Open Access Article
Open Access ArticleIntrahepatic fatty acids composition as a biomarker of NAFLD progression from steatosis to NASH by using 1H-MRS
Aline Xavier *ab,
Flavia Zacconicde,
Constanza Gainzaf,
Daniel Cabreragh,
Marco Arreseg,
Sergio Uribeabi,
Carlos Sing-Longabfj and
Marcelo E. Andiaabi
*ab,
Flavia Zacconicde,
Constanza Gainzaf,
Daniel Cabreragh,
Marco Arreseg,
Sergio Uribeabi,
Carlos Sing-Longabfj and
Marcelo E. Andiaabi
aBiomedical Imaging Center, Pontificia Universidad Católica de Chile, Chile. E-mail: acarvalhodasilva@uc.cl
bMillennium Nucleus for Cardiovascular Magnetic Resonance, Chile
cFaculty of Chemistry and Pharmacy, Pontificia Universidad Católica de Chile, Chile
dResearch Center for Nanotechnology and Advanced Materials CIEN-UC, Pontificia Univesidad Católica de Chile, Chile
eInstitute for Biological and Medical Engineering, Schools of Engineering, Medicine and Biological Sciences, Pontificia Universidad Católica de Chile, Chile
fInstitute for Mathematical and Computational Engineering, Pontificia Universidad Católica de Chile, Chile
gGastroenterology Department, School of Medicine, Pontificia Universidad Católica de Chile, Chile
hDepartment of Chemical and Biological Sciences, Universidad Bernardo O'Higgins, Chile
iRadiology Department, School of Medicine, Pontificia Universidad Católica de Chile, Chile
jMillennium Nucleus Center for the Discovery of Structures in Complex Data, Chile
First published on 19th December 2019
Abstract
Non-alcoholic fatty liver disease (NAFLD) is the most common liver disease in the world and it is becoming one of the most frequent cause of liver transplantation. Unfortunately, the only available method that can reliably determine the stage of this disease is liver biopsy, however, it is invasive and risky for patients. The purpose of this study is to investigate changes in the intracellular composition of the liver fatty acids during the progression of the NAFLD in a mouse model fed with Western diet, with the aim of identify non-invasive biomarkers of NAFLD progression based in 1H-MRS. Our results showed that the intracellular liver fatty acid composition changes as NAFLD progresses from simple steatosis to steatohepatitis (NASH). Using principal component analysis with a clustering method, it was possible to identify the three most relevant clinical groups: normal, steatosis and NASH by using 1H-MRS. These results showed a good agreement with the results obtained by GC-MS and histology. Our results suggest that it would be possible to detect the progression of simple steatosis to NASH using 1H-MRS, that has the potential to be used routinely in clinical application for screening high-risk patients.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is characterized by the accumulation of intracellular fatty acids in the liver in the absence of excessive alcohol consumption. The spectrum of this disease starts with a simple steatosis; it may progress to non-alcoholic steatohepatitis (NASH) with different degrees of inflammation, and with or without fibrosis; and ultimately cirrhosis and hepatocellular carcinoma.1 NAFLD is strongly associate with obesity and metabolic syndrome, which are the most common non-transmissible chronic diseases in Western countries.1,2Research performed in 2015 with a sample of 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 515
515![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 431 patients from 22 countries showed that 25% of the total population in the world suffers from NAFLD.3 According to statistics, about 30–40% of people with steatosis develop NASH. Moreover, 10–30% of them develop cirrhosis, which could progress to hepatocellular carcinoma.4
431 patients from 22 countries showed that 25% of the total population in the world suffers from NAFLD.3 According to statistics, about 30–40% of people with steatosis develop NASH. Moreover, 10–30% of them develop cirrhosis, which could progress to hepatocellular carcinoma.4
Currently, there are no specific biomarkers that can predict this “bad progression” of NAFLD. Most of the current available diagnosis methods mainly focus on estimating the total amount of fat stored in the liver using ultrasound (US), computed tomography (CT) or magnetic resonance imaging (MRI). However, these methods have some drawbacks: US does not work properly in obese patients, CT uses ionizing radiation and none of these methods can recognize either inflammation or early stages of fibrosis. In addition, it is necessary to detect the disease in NASH stages while the disease is reversible.5 Therefore, the only method that can reliably determine the stage of this disease is a biopsy with a histological evaluation, however, it is invasive and risky for patients.6–8
In an attempt to develop biomarkers correlated with the progression of the disease, previous studies have characterized the fatty acids (FA) stored in the intracellular lipids in the liver by using gas chromatography with mass spectrometer (GC-MS), and they have identified some changes in the FA profile when comparing healthy livers, livers with steatosis, and livers with steatohepatitis.9–12 Araya et al. (2004)12 found a decrease in polyunsaturated fatty acids (PUFA), while the saturated fatty acids (SFA) and the monounsaturated fatty acids (MUFA) had no significant changes between healthy patients and patients with steatosis. This study also found a decrease of the long chain polyunsaturated fatty acids between healthy patients and patients with steatosis, and between patients with steatosis and patients with steatohepatitis. Additionally, the same conclusion was found by Puri et al. (2007).11
Although the analysis of the lipid composition opens an interesting field for the diagnosis of fatty liver progression, it still requires a biopsy, so its routine use is limited. On the other hand, magnetic resonance spectroscopy (MRS) provides a non-invasive technique that allows to determine the structure of organic substances.7 The idea of defining a classifier using MRS emerges due to the need to find a way to replace biopsy with a non-invasive method that can classify NAFLD based on the composition of different fatty acids stored in the liver.
