibitors of cell death mechanisms in cancer cells, interact with each other, participate in apoptosis regulation, and could be targeted to induce apoptosis in cancer cells.The anti-apoptotic iASPP protein is a member of the ASPP (apoptosis stimulating proteins of p53) protein family and has an important role in regulating p53 dependent apoptosis.3–5 iASPP also has a role in regulating other important cellular processes such as autophagy and senescence.6,7 iASPP is considered a promising anti-cancer drug target as it is frequently upregulated in many different types of cancers.8,9 Over-expression of iASPP is associated with chemo resistance and cancer malignancy and is correlated with poor prognosis for patients.3,10–13 iASPP, an 828 residue protein, contains an intrinsically disordered Proline rich domain (Pro) at its N terminus, four Ankyrin repeats (Ank) and a Src Homology 3 (SH3) domain at its C terminus (Fig. 1A).5,14,15 iASPP interacts with numerous apoptosis-related proteins such as the p53 protein family14 and NF-κB,16,17 mainly through its Ank–SH3 C-terminal domains. We previously showed that the important residues for iASPP interactions with other proteins are iASPP 739–753 and iASPP 764–778.15 Recently it was shown that the Pro domain of iASPP binds the proteins Keap1 and CEP55.18,19 iASPP PPIs are regulated by an auto-inhibitory interaction between its Pro domain and Ank SH3 domains (Fig. 1A). This interaction is regulated by caspase cleavage and phosphorylation of the Pro domain.15,16,20,21
NAF-1 (nutrient-deprivation autophagy factor-1) belongs to the 2Fe–2S cluster-binding NEET protein family.22,23 NAF-1 is important for the regulation of lifespan, autophagy and apoptosis, and alters reactive oxygen species (ROS), iron and calcium levels in cells.23–28 A homozygous G109C mutation in the CISD2 gene, which causes early termination of the protein, results in the autosomal recessive disorder Wolfram syndrome 2.29 Many types of human cancer cells and tissues show high levels of NAF-1 expression and suppressing the level of NAF-1 using shRNA results in reduced cell proliferation and suppressed tumor growth. High levels of NAF-1 predict poor prognosis of cancer patients.26,30–32 Overexpression of NAF-1 in xenograft breast cancer tumors resulted in increased tumor size compared to control tumors.33 NAF-1 is a homodimeric protein containing a trans-membrane domain and a CDGSH 2Fe–2S domain. NAF-1 binds its cluster by four coordinating residues: Cys99, Cys101, Cys110 and His114 (Fig. 1A).22 The most studied interaction of NAF-1 is with the anti-apoptotic protein Bcl-2, which regulates apoptosis and autophagy. We mapped the interaction between NAF-1 and Bcl-2 and found that NAF-1 binds Bcl-2 through its soluble cytoplasmic region using a groove on its surface and a large number of amino acids near the cluster.27,34 NAF-1 can transfer its cluster to other proteins but the exact role of this transfer is unclear. Altering NAF-1 cluster stability shows multiple effects on the proliferation of cancer cells.33 The mutation H114C in NAF-1, which leads to a highly stable cluster, results in much smaller tumors compared to tumors with wild type NAF-1 protein.33 The Bcl-2 16–30 peptide, derived from the interaction interface between NAF-1 and Bcl-2, destabilizes the NAF-1 cluster.34
Here we show that NAF-1 and iASPP interact in cancer cells during apoptosis. We mapped the binding interfaces in both proteins and found that iASPP residues 764–778 are important for this interaction. A peptide derived from iASPP (iASPP 764–780) stabilized the NAF-1 2Fe–2S cluster, inhibited NAF-1 interaction with iASPP and inhibited apoptosis. The iASPP 764–780 peptide inhibited cell death with an IC50 value of 13 ± 1 μM, and an EC50 value of 7.5 ± 0.6 μM, with maximal inhibition of 80%. This inhibition appeared to be dependent on the level and function of NAF-1 in cells, but not on p53. Because NAF-1 and iASPP were previously shown to be involved in regulating the oxidative stress capacity of cells,18,23 we propose that designing molecules to regulate their PPI could be an important research avenue for controlling cancer cell proliferation, the suppression of apoptosis and the oxidative state of cells.
Results
Binding of NAF-1 to iASPP in cancer cells
To determine whether NAF-1 interacts with iASPP in vivo we conducted a bimolecular fluorescence complementation (BiFC) analysis for this interaction in MCF-7 human epithelial breast cancer cells. Because NAF-1 is a homodimer,23 we could use its own dimerization interaction as a positive control for the BiFC assay. When the split NAF-1–YFPc and NAF-1–YFPn fusion proteins were co-expressed in MCF-7 cells they displayed a BiFC signal that was primarily localized to the ER (Fig. 1B–D). In contrast, when the split NAF-1–YFPc and iASPP–YFPn fusion proteins were co-expressed in MCF-7 cells, no BiFC signal could be detected (Fig. 1B–D). A BiFC signal was however detected in cells co-expressing the split NAF-1–YFPc and iASPP–YFPn fusion proteins one hour following the application of staurosporine (STS, 1 μM), a non-specific kinase inhibitor that activates apoptosis and causes oxidative stress in cells.35 The BiFC signal obtained in STS-treated cells co-expressing NAF-1–YFPc and iASPP–YFPn was at least partially localized to the ER (Fig. 1B–D). Taken together, the findings presented in Fig. 1 demonstrate that NAF-1 interacts with iASPP in cancer cells following the activation of apoptosis by STS, and that this interaction occurs, in part, at the ER.Mapping iASPP Ank–SH3 interactions with NAF-1