Previous studies have shown that it is possible to associate the results of MRS with those obtained from GC-MS. Unfortunately, those studies considered only fatty acids with 14, 16 and 18 carbons and not fatty acids with long chain, which seems to have a big relevance in the NAFLD progression.13–16 Furthermore, studies that emphasize the importance of fatty acids on the liver have analyzed it in vivo with MRS but have not correlated their results with the true amount of fatty acids as no biopsy was performed.17–20
The purpose of this study is to investigate and compare the intracellular composition of the liver fatty acids using histology, GC-MS, 9.4T 1H-MRS during the progression of NAFLD in a mice model fed with Western diet, with the aim of identifying non-invasive biomarkers of NAFLD progression to NASH.
Subjects and methods
All experiments were approved by the Scientific Ethics Committee for the care of animals and the environment of the Pontificia Universidad Católica de Chile (number: 170614002).We fed a group of C57BL/6 male mice with Western diet (AIN76A, Test Diet) for 4 weeks (n = 6), 10 weeks (n = 6) and 24 weeks (n = 6). The Western diet has 4.49 kilocalories per gram, and the calories come from: fat (40%), protein (15.8%), and carbohydrate (44.2%). We also fed a group (n = 6) with a chow diet (500l*, Labdiet) with 3.36 kilocalories per gram, and the calories come from: fat (13.4%), protein (30%), and carbohydrate (57%). The Western diet is a model of “Western fast food” diet characterized by high calories, high cholesterol and high fructose content.21
At the end of the diet intervention, the mice were anesthetized with ketamine/xylosine, and the livers were harvested. A portion of the liver was used for histology analysis. The remaining liver was divided into two portions that were analyzed independently. We extracted the intracellular FA using a protocol adapted by Folch et al.,22 proceeded by an esterification process to obtain fatty acids methyl esters (FAME) since they are more stable (do not form hydrogen bridges). It's important to comment that the quantity of FA is the same as FAME. Finally, those FAME were analyzed using a 9.4 T MRS and GC-MS.
Previous studies were performed and allowed the conclusion that 300 mg of liver was required since, after the extraction in healthy mice, we obtained 5 mg of FAME and that is the minimum amount of sample required to perform a 1H MRS with enough SNR. For the GC-MS, 2 mg of FAME is enough.23
Gas chromatography with mass spectrometer
FAME were analyzed by using gas chromatography with mass spectrometer (PerkinElmer, Clarus 680) equipped with HP-Innowax capillary column (length 25 meters, 0.2 mm internal diameter, 0.2 mm film). Additional configuration of GC-MS is found in Table 1. Data register was in SCAN mode and peak integration was obtained by the TurboMass Training 2016 PRO software.| Injector temp. | Oven ramp | Sample injection | Electron impact |
|---|---|---|---|
| 220 °C | 150 °C × 1 min 15 °C min−1 to 200 °C 200 °C × 5 min 12 °C min−1 to 260 °C | Split mode (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) helium at 1 ml min−1 (constant flow) 20) helium at 1 ml min−1 (constant flow) |
Ionization potential of 70 eV |
FAME were identified by comparing retention times to known standards and by matching them up with the mass spectra from NIST library (National Institute of Standards and Technology, USA).
The mass of each FAME (in µg) was calculated according to the integration area of corresponding peak and the relation with the integration area of the internal standard (C19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) added to the sample (50 µg). Fatty acids composition was defined as the percentage of individual fatty acids in respect of its total.
0) added to the sample (50 µg). Fatty acids composition was defined as the percentage of individual fatty acids in respect of its total.
Magnetic resonance spectroscopy
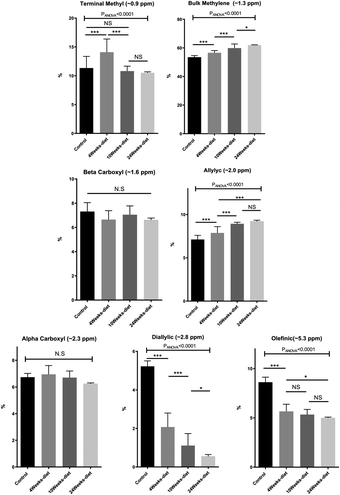
The 1H MRS spectra of the FAME were obtained in a Bruker Avance spectrometer operating at 9.4 Tesla with the acquisition protocol Zg30 (30 degrees in z axis).We took a known value in mg of FAME from each sample and added 700 µl of CDCl3 that has a small proportion of tetramethylsilane (TMS) as an internal reference. This mixture was introduced into a 5 mm diameter tube. The spectra acquisition was made with Topspin V3.0 and the parameters were: spectral width 8012.820 Hz, relaxation delay 1 s, number of scans 16, acquisition time 2.045 s, flip angle 30° to avoid T1 relaxation effects and total acquisition time 48.72 s (number of scans × [time between pulses + acquisition time]). The experiment was conducted at 25 °C. The spectra were analyzed using MestreNova Version 10.0. First, the spectra were centered (TMS in 0 ppm); then we calculated the area under the curve (AUC) of all seven peaks corresponding to fatty acids and finally, we normalized the AUC by the total amount of fatty acids. The fatty acids peaks used were: methyl terminal protons (–CH3, approximately 0.9 ppm); bulk methylene protons (–CH2–, approximately 1.3 ppm); β-methylene protons (COO–CH2–CH2–, approximately 1.6 ppm); allylic protons (–CH2–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH-, approximately 2.0 ppm) α-methylene protons (COO–CH2–CH2–, approximately 2.2 ppm); diallylic protons (
CH-, approximately 2.0 ppm) α-methylene protons (COO–CH2–CH2–, approximately 2.2 ppm); diallylic protons (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–CH2–CH
CH–CH2–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) , approximately 2.8 ppm) and olefinic internal protons (–CH
, approximately 2.8 ppm) and olefinic internal protons (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–, approximately 5.3 ppm).
CH–, approximately 5.3 ppm).
Histology
Liver sections from mice livers were routinely fixed in 10% of formalin and embedded in paraffin. Then 5 µm tissue sections were stained with hematoxylin/eosin, oil red and Picrosirius Red, as described previously.24 Whole slide imaging was obtained using the Aperio Digital Pathology Slide Scanner (Leica Biosystems) allowing us to study the entire left lateral lobe. Histopathological analyses were performed by a blinded pathologist starting from three full cross-sectional slides obtained from distal, medial, and proximal regions derived from the left lateral lobe. The blinded trained pathologist utilized the NAFLD Activity Score (NAS) by using the method proposed by Kleiner et al.25The score of steatoses vary between 0 and 3, the score of ballooning vary between 0 and 2, the score of inflammation vary between 0 and 3, and the score of fibrosis vary between 0 and 4. The NAS is the sum of steatosis, ballooning and inflammation scores. It can vary between 0 and 8. A NAS score less than 3 means no NASH, while a score higher than 4 means NASH. Scores of 3 or 4 means indeterminate.
Correlation between GC-MS and MRS
The MRS allows us to determine the structure of an organic substance. The spectrum shows the signals in a graph of frequency (ppm) versus intensity in arbitrary units. Each set of chemically equivalent protons originates a signal in such a way that the number of signals in a spectrum indicates the amount of different kinds of protons in a substance. The chemical shift is a measure of how far away the signal is from the reference signal of tetramethylsilane (TMS). The greater the displacement with respect to the TMS (0 ppm) is, the closer the proton is to an environment where there is an electronegative group.26The GC is the gold standard to characterize fatty acids. The mixture containing all the fatty acids methyl esters is introduced in a GC-MS where it is warmed, and the fatty acids start to separate from each other by their volatility and polarity.27
There are many types of FA in the liver of a human being. All of them generate just seven peaks corresponding to the chemical equivalent protons. Fig. 1 shows an example of a FAME with 18 carbons and 2 double bonds in the signal of the MR spectroscopy.
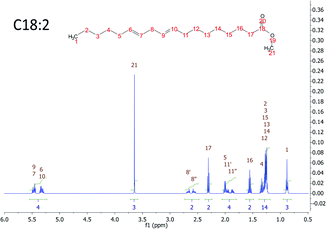 | ||
| Fig. 1 MRS simulation of a FAME with 18 carbons and 2 double bonds. The proton chemically equivalent are shown in numbers. | ||
Therefore, MRS does not detect the quantity of the fatty acids directly, but it is possible to assume that, if we have a change in fatty acids profile, we will have a change in the metabolites detected by MRS.