To validate the protein–protein interaction detected in vivo by BiFc (Fig. 1) we tested whether NAF-1 interacts with iASPP Ank–SH3in vitro using a nickel affinity pulldown assay. NAF-1 interacts with HLT–iASPP Ank–SH3 bound beads and does not interact with controls (Fig. 2A). These results indicate that iASPP Ank–SH3 binds NAF-1 in vitro. To determine which regions of iASPP interact with NAF-1, we used peptide arrays to map the NAF-1 binding sites in iASPP (Fig. 2B and S1†). NAF-1 was screened for binding an array of 91 overlapping peptides derived from iASPP. Out of these candidates, 67 peptides were derived from the iASPP Pro domain and 24 were derived from iASPP Ank–SH3 (Tables S1 and S2†). NAF-1 bound two peptides derived from iASPP Ank–SH3, iASPP 739–753, which is part of the fourth Ankyrin repeat, and iASPP 764–778, which is a part of the iASPP RT loop, connecting the last two beta strands in the SH3 domain (Fig. 2B and E). NAF-1 did not bind any peptides derived from the iASPP Pro domain (Fig. S1†). The mutant NAF-1 H114C, an important control protein, bound only iASPP 764–778 but not iASPP 739–753 (Fig. 2C and E). We therefore focused on studying the iASPP 764–778 interaction with NAF-1. Alanine scanning experiments, in which we systematically replaced each residue of iASPP 764–778 with Alanine and tested their binding to NAF-1, were performed using peptide array screening.36 The results showed that three acidic and one aromatic residues are the most important for iASPP 764–778 interaction with NAF-1: E772, F773, D775, and E776. Altering each of these residues to Alanine abolished iASPP 764–778 binding to NAF-1 (Fig. 2D, Table S3†). We used the binding site identification tool, Fragment-docking and Direct Coupling Analysis (Fd-DCA),37,38 and the PPI docking tool Z-Dock39 to construct a model of the binding complex between NAF-1 and iASPP (Fig. 2F). Based on scanning of the fragment-sized molecular probes, the areas on the surfaces of NAF-1 and iASPP showing the strongest binding affinities were identified (Fig. S2†). A co-evolutionary analysis of multiple sequence alignments (MSAs) generated for the two proteins, further identified pairs of binding sites between them that demonstrated the highest co-evolutionary score. The detailed direct information (DI) values of each pair of residues from the selected pairs of binding sites are listed in Table S4† (Fig. 2G). The residues with the highest DI are the most directly coupled residue–residue interactions. The identified binding interface between the two proteins includes iASPP 764–778 and represents the strongest co-evolutionary interface interaction score. Both DCA and the Alanine scan found iASPP F773 to be important for this interaction. The PPI docking simulation tool Z-Dock was further used to construct the binding complex between NAF-1 and iASPP (Fig. 2F).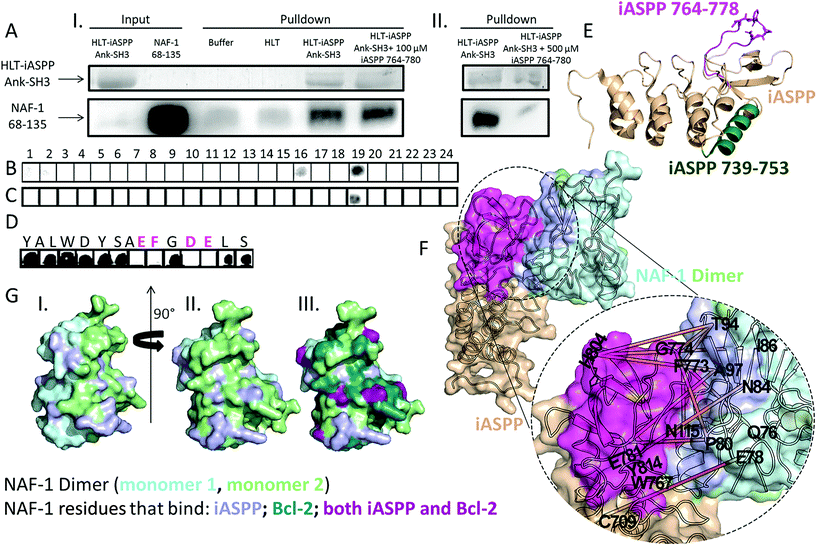 | ||
Fig. 2 Mapping the binding sites between iASPP Ank–SH3 and NAF-1. (A) iASPP Ank–SH3 interacts with NAF-1 68–135 in vitro. Nickel affinity pulldown of NAF-1 by HLT–iASPP Ank–SH3. (I) SDS page gel Coomassie staining results for HLT–iASPP Ank–SH3 and Western blot results for NAF-1. NAF-1 eluted from the Ni beads in the presence of HLT–iASPP Ank–SH3 at a 75% higher intensity than in the controls of Ni beads alone or Ni beads bound to HLT. The presence of 100 μM iASPP 764–780 did not affect iASPP binding to NAF-1 (ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 NAF-1 dimer 2.5 NAF-1 dimer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) iASPP 764–780). (II) The presence of 500 μM iASPP 764–780 inhibited iASPP interaction with NAF-1 and reduced it by 85% (ratio of 1 iASPP 764–780). (II) The presence of 500 μM iASPP 764–780 inhibited iASPP interaction with NAF-1 and reduced it by 85% (ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12.5 NAF-1 dimer 12.5 NAF-1 dimer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) iASPP 764–780); (B–D) peptide array screening. Dark spots indicate binding peptides. (B) Binding of NAF-1 and (C) NAF-1 H114C to an array of peptides derived from iASPP Ank–SH3. NAF-1 bound peptides derived from iASPP Ank–SH3 residues 739–753 and 764–778. The NAF-1 H114C mutant bound only iASPP 764–778. For peptide sequences see Tables S1 and S2.† (D) Binding of NAF-1 to an array of Alanine scan peptides derived from iASPP 764–778. For peptide sequences see Table S3.† Each residue replaced by Alanine is indicated above the relevant spot. The most important residues for NAF-1 interaction with iASPP 764–778 are labelled in magenta. (E) Cartoon view of iASPP Ank–SH3 (PDB 2VGE14). iASPP 739–753 is colored green, iASPP 764–778 colored magenta, and essential residues E772, F773, D775 and E776 are in sticks view; (F) the predicted binding complex between NAF-1 (light cyan and light green) and iASPP (beige). The identified binding site of iASPP, which includes residues 764–778, is highlighted in magenta, and the predicted binding site of NAF-1 is highlighted in light purple. The orange lines represent the most correlated residue pairs across iASPP and NAF-1 identified binding site (Table S4†); (G) surface view (also rotated 90°) (I and II) of NAF-1 69–132 (PDB ID 4OO7 (ref. 54)). The monomers are colored light cyan and light green. NAF-1 residues that bind iASPP in the DCA analysis (Table S4†) are colored light purple. In (III) NAF-1 residues shown to bind Bcl-2 (ref. 34) are colored teal, residues that bind iASPP are colored light purple and residues that bind both proteins are colored purple; (E and G) were generated using PyMOL.55 iASPP 764–780); (B–D) peptide array screening. Dark spots indicate binding peptides. (B) Binding of NAF-1 and (C) NAF-1 H114C to an array of peptides derived from iASPP Ank–SH3. NAF-1 bound peptides derived from iASPP Ank–SH3 residues 739–753 and 764–778. The NAF-1 H114C mutant bound only iASPP 764–778. For peptide sequences see Tables S1 and S2.† (D) Binding of NAF-1 to an array of Alanine scan peptides derived from iASPP 764–778. For peptide sequences see Table S3.† Each residue replaced by Alanine is indicated above the relevant spot. The most important residues for NAF-1 interaction with iASPP 764–778 are labelled in magenta. (E) Cartoon view of iASPP Ank–SH3 (PDB 2VGE14). iASPP 739–753 is colored green, iASPP 764–778 colored magenta, and essential residues E772, F773, D775 and E776 are in sticks view; (F) the predicted binding complex between NAF-1 (light cyan and light green) and iASPP (beige). The identified binding site of iASPP, which includes residues 764–778, is highlighted in magenta, and the predicted binding site of NAF-1 is highlighted in light purple. The orange lines represent the most correlated residue pairs across iASPP and NAF-1 identified binding site (Table S4†); (G) surface view (also rotated 90°) (I and II) of NAF-1 69–132 (PDB ID 4OO7 (ref. 54)). The monomers are colored light cyan and light green. NAF-1 residues that bind iASPP in the DCA analysis (Table S4†) are colored light purple. In (III) NAF-1 residues shown to bind Bcl-2 (ref. 34) are colored teal, residues that bind iASPP are colored light purple and residues that bind both proteins are colored purple; (E and G) were generated using PyMOL.55 | ||
iASPP 764–780 inhibits iASPP interaction with NAF-1 and stabilizes the cluster of NAF-1
We tested the effect of the peptide derived from iASPP 764–780 on the interaction of iASPP with NAF-1 using a pull-down assay. When we incubated NAF-1 with iASPP Ank–SH3 and 500 μM iASPP 764–780 (ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12.5 NAF-1 dimer
12.5 NAF-1 dimer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) iASPP 764–780), the iASPP 764–780 peptide inhibited iASPP binding to NAF-1 by 85% (Fig. 2A). Using CABS-dock, a docking server that performs docking of peptides to proteins without prior knowledge of the binding site,40 a model for iASPP 764–780 mode-of-binding to NAF-1 was generated (Fig. 3A and B). The peptide is found in an extended conformation, tightly bound to NAF-1, with substantial similarity to the binding surface between the full length NAF-1–iASPP, shown in Fig. 2F. The peptide is wrapped around the NAF-1 protein. Molecular interactions between the NAF-1 and the iASPP 764–780 peptide are indicated. These include hydrogen bonds between T106 and D775, Q76 and D768, as well as N79 and W767 (dash lines, Fig. 3A), π–π interactions formed between Y98 and F773, as well as between F107, W102 and Y769. Two other very possible interactions are hydrogen bond between E772 and N84, and a ionic bond between E776 and K95 (Fig. 3A). The interactions that were indicated by the Alanine scan (Fig. 2D; E772, F773, D775 and E776) are circled by a dashed line in the structure. According to the computational mode-of-binding, the iASPP 764–780 protects the NAF-1 2Fe–2S clusters by forming π–π interactions between NAF-1 H114 and iASPP Y764 and iASPP F779. These interactions stabilize H114 and its coordination with the 2Fe–2S clusters (Fig. 3B). This result underscored the importance of aromatic interactions in protecting the integrity of NAF-1, keeping it as holo–NAF-1. To validate the model, and because the stability of the NAF-1 cluster affects cell proliferation,33,34,41 we tested NAF-1 clusters' stability in the absence or presence of the iASPP 764–780 peptide. The iASPP 764–780 peptide stabilized the NAF-1 cluster and in its presence 80% of the clusters were still bound to NAF-1 after 800 min (in contrast, no clusters were found to be bound to NAF-1 following 500 min without the peptide; Fig. 3C).
iASPP 764–780), the iASPP 764–780 peptide inhibited iASPP binding to NAF-1 by 85% (Fig. 2A). Using CABS-dock, a docking server that performs docking of peptides to proteins without prior knowledge of the binding site,40 a model for iASPP 764–780 mode-of-binding to NAF-1 was generated (Fig. 3A and B). The peptide is found in an extended conformation, tightly bound to NAF-1, with substantial similarity to the binding surface between the full length NAF-1–iASPP, shown in Fig. 2F. The peptide is wrapped around the NAF-1 protein. Molecular interactions between the NAF-1 and the iASPP 764–780 peptide are indicated. These include hydrogen bonds between T106 and D775, Q76 and D768, as well as N79 and W767 (dash lines, Fig. 3A), π–π interactions formed between Y98 and F773, as well as between F107, W102 and Y769. Two other very possible interactions are hydrogen bond between E772 and N84, and a ionic bond between E776 and K95 (Fig. 3A). The interactions that were indicated by the Alanine scan (Fig. 2D; E772, F773, D775 and E776) are circled by a dashed line in the structure. According to the computational mode-of-binding, the iASPP 764–780 protects the NAF-1 2Fe–2S clusters by forming π–π interactions between NAF-1 H114 and iASPP Y764 and iASPP F779. These interactions stabilize H114 and its coordination with the 2Fe–2S clusters (Fig. 3B). This result underscored the importance of aromatic interactions in protecting the integrity of NAF-1, keeping it as holo–NAF-1. To validate the model, and because the stability of the NAF-1 cluster affects cell proliferation,33,34,41 we tested NAF-1 clusters' stability in the absence or presence of the iASPP 764–780 peptide. The iASPP 764–780 peptide stabilized the NAF-1 cluster and in its presence 80% of the clusters were still bound to NAF-1 after 800 min (in contrast, no clusters were found to be bound to NAF-1 following 500 min without the peptide; Fig. 3C).