Statistical analyses
All statistical analyses were performed using Prism 6 (GraphPad Software Inc, La Jolla, CA). All graphs were plotted as mean ± standard deviation or boxplot. To compare between 3 or more groups, the ANOVA test was performed followed by a Bonferroni post-hoc test. A P-value < 0.05 was considered to indicate statistical significance.Principal component analysis and clustering method
Even though mice are fed with the same amount of diet for a fixed period, the disease may progress differently in each mouse. In order to identify changes in fatty acids and metabolites, we have used principal component analysis (PCA) to visualize the stage of the disease in each mouse, and a clustering method to group the mice according to their GC-MS and MRS measurements.PCA is a data analysis technique that is used to represent a large set of data samples with a reduced set of features that explain most of the variability in the original set. These features, called principal components, are determined by finding linear combinations of the data samples that explain the largest variability in the dataset. Therefore, the first principal component corresponds to the direction of maximum possible variance, whereas the second explains the maximum remaining variance and so on.28 By considering a small number of principal components, we can visualize the data and determine if there is a natural distribution of the measurements performed on each mouse that correlates to their disease progression.
To determine if the distribution of GC-MS and 1H-MRS data has itself some structure from which we can infer the presence or progression of the disease, an agglomerative hierarchical clustering method was applied. Agglomerative hierarchical clustering is a “bottom-up” approach; each data point is initially considered to be a cluster, and data points are grouped together at each step according to some criterion. At the end of the procedure we obtain several potential groupings of the data, with a decreasing number of clusters.28 For our analysis, the data points are grouped according to their Euclidean distance and Ward's linkage function.29 This choice promotes that at each step, among all possible choices, pairs of clusters are merged in order to minimize the variation within the resulting cluster with respect to all other possible choices.
Results
Mice fed with Western diet increased their weight and accumulated fat in the liver. Table 2 shows the mice weight, liver weight and the total liver FAME content (mass of FAME/total liver sample, %) for each group at 4 time points: 0, 4, 10 and 24 weeks since diet intervention. By comparing the total amount of fatty acid in two portions of the same liver (that were analyzed independently before averaging), we identified a maximum difference of 5%.| Mice group | Mice age (weeks) | Mice weight (g) | Liver weight (g) | Weight of FAME/total liver sample, % |
|---|---|---|---|---|
| Chow-diet (control) | 16 | 26.63 ± 1.33 | 1.22 ± 0.07 | 3.07 ± 0.65 |
| Western diet for 4 weeks | 16 | 28.27 ± 2.76 | 1.14 ± 0.08 | 5.84 ± 2.11 |
| Western diet for 10 weeks | 22 | 36.23 ± 6.24 | 1.61 ± 0.56 | 12.68 ± 4.28 |
| Western diet for 24 weeks | 36 | 46.23 ± 2.12 | 3.32 ± 0.48 | 23.62 ± 2.54 |
Histology
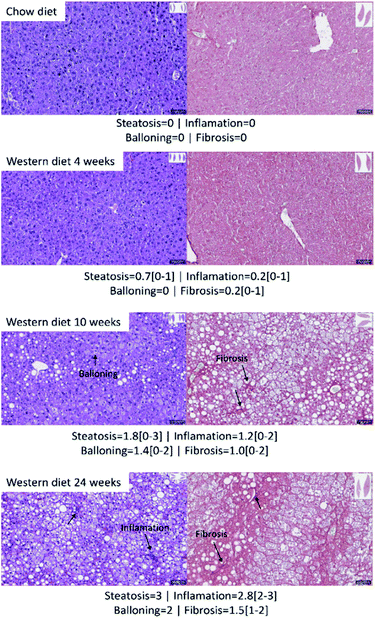
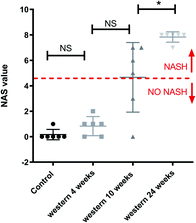
Fig. 2 shows the results of histology for each group. Besides that, the average score and the range of steatosis, ballooning, inflammation and fibrosis for each group is shown below the image. The NAFLD activity score (NAS) for each group was, in average, 0 (zero) for control group with chow diet, 0.8 for the group of mice with Western diet for 4 weeks, 4.4 for the group of mice with Western diet during 10 weeks and 7.8 for the group of mice with Western diet during 24 weeks.Fig. 3 shows a boxplot with individual values of the NAS score. The control group, mice fed with Western diet for 4 weeks and two mice fed with Western diet for 10 weeks have no NASH, while the mice fed with Western diet for 24 weeks and four mice fed with Western diet for 10 weeks have NASH.