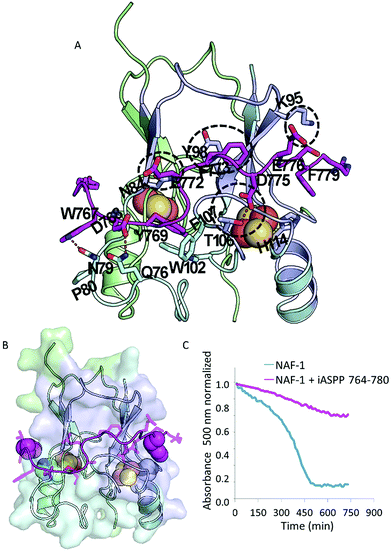 | ||
| Fig. 3 The iASPP 764–780 peptide stabilizes the NAF-1 clusters. (A + B) The mode-of-binding of the iASPP 764–780 peptide to the NAF-1 protein as found by computational docking. iASPP 764–780 is shown in magenta as an extended molecule that wraps across both 2Fe–2S cluster motifs. The NAF-1 dimer is presented in light green and light cyan. The identified protein–protein interaction region of NAF-1 with iASPP is highlighted in light purple. The iron–sulfur clusters are shown as orange and yellow spheres. (A) Cartoon and stick views indicating the interactions between iASPP 764–780 and NAF-1. The important interaction identified by the Alanine scan (Fig. 2D) are circled in dashed lines. (B) Cartoon and surface view of NAF-1. The critical Histidine cluster-coordinating ligands of NAF-1 are shown in white and blue spheres, the aromatic groups (magenta spheres) of the iASPP peptide interact with the Histidines (H114) of NAF-1. Such interactions potentially stabilize/protect the clusters from detachment from the NAF-1 protein. (C) Cluster stability assay of NAF-1 in the presence of 8 μM iASPP 764–780 (magenta), compared to the cluster stability without any peptide added (blue). Adding the iASPP 764–780 peptide significantly stabilized the NAF-1 holo protein. | ||
iASPP 764–780 inhibits apoptosis in cancer cells in a NAF-1-dependent manner
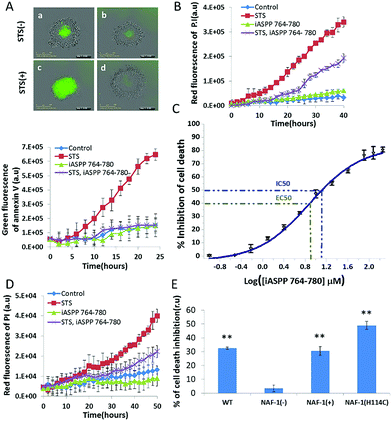
Both iASPP and NAF-1 inhibit apoptosis in cancer cells. As shown above, iASPP and NAF-1 are induced to interact upon activation of apoptosis (Fig. 1). Since iASPP 764–780 inhibits the interaction of iASPP with NAF-1 in vitro (Fig. 2A), we tested whether the iASPP 764–780 peptide affects apoptosis activation in cancer cells. The iASPP 764–780 peptide penetrated MDA-MB-231 cells but did not kill them (Fig. S3†). The iASPP 764–780 peptide did however inhibit STS-induced apoptosis of MDA-MB-231 spheroids (Fig. 4A and B). The IC50 value of iASPP 764–780 for inhibiting cell death in MDA-MB-231 cells was determined to be 13 ± 1 μM, with an EC50 of 7.5 ± 0.6 μM and maximal inhibition reaching about 80% (Fig. 4C). Since the peptide is derived from iASPP, which also binds and inhibits p53 induced apoptosis,3 we introduced the peptide into p53 null PC-3 prostate cancer cells. The peptide significantly inhibited the STS-induced cell death of PC-3 spheroids (Fig. 4D). To test whether apoptosis inhibition by the peptide is associated with NAF-1 in cancer cells, we used four different MDA-MB-231 breast cancer cell lines with altered expression of NAF-1: wild type, NAF-1 suppressed or over-expressed, and H114C mutant over-expressed.33 The iASPP 764–780 peptide (30 μM) inhibited cell death of MDA-MB-231 spheroids with WT or over-expressed levels of NAF-1 by ca. 30%. In spheroids with over-expressed H114C mutant the inhibition was ca. 50%. In contrast, no inhibition was observed in MDA-MB-231 cells in which NAF-1 expression was suppressed (Fig. 4E and S4†). These results are consistent with the hypothesis that the iASPP 764–780 peptide function in cells is associated with NAF-1 presence and/or function.Discussion
NAF-1 and iASPP were both recently discovered,3,29 with new details regarding their role in regulating multiple pathways in cells still emerging.9,18,33,42 Here we show for the first time that iASPP and NAF-1 interact with each other in vitro and in cells during STS-induced apoptosis. Our results suggest that at least some of the NAF-1–iASPP interactions that occur in cells during apoptosis activation occur at the ER.The surfaces at which NAF-1 binds to iASPP are known to be important for regulatory interactions between iASPP Ank–SH3 and iASPP Pro.15 The Alanine scan of iASPP 764–780 binding to NAF-1 identified iASPP E772, F773, D775, and E776 as important residues for the peptide–protein interactions. The iASPP 764–780 peptide – NAF-1 protein model indeed identified these residues as iASPP binding residues forming hydrogen, ionic, and π–π interactions with NAF-1. The important binding interface between NAF-1 and iASPP at the protein level, identified by our Fd-DCA method, which also found F773 important for the iASPP–NAF-1 interaction, overlaps with the model at the peptide level of iASPP 764–780 bound to NAF-1. The peptide–protein model suggests that the peptide has an extended conformation and can interact with both 2Fe–2S cluster domains, while the full protein can interact with only one. According to the DCA analysis, 11 out of the 20 most correlated residue pairs (highest DI) between NAF-1 and iASPP overlapped with the identified binding sites of the two proteins, i.e., site 2 of iASPP and site 1 of NAF-1 (Fig. 2S,† highlighted in red in Table S4†). The only previously mapped interaction of NAF-1 is with Bcl-2. Some of the NAF-1 residues that bind iASPP, were found to bind Bcl-2,34 these included P80, N84, T94, T106, and N115. The binding surface between NAF-1 and Bcl-2 is much larger than the binding surface between NAF-1 and iASPP and contains 23 residues (Fig. 2G). Our findings suggest that this part of the surface of NAF-1 is important for its cell death regulation activity.
The iASPP 764–780 peptide inhibited the interaction of iASPP with NAF-1 and stabilized the cluster of NAF-1. Altering the stability of the NAF-1 clusters was shown to result in suppressed cell proliferation or death of cancer cells.33,41 In the current study the iASPP 764–780 peptide inhibited apoptosis of cancer cells, possibly by inhibiting the NAF-1–iASPP interaction. The iASPP 764–780 peptide inhibited STS-induced cell death of both MDA-MB-231 and PC-3 cancer cells. The peptide did not act by inhibiting p53, the known target of iASPP, since p53 in the PC-3 cells is inactive due to a frame shift mutation.43 Furthermore, iASPP 764–780 inhibited cell death in PC-3 cells more than in MDA-MB-231 cells. The IC50 value for cell death inhibition of iASPP 764–780 in MDA-MB-231 was found to be 13 ± 1 μM, with EC50 value of 7.5 ± 0.6 μM and a maximal efficacy of about 80%. iASPP 764–780 showed different levels of cell death inhibition in cells with different NAF-1 levels or function, and specifically did not display cell death inhibition of cells that have suppressed expression of NAF-1, suggesting that the mode of iASPP 764–780 action is associated with NAF-1.