Gas chromatography with mass spectrometer
We identified 12 main fatty acids using the CG-MS, however, 5 of them had less than 1% of participation, therefore, we only considered 7 FA for the analysis (C16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, C18
0, C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, C16
0, C16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, C18
1, C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, C18
1, C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, C20
2, C20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, C22
4, C22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6). Two of the FA changed significantly between all the groups (control, 4 weeks-diet, 10 weeks-diet and 24 weeks-diet) corresponding to the progression of NAFLD: C18
6). Two of the FA changed significantly between all the groups (control, 4 weeks-diet, 10 weeks-diet and 24 weeks-diet) corresponding to the progression of NAFLD: C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 increased, while C18
1 increased, while C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 decreased. Other two FA (C16
2 decreased. Other two FA (C16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and, C22
1 and, C22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) changed significantly between 3 of the 4 groups, C16
6) changed significantly between 3 of the 4 groups, C16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 increased, while C22
1 increased, while C22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
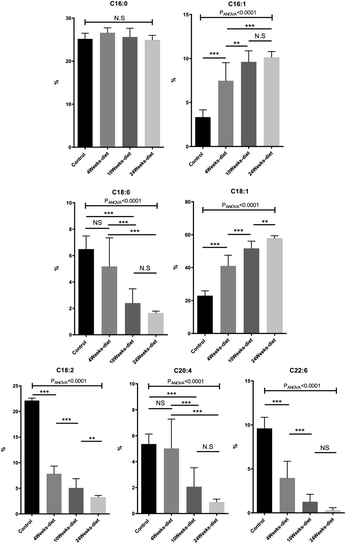
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 decreased (Fig. 4). Additionally, by comparing the composition of the fatty acids in two portions of the same liver, we identified a maximum difference of 2.5%, most of them less than 1%.
6 decreased (Fig. 4). Additionally, by comparing the composition of the fatty acids in two portions of the same liver, we identified a maximum difference of 2.5%, most of them less than 1%.
All the FA can be grouped into three main categories: saturated fatty acids (SFA), monounsaturated fatty acids (MUFA) and polyunsaturated fatty acids (PUFA). The SFA have no double bonds (i.e., C14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, C15
0, C15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, C16
0, C16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, C18
0, C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), while the MUFA have one double bond (i.e., C16
0), while the MUFA have one double bond (i.e., C16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, C18
1, C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and the PUFA have two or more double bonds (i.e., C18
1) and the PUFA have two or more double bonds (i.e., C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, C18
2, C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, C20
3, C20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, C20
3, C20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, C20
4, C20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, C22
5, C22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6).
6).
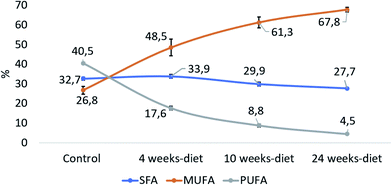
Fig. 5 shows that PUFA liver content decreased with the progression of the disease from 40.5% in control group to 4.5% at 24 weeks (PANOVA < 0.001). In contrast, MUFA liver content increased from 26.8% in control group to 67.8% at 24 weeks (PANOVA < 0.001), while SFA remain constant.
Magnetic resonance spectroscopy
We have identified seven metabolite peaks in the MRS spectra that correspond to the FA and the peak of methyl ester (3.6 ppm). Fig. 6 shows a visual comparison between a representative spectrum of one control group mouse and the spectrum of a mouse fed Western diet for 24 weeks. Five peaks (terminal methyl, bulk methylene, allylic, diallylic and olefinic) showed significant differences with the progression of the disease (Fig. 7). However, the terminal methyl group should not change since this peak is present in all FA, with the same quantity of protons. The alfa and beta methylene should be present in all FA with the same number of protons; therefore, we shouldn't expect any significant differences.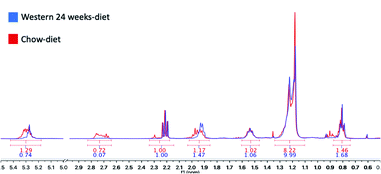 | ||
| Fig. 6 Areas under the curve (AUC) calculated with MestreNova V10.0. In red, the results of a control mouse with a chow-diet and in blue, a mouse with a Western diet for 24 weeks. | ||
Principal component analysis
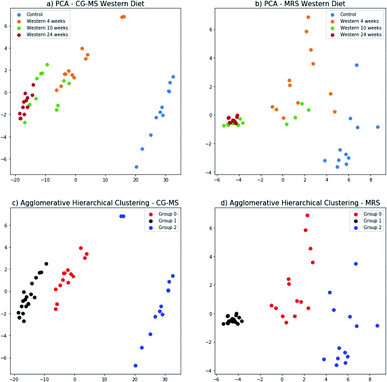
The principal components for the FA composition measured with GC-MS and metabolites identified in the 1H-MRS were computed separately; and the data projected onto the space generated by their corresponding first two principal components (Fig. 8a and b). In this 2-dimensional representation it is possible to identify changes in the GC-MS measurements as the time of diet intervention increases and the disease progresses (Fig. 8a). In particular, the control group and the mice fed for 24 weeks are well-separated. This behavior is replicated for the 1H-MRS measurements (Fig. 8b) indicating the potential for 1H-MRS to be used both as a biomarker for the progression of the disease and as a surrogate for GC-MS.The three groups that can be visually identified show a rough correlation with the number of weeks each mouse has been fed (Fig. 8a and b). Interestingly, the group of mice fed for 10 weeks has been split in two, with some of them grouped with mice fed for 4 weeks, and the rest grouped with mice fed for 24 weeks. Those grouped with the mice fed for 4 weeks are those with no NASH whereas those grouped with the mice fed for 24 weeks are precisely those that have NASH (Fig. 3).
We used agglomerative hierarchical clustering to group both the GC-MS and 1H-MRS data (Fig. 8c and d). Interestly, the clusters obtained correspond to three stages of NAFLD: the first one with a low NAS score, normal; the second one, with an intermedium NAS score, low to medium steatosis; and the third one with high NAS score with inflammation and early stages of fibrosis.
Furthermore, the mouse whose result is at the top of the GC-MS graph (Fig. 8a) was grouped as a control mouse, even though it was fed with Western diet for 4 weeks. However, its NAS value was 0. Fig. 9 shows the mean of NAS score for each the cluster identified (control, No NASH, NASH).
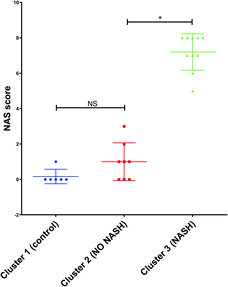 | ||
| Fig. 9 NAS score for each cluster found by PCA with the data from GC-MS and MRS. *p < 0.05 (significant difference between groups) and NS (no significant difference between groups). | ||
Discussion
We have analysed the effects of a diet intervention with a fast food-like model (i.e., Western diet) in mice for 24 weeks. During the progression of the disease, the mice consecutively develop steatosis, steatohepatitis and liver fibrosis, with a progressive increase in the NAS score. Simultaneously, Western diet produced numerous changes in liver fatty acids composition, which could provide new evidence to understand the progression of the disease and identify biomarkers to predict the progression of steatosis and NASH.GC-MS analysis showed that the fat in the liver is composed of, at least, 12 different fatty acids, which have different evolution during the NAFLD progression. Besides that, the analysis identifies a pattern of fatty acids composition during NAFLD progression: although the total amount of fatty acids increased during the NAFLD progression, not all the fatty acids progressed in the same way. The PUFA decreased, while the MUFA increased and the SFA remained the same during the progression of the disease. Similar results were found by Levant et al. (2013) in mice with a high-fat diet for 8 weeks.30 The decrease in the PUFA can be explained since the arachidonic (C20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4); eicosapentaenoic acid (20
4); eicosapentaenoic acid (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) and docosahexaenoic acids (C22
5) and docosahexaenoic acids (C22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) are precursors for a variety of anti- and pro-inflammatory mediators.31,32
6) are precursors for a variety of anti- and pro-inflammatory mediators.31,32
MRS analysis found all the seven peaks related to fatty acids. The diallylic peak (2.8 ppm), corresponding to FA with two or more double bonds, also decreased in good agreement with the PUFA results found by CG-MS. The olefinic (5.3 ppm), allylic (2.0 ppm) and bulk methylene (1.3 ppm) peaks are somehow related to this change in PUFA and MUFA, because they are related to double bonds.
As mentioned by Leporq et al. (2014), who used only theoretical values calculated from oil mass composition for validation, one of the limitations of their work was not to have the gas chromatography analysis as gold standard to characterize those FA.33 Opposite to this, our study overcomes this limitation with the GC-MS analysis.
Our results, as evidenced by PCA, showed that the liver fatty acid composition changes as NAFLD progresses. In addition, by using agglomerative hierarchical clustering it was possible to identify the 3 most relevant clinical groups: normal, steatosis and NASH. In essence, the NAS score of the entire population was reproduced by clustering the GC-MS data in three groups. In addition, applying the same analysis to the 1H-MRS data shows it is possible to identify the same 3 groups using ex-vivo MRS, which provide some evidence that the proposed methodology could be used in vivo and non-invasive, however further studies has to be done to prove this hypothesis.
Some limitations of our studies are related to the possibility that our results may vary if the mice were fed with different diets, although we have used the diet most like human diet.34,35 However, this limitation might explain differences in the NAFLD progression among patients, in which some of them evolve to NASH and cirrhosis, whereas others remain in the early stages of NAFLD for a long time.
Additionally, the MRS acquisition was performed only over the extracted liver fatty acids, so that it is now necessary to validate our results with in vivo whole liver MRS acquisition. The transition to an in vivo measurement has some challenges since it may account with a series of extra parameters like respiratory and cardiac motion, presence of water component, the correlation between acquisition time, NSA, voxel size and SNR to assure the comfort of the patient, but also the spectra quality.
Preliminary results showed that it would be possible to identify the same 7 peaks in human and mice livers in vivo at 7.0 T (ref. 36) and 9.4 T (ref. 37), respectively (Fig. 10). Although the 7.0 T is not available for clinical use yet, research could be made in this equipment by benefitting from its good spectral resolution to validate the method.
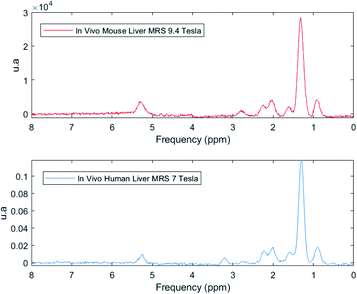 | ||
| Fig. 10 In vivo MR spectra from a 9.4 Tesla in a mouse (red) and 7 Tesla in human (blue) showing that it is possible to identify all the seven peaks correspondent to the fat spectrum in the liver. | ||
Conclusions
To conclude, our study compares different stages of the NAFLD disease induced by a Western diet. The differences in the FA composition found by GC-MS, which is also reflected in the MR spectrum, could have clinical potential for monitoring NAFLD progression from steatosis to NASH in a non-invasive way.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This publication has received funding from Millennium Science Initiative of the Ministry of Economy, Development and Tourism, grant Nucleus for Cardiovascular Magnetic Resonance and grant Millennium Nucleus Center for Discovery of Structures in Complex Data, FONDECYT 1180525, 11171001 and 1191145 to MA, DC and MA respectively. CONICYT-PCHA/Doctorado Nacional/2016-21160835. CSL was partially funded by FONDECYT 11160728. FZ is grateful to FONDEQUIP EQM120021 and EQM150020 from Pontificia Universidad Católica de Chile.References
- S. L. Friedman, B. A. Neuschwander-Tetri, M. Rinella and A. J. Sanyal, Nat. Med., 2018, 24(7), 908–922 CrossRef CAS PubMed.
- R. Wiliams and S. D. Taylor-Robinson, Clinical Dilemmas in Non-Alcoholic Fatty Liver Disease, 1st edn, 2016 Search PubMed.
- Z. M. Younossi, A. B. Koenig, D. Abdelatif, Y. Fazel, L. Henry and M. Wymer, Hepatology, 2016, 64(1), 73–84 CrossRef PubMed.
- M. G. Neumann and L. B. Cohem, Can. J. Gastroenterol. Hepatol., 2014, 28(11), 607–618 CrossRef PubMed.
- R. J. Perry, D. Zhang, X. M. Zhang, J. L Boyer and G. I. Shulman, Science, 2015, 347, 1253–1256 CrossRef CAS PubMed.
- M. Ahmed M, World J. Hepatol., 2015, 7(11), 1450–1459 CrossRef PubMed.
- Z. Permutt, T.-A. Le, M. R. Peterson, E. Seki, D. A. Brenner, C. Sirlin and R. Loomba, Aliment. Pharmacol. Ther., 2012, 36(1), 22–29 CrossRef CAS PubMed.
- S. B. Reeder, I. Cruite, G. Hamilton and C. B. Sirlin, J. Magn. Reson. Imaging, 2011, 34(4), 729–749 CrossRef PubMed.
- K. Yamada, E. Mizukoshi, H. Sunagozaka, K. Arai, T. Yamashita, Y. Takeshita, H. Misu, T. Takamura, S. Kitamura, Y. Zen, Y. Nakanuma, M. Honda and S. Kaneko, Liver Int., 2015, 35(2), 582–590 CrossRef CAS PubMed.
- G. Guo and X. Zhang, Lipids in healthy and disease, 2011, vol. 10, p. 234 Search PubMed.
- P. Puri, R. A. Baillie, M. M. Wiest, F. Mirshahi, J. Choudhury, O. Cheung, C. Sargeant, M. J. Contos and A. J. Sanyal, Hepatology, 2007, 46(4), 1081–1090 CrossRef CAS PubMed.
- J. Araya, R. Rodrigo, L. A. Videla, L. Thielemannt, M. Orellana, P. Pettinelli and J. Poniachik, Clin. Sci., 2004, 106, 635–643 CrossRef CAS PubMed.
- G. Knote and J. Kenar, Eur. J. Lipid Sci. Technol., 2004, 106(2), 88–96 CrossRef.
- M. D. Guillen and A. Ruiz, J. Sci. Food Agric., 2003, 83(4), 338–346 CrossRef CAS.
- Y. Miyake, K. Yokomizo and N. Matsuzaki, J. Am. Oil Chem. Soc., 1998, 75, 15–19 CrossRef CAS.
- J. Ren, I. Dimitrov, A. D. Sherry and C. R. Malloy, J. Lipid Res., 2008, 49, 2055–2062 CrossRef CAS PubMed.
- G. Hamilton, T. Yokoo, M. Bydder, I. Cruite, M. E. Schroeder, C. B. Sirlin and M. S. Middleton, NMR Biomed., 2011, 24, 784–790 CrossRef PubMed.
- R. Longo, P. Pollesello, C. Ricci, F. Masutti, B. J. Kvam, L. Bercich, L. S. Croce, P. Grigolato, S. Paoletti, B. de Bernard, C. Tiribelli and L. D. Palma, J. Magn. Reson. Imaging, 1995, 5, 281–285 CrossRef CAS PubMed.
- N. A. Johnson, D. W. Walton, T. Sachinwalla, C. H. Thompson, K. Smith, P. A. Ruell, S. R. Stannard and J. George, Hepatology, 2008, 47, 1513–1523 CrossRef CAS PubMed.
- Y. Lee, H.-J. Jee, H. Noh, G.-H. Kang, J. Park, J. Cho and H. Kim, Magn. Reson. Med., 2013, 70(3), 620–629 CrossRef CAS PubMed.
- A. Krishnan, T. S. Abdullah, T. Mounajjed, S. Hartono, A. McConico, T. White, N. LeBrasseur, I. Lanza, S. Nair, G. Gores and M. Charlton, Am. J. Physiol.: Gastrointest. Liver Physiol., 2017, 312, G666–G680 CrossRef PubMed.
- J. Folch, M. Lees and G. H. Sloane Stanley, J. Biol. Chem., 1957, 226(1), 497–509 CAS.
- A. Xavier, F. Zacconi, D. Cabrera, K. Fuenzalida and M. E. Andia, IFMBE Proceedings, 2019, vol. 70, (1), pp. 51–56 Search PubMed.
- D. Cabrera, A. Wree, D. Povero, N. Solís, A. Hernandez, M. Pizarro, H. Moshage, J. Torres, A. E. Feldstein, C. Cabello-Verrugio, E. Brandan, F. Barrera, J. P. Arab and M. Arrese, Sci. Rep., 2017, 7(1), 3491 CrossRef PubMed.
- D. E. Kleiner, E. M. Brunt, M. Van Natta, C. Behling, M. J. Contos, O. W. Cummings, L. D. Ferrell, Y. C. Liu, M. S. Torbenson, A. Unalp-Arida, M. Yeh, A. J. McCulloughJ and A. J. Sanyal, Hepatology, 2005, 41(6), 1313–1321 CrossRef PubMed.
- P. B. Yurkanis, Química orgânica, 5th edn, 2008 Search PubMed.
- A. Di Giulio and A. Bonamore, Globins and Other Nitric Oxide-Reactive Proteins Part A, 2008, pp. 239–253 Search PubMed.
- G. James, D. Witten, T. Hastie and R. Tibshirani, An Introduction to Statistical Learning, 2013, ch. 6 and 10 Search PubMed.
- J. H. Ward Jr, J. Am. Stat. Assoc., 1963, 58(301), 236–244 CrossRef.
- B. Levant, M. K. Ozias, B. L. Guilford and D. E. Wright, Streptozotocin-induced diabetes partially attenuates the effects of a high-fat diet on liver and brain fatty acid compo-sition in mice, Lipids, 2013, 48, 939–948 CrossRef CAS PubMed.
- N. G. Bazan, Brain Pathol., 2005, 15, 159–166 CrossRef CAS PubMed.
- G. Bannenberg, M. Arita and C. N. Serhan, Sci. World J., 2007, 7, 1440–1462 CrossRef CAS PubMed.
- B. Leporq, S. A. Lambert, M. Ronot, V. Vilgrain and B. E. Van Beers, NMR Biomed., 2014, 27, 1211–1221 CrossRef CAS PubMed.
- L. H. Tetri, M. Basaranoglu, E. M. Brunt, L. M Yerian and B. A. Neuschwander-Tetri, Am. J. Physiol.: Gastrointest. Liver Physiol., 2008, 295, G987–G995 CrossRef CAS PubMed.
- L. Longato, Fibrog. Tissue Repair, 2013, 6, 14 CrossRef CAS PubMed.
- A. Filmer, C. A. Castro, A. Xavier, P. R. Luijten, D. W. Klomp and J. J. Prompers, ISMRM proceeding, 2019, p. 4232 Search PubMed.
- A. Xavier, F. Zacconi, T. Eykyn, B. Plaza, A. Phinikaridou and M. E. Andia. MAGMA: Book of abstract ESMRMB, 2019, vol. 32, (1), p. S55 Search PubMed.
| This journal is © The Royal Society of Chemistry 2019 |