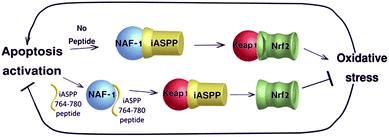
Both iASPP and NAF-1 inhibit apoptosis and are involved in regulating oxidative stress and both are associated with the anti-oxidative protein Nrf2.18,25,44 Nrf2 is one of the master regulators of oxidative stress responses that can function as a driver or suppressor of cancer cell proliferation.45 Nrf2 activity is regulated by Keap1 that induces ubiquitin–proteasome degradation of Nrf2. Nrf2 degradation is quickly reduced upon oxidative stress, leading to Nrf2 accumulation and nuclear translocation.46 iASPP was found to activate Nrf2 by binding to its inhibitor Keap1, resulting in lower ROS, decrease in cell death and higher drug resistance of renal cell carcinoma.18 Cells with low NAF-1 expression showed increased ROS47 and high levels of Nrf2 protein and activity.25,44 As both proteins are related to Nrf2, we speculate that the newly discovered interaction described here may regulate Nrf2 activity. By doing so, the iASPP–NAF-1 interaction may allow the cell to undergo or resist apoptosis in an oxidative stress-dependent manner. When NAF-1 binds iASPP, Keap1 is free to inhibit Nrf-2 activity resulting in enhanced oxidative stress. In the presence of the iASPP 764–780 peptide, iASPP interaction with NAF-1 is inhibited and iASPP is free to bind Keap1, which results in Nrf2 accumulation and activation, decreased ROS levels and inhibition of STS-induced apoptosis (Fig. 5).
Conclusions
Although both iASPP and NAF-1 are important inhibitors of apoptosis, their interaction that drives apoptosis in cancer cells, reveals another mechanism within the highly complex network of cell death regulation. Compounds that inhibit NAF-1–iASPP interaction could therefore act as lead compounds against diseases that are caused by excessive apoptosis of cells, such as neurodegenerative diseases.48Methods
BiFC
The C-terminal (aa 175-239) and N-terminal (aa 1-174) fragments of YFP were PCR amplified from pEYFP-Pds1Δdestruction box (383) which was a gift from Jonathon Pines (Addgene plasmid # 39848), sequenced, and cloned as an in-frame fusion to pEGFP-N1 containing NAF-1 or iASPP.49 The linker sequence DPRSIAT was used for connecting the NAF-1 or iASPP proteins to the YFPc/YFPn fragments. MCF-7 cells plated in RPMI medium 1640 (11875-093, gibco) with 10% FBS and 100 U mL−1 penicillin, 100 μg mL−1 streptomycin, and 0.25 μg mL−1 amphotericin B on coverslips coated with poly-L-lysine solution (p4707-50 mL, sigma) were co-transfected with the different fusion protein constructs, using the GeneJuice Transfection Reagent (70967-3, Novagen; 49). Following staining with the ER-Tracker Red Dye (E34250, Invitrogen), complete FluoroBrite DMEM (A18967-01, Gibco) medium was used to replace the RPMI medium 1640, and confocal images were obtained using a Zeiss LSM 710 confocal microscope. YFP, ER-Tracker and DAPI were excited by a 415 nm Argon, 561 nm diode and 405 nm diode laser respectively, and their fluorescence was detected at 508–604 nm, 585–700 nm and 400–580 nm, respectively.Nickel affinity pulldown assay
30 μL nickel–NTA beads (Qiagen) beads were incubated with: (1) 500 μL buffer A (20 mM Tris pH = 8, 100 mM NaCl, 60 mM Imidazo); (2) 400 μL 27 μM HLT (in buffer A) or (3) 175 μL 38 μM HLT–iASPP Ank–SH3 (in buffer A) for 1 hour with gentle mixing at 4 °C. The samples were centrifuged (2 min 3500 RPM), and washed with buffer A (5 min, 4 °C, gentle mixing). 350 μL of 40 μM NAF-1 68–135 dimer in buffer A were added to all of the samples. In the samples that were incubated with the peptide, the 350 μL NAF-1 also contained iASPP 764–780 in a concentration of 100 μM or 500 μM. The nickel–NTA beads and NAF-1 68–135 were gently mixed for 2 hours at 4 °C. The samples were centrifuged, and the supernatants were collected. The samples were washed three times with 1 mL buffer A and the third washes were kept and analyzed. The beads were eluted by boiling them in SDS sample solution. Samples were analyzed on a 12% SDS-PAGE gel. iASPP Ank–SH3 was detected using Coomassie blue dye. NAF-1 68–135 was detected using western blot with rabbit anti-NAF-1 primary antibody and HRP conjugated mouse anti-rabbit secondary antibody. Western blots were analyzed using ImageJ software.Peptide array
The CelluSpotsTM peptide micro-arrays were synthesized by INTAVIS Bioanalytical Instruments AG, Köln, Germany. The 15 residue peptides were acetylated at their N-termini and attached to a cellulose membrane via their C termini through an amide bond. For screening the binding of the array to His6–NAF-1 the array was first washed for 4 h at room temperature with 50 mM Tris–HCl pH = 8, 150 mM NaCl, 0.05% Tween 20 (TBST) and 2.5% (W/V) skimmed milk (blocking buffer) for blocking unspecific binding. 10 μM of His6–NAF-1 68–135/His6–NAF-1 68–135 H114C were dissolved in blocking buffer. 5 mL of the proteins were incubated with the arrays at 4 °C with shaking overnight. After three washes with TBST, the array was incubated with anti His HRP conjugated antibody at room temperature for 1 h. The antibody was dissolved in blocking buffer. The array was then washed again three times with TBST. Immunodetection was performed using chemiluminescence (ECL reagents). For screening iASPP binding to peptides derived from NAF-1, 5 μM of HLT–iASPP Ank–SH3 were dissolved in TBST with 3% (w/v) skimmed milk.Computational reconstruction of the NAF-1–iASSP binding complex
To reconstruct the NAF-1–iASPP binding complex, our in-house protein–protein interaction prediction method Fd-DCA (Fragment docking-Direct Coupling Analysis;50) was used to identify the binding sites for NAF-1 and iASPP. Based on the screening of the molecular probes on the surface of iASPP (PDB code: 2VGE) and NAF-1 (PDB code: 4OO7), four different candidate binding sites with diverse shapes and sizes were found for iASPP, and one site was found on the surface of the soluble domain of NAF-1 (Fig. S3†). Site 2 of iASPP is the site which includes the sequence of residues 764–780. DCA analysis of the two proteins further identified the most correlated 20 residue pairs (highest DI; Table S4†). Site 2 of iASPP and site 1 of NAF-1 presented the strongest co-evolutionary interface interaction score, suggesting that NAF-1 may interact with the identified site 2 of iASPP via its site 1. To further obtain the molecular binding complex for these two proteins, we used the correlated residue pairs, which overlap with the residues on the confirmed binding sites (highlight in Table S3†), as the index of the contacting residues and performed a protein–protein docking simulation by using Z-Dock (version 3.0.2; 51). The parameters were set as default. As a result, the highest ranked binding complex of NAF1/iASPP was obtained as shown in Fig. 2F.Peptides synthesis labeling and purification
Peptides were synthesized on a Liberty Blue Microwave Assisted Peptide Synthesizer (CEM) using standard Fmoc chemistry and DIC/Oxyma as coupling reagents. The peptides were labeled with 5(6)-carboxyfluorescein at their N termini as described51 and cleaved from the resin as described.52 The peptides were purified on a MERCK-Hitachi HPLC using a reverse-phase C8 semi-preparative column with a gradient of ACN/TDW. ESI mass spectrometry and analytical HPLC were used to verify the identity and purity of the peptides (Fig. S5†).Cluster stability assay
Cluster stability was performed as described in ref. 34. Specifically 8 μM of NAF-1 and 8 μM of iASPP 764–780 were used.Binding mode prediction for iASPP 764–780 with NAF-1
To predict the binding of iASPP 764–780 with NAF-1, the flexible peptide-docking online tool CABS-dock (http://biocomp.chem.uw.edu.pl/CABSdock)53 was adopted to perform the simulation. NAF-1 structure (PDB code: 4OO7) and the sequence of iASPP 764–780 (YALWDYSAEFGDELSFR) were submitted as input, and the other parameters were kept as default.Cell culture
Malignant breast epithelial cells (MDA-MB-231) and human prostate cancer cells (PC-3) were grown at 37 °C under 5% CO2 in RPMI 1640 medium supplemented with 10% fetal calf serum, L-glutamine, and antibiotics containing penicillin, streptomycin, and amphotericin B (Biological Industries). Both cell types were plated at a density of 2 × 103 in a 96 well ULA plate (Corning 7007) for 3D-spheroid experiments. To form 3D-spheroids, the cells were further cultured for 3 days.Cells death by STS experiments
Staurosporine (STS)-induced cytotoxicity of 3D-spheroids of MDA-MB-231 cancer cells was evaluated using the IncuCyte Zoom system (Essen Bioscience). MDA-MB-231 cancer cells were seeded in a 96 well ULA plate (Corning 7007) at a density of 2 × 103, and allowed to culture for an additional 3 days to form the spheroids. The resulting spheroids were then treated with STS and the annexin V FITC reagent (abcam) to detect apoptosis, or propidium iodide reagent (abcam) to detect cell death. 30 μM of peptide was added in each experiment (iASPP 764–780). Cell images were recorded every two hours, and the resulting green color indicating apoptosis and red color, reflecting cell death, were analyzed with the IncuCyte software. This allowed the evaluation of the time-dependent apoptosis and cell death of cells.Determination of IC50 and EC50
MDA-MB-231 cells were seeded at a density of 10 × 103 in a 96 well Nunc plate and treated with iASPP 764–780 at the series of concentrations (0 μM, 0.1 μM, 0.3 μM, 0.6 μM, 1.25 μM, 2.5 μM, 5 μM, 10 μM, 20 μM, 40 μM, 80 μM, 120 μM, 180 μM), together with STS (staurosporine, 1μM) and PI probes (propidium iodide, abcam) for tracing STS induced cell death. Images were recorded every two hours using the IncuCyte Zoom System and analysed with the IncuCyte software. The presented results were taken following 24 hours incubation. % of cell death inhibition was calculated in reference to cells without the peptide. The % of cell death inhibition was plotted against log of the peptide concentration. The IC50 and EC50 values were calculated by fitting the plot to the Hill equation:| Y = B + ((T − B)/(1 + 10(log(EC50) − X) × Hill slope)) |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Science Foundation NSF-BSF funding NSF-MCB-1613462 (R. M.) and BSF (Binational Science Foundation (BSF) Grant no. 2015831 (R. N.). R. N. and A. F. acknowledge the support of the MINERVA Center for Bio-hybrid Complex Systems. A. F. was also supported by grants from the Israel Science Foundation and the Israel Cancer Research Foundation (ICRF). A. I. was supported by the Dalia and Dan Maydan Fellowship for advanced degree students at the Hebrew University of Jerusalem. Work at the laboratory of PAJ is supported by the National Institutes of Health Grant GM101467. J. N. O. was supported by the Cancer Prevention and Research Institute of Texas (CPRIT-grant R1110), by the Center for Theoretical Biological Physics sponsored by the NSF (Grant PHY-1427654) and by NSF-CHE 1614101. The funders had no role in the design, data collection, analysis, decision to publish, or preparation of the manuscript.References
- E. M. Phizicky and S. Fields, Microbiol. Rev., 1995, 59, 94–123 CAS.
- N. London, B. Raveh and O. Schueler-Furman, Curr. Opin. Chem. Biol., 2013, 17, 952–959 CrossRef CAS PubMed.
- D. Bergamaschi, Y. Samuels, N. J. O'Neil, G. Trigiante, T. Crook, J.-K. K. Hsieh, D. J. O'Connor, S. Zhong, I. Campargue, M. L. Tomlinson, P. E. Kuwabara and X. Lu, Nat. Genet., 2003, 33, 162–167 CrossRef CAS PubMed.
- A. Iosub-Amir and A. Friedler, MedChemComm, 2014, 5, 1435–1443 RSC.
- E. A. Slee, S. Gillotin, D. Bergamaschi, C. Royer, S. Llanos, S. Ali, B. Jin, G. Trigiante and X. Lu, Oncogene, 2004, 23, 9007–9016 CrossRef CAS PubMed.
- M. Notari, Y. Hu, S. Koch, M. Lu, I. Ratnayaka, S. Zhong, C. Baer, A. Pagotto, R. Goldin, V. Salter, E. Candi, G. Melino and X. Lu, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16645–16650 CrossRef CAS PubMed.
- A. Chikh, P. Sanzà, C. Raimondi, O. Akinduro, G. Warnes, G. Chiorino, C. Byrne, C. A. Harwood and D. Bergamaschi, J. Cell Sci., 2014, 127, 3079–3093 CrossRef CAS PubMed.
- H. S. Bell and K. M. Ryan, Cancer Res., 2008, 68, 4959–4962 CrossRef CAS PubMed.
- P. Dong, K. Ihira, J. Hamada, H. Watari, T. Yamada, M. Hosaka, S. J. B. Hanley, M. Kudo and N. Sakuragi, OncoTargets Ther., 2015, 6, 19968–19975 Search PubMed.
- X. Zhang, M. Wang, C. Zhou, S. Chen and J. Wang, Leuk. Res., 2005, 29, 179–183 CrossRef CAS PubMed.
- L. Jiang, M. K. Y. Siu, O. G. W. Wong, K.-F. F. Tam, X. Lu, E. W.-F. Lam, H. Y. S. Ngan, X.-F. F. Le, E. S. Y. Wong, L. J. Monteiro, H.-Y. Y. Chan and A. N. Y. Cheung, Clin. Cancer Res., 2011, 17, 6924–6933 CrossRef CAS PubMed.
- Z. Liu, X. Zhang, D. Huang, Y. Liu, X. Zhang, L. Liu, G. Li, Y. Dai, H. Tan, J. Xiao and Y. Tian, Med. Oncol., 2012, 29, 3381–3388 CrossRef CAS PubMed.
- P. Dong, Y. Xiong, H. Watari, S. J. Hanley, Y. Konno, K. Ihira, F. Suzuki, T. Yamada, M. Kudo, J. Yue and N. Sakuragi, Sci. Rep., 2016, 6, 35480 CrossRef CAS PubMed.
- R. A. Robinson, X. Lu, E. Y. Jones and C. Siebold, Structure, 2008, 16, 259–268 CrossRef CAS PubMed.
- A. Iosub-Amir, M. van Rosmalen, G. Mayer, M. Lebendiker, T. Danieli and A. Friedler, Sci. Rep., 2015, 5, 11629 CrossRef PubMed.
- Y. Hu, W. Ge, X. Wang, G. Sutendra, K. Zhao, Z. Dedeić, E. A. Slee, C. Baer and X. Lu, OncoTargets Ther., 2015, 6, 42478–42490 Search PubMed.
- J. Yang, M. Hori, T. Sanda and T. Okamoto, J. Biol. Chem., 1999, 274, 15662–15670 CrossRef CAS PubMed.
- W. Ge, K. Zhao, X. Wang, H. Li, M. Yu, M. He, X. Xue, Y. Zhu, C. Zhang, Y. Cheng, S. Jiang and Y. Hu, Cancer Cell, 2017, 32, 561–573 CrossRef CAS PubMed.
- K. Gao, Y. Zhang, Q. Shi, J. Zhang, L. Zhang, H. Sun, D. Jiao, X. Zhao, H. Tao, Y. Wei, Y. Wang, H. Saiyin, S. M. Zhao, Y. Li, P. Zhang and C. Wang, Cell Death Dis., 2018, 9, 528 CrossRef PubMed.
- M. Lu, H. Breyssens, V. Salter, S. Zhong, Y. Hu, C. Baer, I. Ratnayaka, A. Sullivan, N. R. Brown, J. Endicott, S. Knapp, B. M. Kessler, M. R. Middleton, C. Siebold, E. Y. Jones, E. V. Sviderskaya, J. Cebon, T. John, O. L. Caballero, C. R. Goding and X. Lu, Cancer Cell, 2013, 23, 618–633 CrossRef CAS PubMed.
- H. Benyamini and A. Friedler, J. Mol. Recognit., 2011, 24, 266–274 CrossRef CAS PubMed.
- A. R. Conlan, H. L. Axelrod, A. E. Cohen, E. C. Abresch, J. Zuris, D. Yee, R. Nechushtai, P. A. Jennings and M. L. Paddock, J. Mol. Biol., 2009, 11, 143–153 CrossRef PubMed.
- S. Tamir, M. L. Paddock, M. Darash-Yahana-Baram, S. H. Holt, Y. S. Sohn, L. Agranat, D. Michaeli, J. T. Stofleth, C. H. Lipper, F. Morcos, I. Z. Cabantchik, J. N. Onuchic, P. a. Jennings, R. Mittler and R. Nechushtai, Biochim. Biophys. Acta, Mol. Cell Res., 2014, 1853, 1294–1315 CrossRef PubMed.
- Y. F. Chen, C. H. Kao, R. Kirby and T. F. Tsai, Autophagy, 2009, 5, 1043–1045 CrossRef CAS PubMed.
- S. H. Holt, M. Darash-Yahana, Y. S. Sohn, L. Song, O. Karmi, S. Tamir, D. Michaeli, Y. Luo, M. L. Paddock, P. A. Jennings, J. N. Onuchic, R. K. Azad, E. Pikarsky, I. Z. Cabantchik, R. Nechushtai and R. Mittler, J. Cell Sci., 2016, 129, 155–165 CrossRef CAS PubMed.
- S.-M. Li, C.-H. Chen, Y.-W. Chen, Y.-C. Yen, W.-T. Fang, F.-Y. Tsai, J.-L. Chang, Y.-Y. Shen, S.-F. Huang, C.-P. Chuu, I.-S. Chang, C. A. Hsiung and S. S. Jiang, Sci. Rep., 2017, 7, 11893 CrossRef PubMed.
- N. C. Chang, M. Nguyen, M. Germain and G. C. Shore, EMBO J., 2010, 29, 606–618 CrossRef CAS PubMed.
- C.-H. Wang, Y.-F. Chen, C.-Y. Wu, P.-C. Wu, Y.-L. Huang, C.-H. Kao, C.-H. Lin, L.-S. Kao, T.-F. Tsai and Y.-H. Wei, Hum. Mol. Genet., 2014, 23, 4770–4785 CrossRef CAS PubMed.
- S. Amr, C. Heisey, M. Zhang, X.-J. Xia, K. H. Shows, K. Ajlouni, A. Pandya, L. S. Satin, H. El-Shanti and R. Shiang, Am. J. Hum. Genet., 2007, 81, 673–683 CrossRef CAS PubMed.
- Y.-S. Sohn, S. Tamir, L. Song, D. Michaeli, I. Matouk, A. R. Conlan, Y. Harir, S. H. Holt, V. Shulaev, M. L. Paddock, A. Hochberg, I. Z. Cabanchick, J. N. Onuchic, P. A. Jennings, R. Nechushtai and R. Mittler, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 14676–14681 CrossRef CAS PubMed.
- B. Chen, S. Shen, J. Wu, Y. Hua, M. Kuang, S. Li and B. Peng, Int. J. Clin. Exp. Pathol., 2015, 8, 13725–13738 CAS.
- L. Liu, M. Xia, J. Wang, W. Zhang, Y. Zhang and M. He, Med. Oncol., 2014, 31, 183 CrossRef PubMed.
- M. Darash-Yahana, Y. Pozniak, M. Lu, Y.-S. Sohn, O. Karmi, S. Tamir, F. Bai, L. Song, P. A. Jennings, E. Pikarsky, T. Geiger, J. N. Onuchic, R. Mittler and R. Nechushtai, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 10890–10895 CrossRef CAS PubMed.
- S. Tamir, S. Rotem-Bamberger, C. Katz, F. Morcos, K. L. Hailey, J. A. Zuris, C. Wang, A. R. Conlan, C. H. Lipper, M. L. Paddock, R. Mittler, J. N. Onuchic, P. A. Jennings, A. Friedler and R. Nechushtai, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 5177–5182 CrossRef CAS PubMed.
- C. A. Belmokhtar, J. Hillion and E. Ségal-Bendirdjian, Oncogene, 2001, 20, 3354–3362 CrossRef CAS PubMed.
- R. Gabizon, O. Faust, H. Benyamini, S. Nir, A. Loyter and A. Friedler, MedChemComm, 2013, 4, 252–259 RSC.
- F. Morcos, A. Pagnani, B. Lunt, A. Bertolino, D. S. Marks, C. Sander, R. Zecchina, J. N. Onuchic, T. Hwa and M. Weigt, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, E1293–E1301 CrossRef CAS PubMed.
- F. Bai, F. Morcos, R. R. Cheng, H. Jiang and J. N. Onuchic, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E8051–E8058 CrossRef CAS PubMed.
- B. G. Pierce, K. Wiehe, H. Hwang, B.-H. Kim, T. Vreven and Z. Weng, Bioinformatics, 2014, 30, 1771–1773 CrossRef CAS PubMed.
- M. Kurcinski, M. Jamroz, M. Blaszczyk, A. Kolinski and S. Kmiecik, Nucleic Acids Res., 2015, 43, W419–W424 CrossRef CAS PubMed.
- F. Bai, F. Morcos, Y.-S. Sohn, M. Darash-Yahana, C. O. Rezende, C. H. Lipper, M. L. Paddock, L. Song, Y. Luo, S. H. Holt, S. Tamir, E. A. Theodorakis, P. A. Jennings, J. N. Onuchic, R. Mittler and R. Nechushtai, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 3698–3703 CAS.
- C.-Y. Y. Wu, Y.-F. F. Chen, C.-H. H. Wang, C.-H. H. Kao, H.-W. W. Zhuang, C.-C. C. Chen, L.-K. K. Chen, R. Kirby, Y.-H. H. Wei, S.-F. F. Tsai and T.-F. F. Tsai, Hum. Mol. Genet., 2012, 21, 3956–3968 CrossRef CAS PubMed.
- W. B. Isaacs, B. S. Carter and C. M. Ewing, Cancer Res., 1991, 51, 4716–4720 CAS.
- L. Cheng, B. Yan, K. Chen, Z. Jiang, C. Zhou, J. Cao, W. Qian, J. Li, L. Sun, J. Ma, Q. Ma and H. Sha, Oxid. Med. Cell. Longevity, 2018, 9482018 Search PubMed.
- M. Rojo de la Vega, E. Chapman and D. D. Zhang, Cancer Cell, 2018, 34, 21–43 CrossRef CAS PubMed.
- A. Kobayashi, M.-I. Kang, H. Okawa, M. Ohtsuji, Y. Zenke, T. Chiba, K. Igarashi and M. Yamamoto, Mol. Cell. Biol., 2004, 24, 7130–7139 CrossRef CAS PubMed.
- S. E. Wiley, A. Y. Andreyev, A. S. Divakaruni, R. Karisch, G. Perkins, E. A. Wall, P. van der Geer, Y. F. Chen, T. F. Tsai, M. I. Simon, B. G. Neel, J. E. Dixon and A. N. Murphy, EMBO Mol. Med., 2013, 5, 904–918 CrossRef CAS PubMed.
- B. Favaloro, N. Allocati, V. Graziano, C. Di Ilio and V. De Laurenzi, Aging, 2012, 4, 330–349 CAS.
- O. Karmi, S. H. Holt, L. Song, S. Tamir, Y. Luo, F. Bai, A. Adenwalla, M. Darash-Yahana, Y.-S. Sohn, P. A. Jennings, R. K. Azad, J. N. Onuchic, F. Morcos, R. Nechushtai and R. Mittler, PLoS One, 2017, 12, e0175796 CrossRef PubMed.
- R. Chen, L. Li and Z. Weng, Proteins: Struct., Funct., Genet., 2003, 52, 80–87 CrossRef CAS PubMed.
- P. J. Weber, J. E. Bader, G. Folkers and A. G. Beck-Sickinger, Bioorg. Med. Chem. Lett., 1998, 8, 597–600 CrossRef CAS PubMed.
- R. Gabizon, M. Mor, M. M. Rosenberg, L. Britan, Z. Hayouka, M. Kotler, D. E. Shalev and A. Friedler, Biopolymers, 2008, 90, 105–116 CrossRef CAS PubMed.
- M. Kurcinski, M. Jamroz, M. Blaszczyk, A. Kolinski and S. Kmiecik, Nucleic Acids Res., 2015, 43, W419–W424 CrossRef CAS PubMed.
- S. Tamir, Y. Eisenberg-Domovich, A. R. Conlan, J. T. Stofleth, C. H. Lipper, M. L. Paddock, R. Mittler, P. A. Jennings, O. Livnah and R. Nechushtai, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2014, 70, 1572–1578 CrossRef CAS PubMed.
- The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental section Fig. S1–S4 and Tables S1–S4. See DOI: 10.1039/c8sc03390k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |