Open Access Article
Open Access ArticleAnticancer and antimicrobial properties of novel η6-p-cymene ruthenium(II) complexes containing a N,S-type ligand, their structural and theoretical characterization†
Ewelina Namiecińskaa,
Beata Sadowskab,
Marzena Więckowska-Szakielb,
Anna Dołęga c,
Beata Pasternakd,
Magdalena Grazule and
Elzbieta Budzisz
c,
Beata Pasternakd,
Magdalena Grazule and
Elzbieta Budzisz *a
*a
aDepartment of Cosmetic Raw Materials Chemistry, Medical University of Lodz, Muszynskiego 1, 90-151 Lodz, Poland. E-mail: elzbieta.budzisz@umed.lodz.pl
bDepartment of Immunology and Infectious Biology, Institute of Microbiology, Biotechnology and Immunology, Faculty of Biology and Environmental Protection, University of Lodz, Banacha 12/16, 90-237 Lodz, Poland
cDepartment of Inorganic Chemistry, Faculty of Chemistry, Gdansk University of Technology, Narutowicza 11/12, 80-233 Gdansk, Poland
dDepartment of Organic Chemistry, Faculty of Chemistry, University of Lodz, Tamka 12, 91-403 Lodz, Poland
eDepartment of Pharmaceutical Microbiology and Microbiological Diagnostics, Faculty of Pharmacy, Medical University of Lodz, Pomorska 137 Street, 90-235 Lodz, Poland
First published on 27th November 2019
Abstract
Ruthenium(II) complexes are lately of great scientific interest due to their chemotherapeutic potential as anticancer and antimicrobial agents. Here we present the synthesis of new pyrazole carbothioamide derivatives and their four arene–ruthenium complexes. The title compounds were characterized with the application of IR, NMR, mass spectrometry, elemental analysis and X-ray diffraction. Additionally, for new complexes DFT calculations were done. Their antimicrobial activity (MIC, MBC/MFC) was examined in vitro against Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecalis, Pseudomonas aeruginosa, Proteus vulgaris and Candida albicans. Their cytotoxic effects, using the MTT assay, against three cancer cell lines: HL-60, NALM-6, WM-115 and normal human foreskin fibroblasts (HFF-1) were also investigated. The influence of the new arene–ruthenium(II) complexes on the DNA structure was also tested. From our results, compound 2d showed higher cytotoxicity against melanoma cell line WM-115 than cisplatin. Strong biostatic and biocidal activity of the tested complexes against Gram-positive bacteria, including S. aureus, S. epidermidis and E. faecalis was demonstrated. The new arene–ruthenium(II) compounds could not only inhibit proliferation of cancer cells, but also protect patients against malignant wound infections.
1. Introduction
The half-sandwich arene–ruthenium(II) complexes with different co-ligands have been recognized as chemotherapeutics agents and promising alternatives to cisplatin and its oxaliplatin analogs.1,2 Cisplatin was the first metal-ion complex used in chemotherapy, but unfortunately drug resistance and side effects have limited its clinical utility. The ruthenium complexes could be a valuable alternative to cisplatin. Some of the most investigated classes of organometallic compounds are RAPTA-C [(Ru-p-cymene)(1.3.5-triaza-7-phosphaadamantane)Cl2], NAMI-A (imidazolium trans-[tetrachloro(dimethylsulfoxide)(1H-imidazole)ruthenate(III)]), KP1019 (indazolium trans-[tetrachlorobis(1H-indazole)-ruthenate(III)]) complexes. Currently, most of them are undergoing clinical trials (Scheme 1).3–6 An arene substituent is relatively inert towards displacement and is known to stabilize ruthenium complexes in the oxidation state +2 under physiological conditions. Corresponding Ru(II) complexes are kinetically similar to Pt(II) complexes.7 Chelating ligands containing sulfur and nitrogen atoms have been used by many researchers to achieve a limited lability of the complexes, in order to enhance their biological activity. N,S-Ligands coordinate to metal ion center via the sulfur atom and the pyridine-like nitrogen of pyrazole moiety with the formation of a stable five-membered ring.8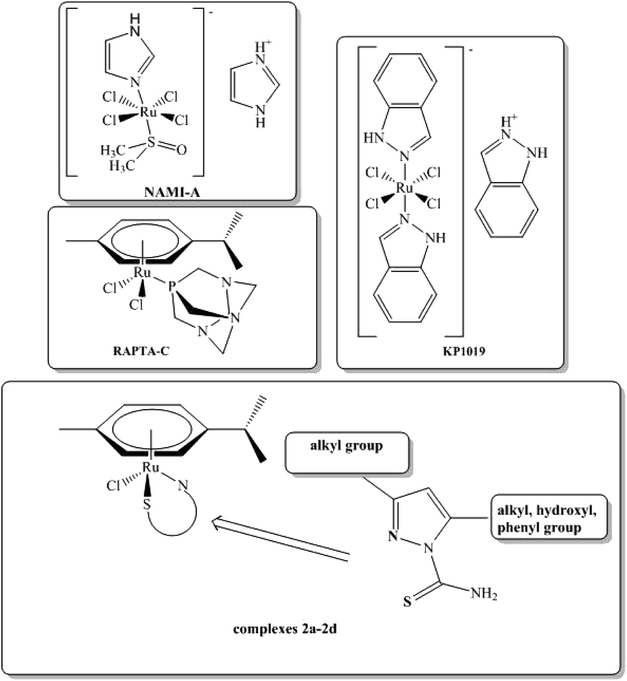 | ||
| Scheme 1 Structures of anticancer NAMI-A, KP1019, RAPTA-C compounds and newly synthesized 2a–2d complexes. | ||
Both the size and hydrophobicity of the coordinated arene substituents as well as the structure of the mono- or bidentate co-ligands have an influence on biological activity of arene–ruthenium(II) complexes.9 The metal center in arene–ruthenium(II) complexes is pseudotetrahedral. Thus, ligands can occupy maximum three coordination sites. This structural feature of the piano-stool complexes allows to create diverse coordination complexes using a variety of N-, O-, S- and P-donors as ligands. A large number of possibilities of coordination allows to manipulate pharmacological properties of half-sandwich arene–Ru(II) complexes making them ideal candidates for preparing multifunctional drugs.10,11
Pyrazole and its derivatives demonstrate various biological activities, e.g. antibacterial, antifungal, antiviral, anticancer, anticonvulsant, analgesic, anti-inflammatory, anti-diabetic, sedative, anti-rheumatic. Moreover, they are applied as dyes and agrochemicals.12–16 So far, scientific interest has been focused mainly on the anticancer activity of the complexes of pyrazole carbothioamide derivatives with various metal ions, e.g. Cu(II), Zn(II), Co(II) and Ni(II) against HL-60, NALM-6, WM-15 and HUVECs cell lines.17–20 It is widely known that melanoma is one of the most difficult diseases to cure due to its high resistance to anticancer drugs. It has been proved that arene–ruthenium(II) complexes display anti-melanoma activity, which is probably based on their interference with mTOR and independent from EGFR (epidermal growth factor receptors) inhibition.
Due to the fact that human microbiome plays an important role not only by providing proper functions of skin and mucous membrane barriers, but also in the etiology of many pathological processes (e.g. wound infections, cancer, ulcerations), the antimicrobial activity of potential anticancer drugs should be tested. Wound infections are quite common in patients with cancer, especially with breast, head, neck or melanoma cancers. Interestingly, it has been proved that metal complexes have antimicrobial effect; therefore their application on the skin could diminish the development of melanoma cells and at the same time protect against wound infections. Anyway, it has to be pointed out that the power of antimicrobial activity of such compounds strongly depends on their structure and composition. Thus, it is a good practice to test antimicrobial activity of each new anticancer drug.
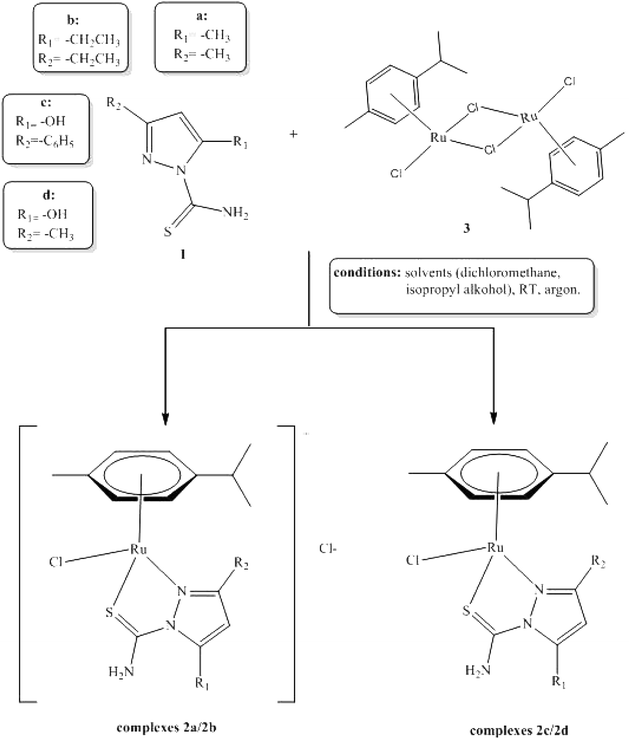
The aim of this study was to synthesize new arene–ruthenium(II) complexes with selected pyrazole derivatives and to evaluate their anticancer and antimicrobial activity. All the complexes were obtained in reactions between ligands containing various substituents (methyl, ethyl, carbonyl, phenyl) in C-3 and C-5 position of the pyrazole ring and [RuCl2(p-cymene)2]2 dimer. Next, they were characterized with the use of several physico-chemical methods, including X-ray diffraction and DFT calculations.
2. Experimental
2.1. Materials and methods
All starting reagents: [RuCl2(p-cymene)2]2 dimer, anhydrous solvents: dichloromethane, isopropyl alcohol, diethyl ether were purchased from Sigma-Aldrich and Avantor Performance and they were commercially available. Synthesis of complexes was carried out in the argon atmosphere conditions using Schlenk techniques. Ligands 1a, 1c and 1d were prepared with the use of methods described before.20,23,24 The 1H NMR and 13C NMR spectra were registered in DMSO-d6 (for all complexes) and CDCl3-d (for ligand 1b) using Bruker Avance III 600 MHz spectrometer. The IR spectra were recorded in KBr on the FTIR-8400S Shimadzu Spectrophotometer. The ESI-MS spectra were measured on Varian 500-MS LC Ion Trap. An elemental analysis was performed at the Faculty of Chemistry (University of Lodz) using a Vario Micro Cube by Elemental analyzer. Melting points of all compounds were determined in open capillary tubes on Büchii apparatus B-540 and were uncorrected. Anticancer activity was tested against three cancer cell lines: HL-60, NALM-6 and WM-115 in the Department of Bioorganic Chemistry at the Medical University of Lodz (anticancer), while antimicrobial activity was carried out on selected bacteria and fungi strains were done in the Department of Immunology and Infectious Biology at the University of Lodz. The MTT method was used to determine cells viability.2.2. Synthetic procedures
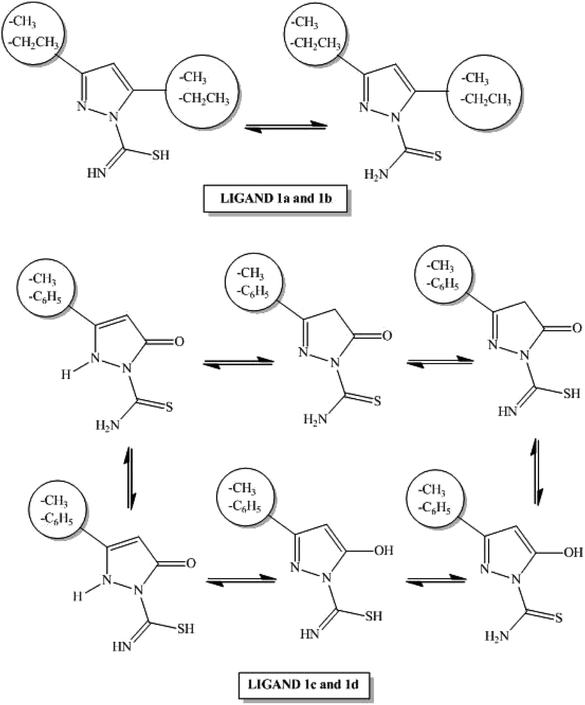
Synthesis of ligand 1-[amino(thioxo)methyl]-3,5-dimethyl-1H-pyrazole (1a), 1-[amino(thioxo)methyl]-5-hydroxy-3-phenyl-1H-pyrazole (1c) and 1-[amino(thioxo)methyl]-5-hydroxy-3-methyl-1H-pyrazole (1d) was executed as described before (Sobiesiak et al. and Grazul et al.)20,23,24 with some modifications.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1594, 1571; ν (C–N) 1368; ν (N–N) 1026; ν (C
N) 1594, 1571; ν (C–N) 1368; ν (N–N) 1026; ν (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 886. Elemental analysis for C8H13N3S: C 52.42(52.35), H 7.15(7.17), N 22.93(22.99), S 17.50(17.42). 1H NMR (600 MHz, CDCl3-d) δ (ppm): 1.22 (t, 3H, –CH2CH3, 3JHH = 6.0 Hz), 1.27 (t, 3H, –CH2CH3, 3JHH = 6.0 Hz), 2.57 (q, 2H, 3JHH = 6.0 Hz, –CH2CH3), 3.27 (q, 2H, 3JHH = 6.0 Hz –CH2CH3), 6.07 (s, 1H, –CH pyrazole), 7.01, 8.64 (2 br. s., 2H, NH2).13C NMR (600 MHz, CDCl3-d) δ (ppm): 179.4 (C
S) 886. Elemental analysis for C8H13N3S: C 52.42(52.35), H 7.15(7.17), N 22.93(22.99), S 17.50(17.42). 1H NMR (600 MHz, CDCl3-d) δ (ppm): 1.22 (t, 3H, –CH2CH3, 3JHH = 6.0 Hz), 1.27 (t, 3H, –CH2CH3, 3JHH = 6.0 Hz), 2.57 (q, 2H, 3JHH = 6.0 Hz, –CH2CH3), 3.27 (q, 2H, 3JHH = 6.0 Hz –CH2CH3), 6.07 (s, 1H, –CH pyrazole), 7.01, 8.64 (2 br. s., 2H, NH2).13C NMR (600 MHz, CDCl3-d) δ (ppm): 179.4 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 155.6, 152.50 (2C pyrazole), 109.0 (CH pyrazole), 24.33, 21.45 (2CH2), 12.80, 12.77 (2CH3). ESI-MS (m/z): 182.1 (100%); 184.1(25%).
S), 155.6, 152.50 (2C pyrazole), 109.0 (CH pyrazole), 24.33, 21.45 (2CH2), 12.80, 12.77 (2CH3). ESI-MS (m/z): 182.1 (100%); 184.1(25%).2.2.2.1. Synthesis of [(η6-p-cymene)Ru(1-[amino(thioxo)methyl]-3,5-dimethyl-1H-pyrazole)Cl2 (2a). The solution of 1-[amino(thioxo)methyl]-3,5-dimethyl-1H-pyrazole (47.50 mg, 0.3 mmol) in 6 ml of dry anhydrous dichloromethane was dropped slowly to the solution of dichloro(p-cymene)ruthenium(II)dimer (92.06 mg, 0.15 mmol) in 5 ml of dry anhydrous dichloromethane. Next, the mixture was stirred out under atmospheric argon condition at room temperature in Schlenk tube. After 24 hour heating, the mixture was concentrated under reduced pressure and then diethyl ether was added. The precipitation was filtered off as orange solid. Single crystals of 2a for X-ray analysis were obtained from recrystallization of precipitated product (diffusion method in solvents system: dichloromethane/hexane) Yield: (127.76 mg, 88.83%), orange crystals, mp 131.5–132.3 °C. FTIR (KBr cm−1): ν (N–H) 3379; ν (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1622; ν (C
N) 1622; ν (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1470; ν (C–N) 1353; ν (C–H) 1157; ν (N–N) 1052, ν (C
C) 1470; ν (C–N) 1353; ν (C–H) 1157; ν (N–N) 1052, ν (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 872. Anal. calcd for C16H24ClN3SRu 0.5CH2Cl2 (M = 479.42 g mol−1): C 40.93 (40.96), H 5.15 (5.55), N 8.95(8.97), S 6.83(6.85). 1H NMR (600 MHz, DMSO-d6) δ (ppm): 1.02, 1.15 (2d, 3JHH = 12.0 Hz, 6H, CH(CH3)2 p-cymene), 2.08 (s, 3H, CH3 p-cymene), 2.62 (septet, 3JHH = 6.0 Hz, 1H, CH(CH3)2 p-cymene), 2.70 (2 s, 6H, 2CH3 pyrazole), 5.70, 6.05, 6.12, 6.16 (4d, 3JHH = 6.0 Hz, 4Ar CH), 6.70 (s, 1H, CH pyrazole), 9.15 (s, 1H, –NH), 11.41 (s, 1H, –SH pyrazole). 13C NMR (600 MHz, DMSO-d6) δ (ppm): 14.22, 17.11, 18.35, 18.73, 21.60 (5CH3), 23.11 (CH), 30.90 (CH pyrazole), 80.95, 83.14, 85.98, 86.83, 86.95, 87.84 (6CHAr p-cymene), 115.33(C–NH), 145.70 (C
S) 872. Anal. calcd for C16H24ClN3SRu 0.5CH2Cl2 (M = 479.42 g mol−1): C 40.93 (40.96), H 5.15 (5.55), N 8.95(8.97), S 6.83(6.85). 1H NMR (600 MHz, DMSO-d6) δ (ppm): 1.02, 1.15 (2d, 3JHH = 12.0 Hz, 6H, CH(CH3)2 p-cymene), 2.08 (s, 3H, CH3 p-cymene), 2.62 (septet, 3JHH = 6.0 Hz, 1H, CH(CH3)2 p-cymene), 2.70 (2 s, 6H, 2CH3 pyrazole), 5.70, 6.05, 6.12, 6.16 (4d, 3JHH = 6.0 Hz, 4Ar CH), 6.70 (s, 1H, CH pyrazole), 9.15 (s, 1H, –NH), 11.41 (s, 1H, –SH pyrazole). 13C NMR (600 MHz, DMSO-d6) δ (ppm): 14.22, 17.11, 18.35, 18.73, 21.60 (5CH3), 23.11 (CH), 30.90 (CH pyrazole), 80.95, 83.14, 85.98, 86.83, 86.95, 87.84 (6CHAr p-cymene), 115.33(C–NH), 145.70 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). ESI-MS (m/z): 423.1(42%) 425.0(69%) 426.1(100%) 427.1(36%) 428.0(76%).
S). ESI-MS (m/z): 423.1(42%) 425.0(69%) 426.1(100%) 427.1(36%) 428.0(76%).
2.2.2.2. Synthesis of [(η6-p-cymene)Ru(1-[amino(thioxo)methyl]-3,5-diethyl-1H-pyrazole)Cl2 (2b). The solution of 1-[amino(thioxo)methyl]-3,5-diethyl-1H-pyrazole (55.0 mg, 0.3 mmol) in 6 ml of dry anhydrous isopropanol and 3 ml anhydrous dichloromethane was dropped slowly to the solution of dichloro(p-cymene)ruthenium(II)dimer (92.0 mg, 0.15 mmol) in 5 ml of dry anhydrous dichloromethane. Next, the mixture was stirred under atmospheric argon at room temperature in Schlenk tube. After 24 h of heating, the mixture was concentrated under reduced pressure and then diethyl ether was added. The precipitation was filtered as orange solid. Single crystals of 2b from X-ray analysis were obtained from recrystallization of precipitated product (diffusion method in solvents system: dichloromethane/hexane). Yield: (109.79 mg, 82.1%), orange crystals, mp 147.9–148.6 °C. FTIR (KBr cm−1): ν (N–H) 3417; ν (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1606; 1486 ν (C
N) 1606; 1486 ν (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1470; ν (C–N) 1363; ν (C–H) 1166; ν (N–N) 1046, ν (C
C) 1470; ν (C–N) 1363; ν (C–H) 1166; ν (N–N) 1046, ν (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 872. Anal. calcd for C18H27ClN3SRu (M = 496.51 g mol−1): C 43.63(43.60), H 5.29(5.64), N 8.48(8.16), S 6.47(7.03).
S) 872. Anal. calcd for C18H27ClN3SRu (M = 496.51 g mol−1): C 43.63(43.60), H 5.29(5.64), N 8.48(8.16), S 6.47(7.03).1HNMR (600 MHz, DMSO-d6) δ (ppm): 1.04, 1.17 (2d, 3JHH = 6.0 Hz, 6H, CH(CH3)2), 1.26, 1.37 (2t, 3JHH = 18.0 Hz, 6H, CH2CH3), 2.16 (s, 3H, CH3 p-cymene), 2.62 (septet, 3JHH = 6.0 Hz, 1H, CH(CH3)2), 3.16 (m, 4H, CH2CH3), 5.72, 6.04, 6.14, 6.19 (4d, 3JHH = 6.0 Hz, 4Ar CH), 6.84 (s, CH pyrazole), 11.40 (s, 1H, –NH pyrazole).13C NMR (600 MHz, DMSO-d6) δ (ppm): 11.83, 12.94, 18.29, 18.76, 21.21, 21.44, 23.05 (7CH3), 24.30 (CH), 30.86 (CH pyrazole), 81.03, 83.27, 86.01, 86.81, 87.28, 88.08 (CHAr p-cymene), 111.11(C–NH), 152.01 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). ESI-MS (m/z): 415.1(45%) 416.1(49%) 417.1(59%) 418.1(100%) 419.1(33%) 420.0(74%).
S). ESI-MS (m/z): 415.1(45%) 416.1(49%) 417.1(59%) 418.1(100%) 419.1(33%) 420.0(74%).
2.2.2.3. Synthesis of [(η6-p-cymene)Ru(1-[amino(thioxo)methyl]-5-hydroxy-3-phenyl-1H-pyrazole)Cl2 (2c). The solution of 1-[amino(thioxo)methyl]-5-hydroxy-3-phenyl-1H-pyrazole (67.52 mg, 0.3 mmol) in 8 ml of dry anhydrous dichloromethane and 2.5 ml anhydrous isopropanol was dropped slowly to the solution of dichloro(p-cymene)ruthenium(II)dimer (92.46 mg, 0.15 mmol) in 6 ml of dry anhydrous dichloromethane. Next, the mixture was stirred under atmospheric argon at room temperature in Schlenk tube. After 24 h of heating, the mixture was concentrated under reduced pressure and then diethyl ether was added. The precipitation was filtered off as orange solid. Single crystals of 2c for X-ray analysis were obtained from recrystallization of precipitated product (diffusion method in solvents system: dichloromethane/hexane). Yield: (137.31 mg, 84.29%), bright orange crystals, mp (dec. > 274 °C). FTIR (KBr cm−1): ν (OH) 3237; ν (N–H) 3072; ν (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1651; ν (C
N) 1651; ν (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) 1609; 1458 (C
O) 1609; 1458 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1470; ν (C–N) 1391; ν (N–N) 1087, ν (C
C) 1470; ν (C–N) 1391; ν (N–N) 1087, ν (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 872. Anal. calcd for C20H25ClN3OSRu·1.3H2O (M = 528.5 g mol−1) C 45.45(45.92), H 4.73 (4.93), N 7.95(7.93), S 6.06(5.34).1H NMR (600 MHz, DMSO-d6) δ (ppm): 0.93, 1.00 (2d, 3JHH = 6.0 Hz, CH(CH3)2), 2.76 (s, CH3 p-cymene), 2.43 (septet, 3JHH = 6.0 Hz, CH(CH3)2), 4.71, 4.80, 5.13, 5.63 (4d, 3JHH = 6.0 Hz, 4Ar CH), 5.20 (s, CH pyrazole), 7.65 (m, 3H Ar CH), 8.02 (m, 2H Ar CH), 10.35 (s, 1H, –OH pyrazole), 10.80 (s, 1H, –NH pyrazole). 13C NMR (600 MHz, DMSO-d6) δ (ppm): 18.64, 21.24, 21.91, 22.97 (4CH3), 30.54 (CH p-cymene), 80.46, 83.36, 85.64, 85.86, 86.37, 87.93 (12CHAr), 100.47 (CH), 66.94 (C
S) 872. Anal. calcd for C20H25ClN3OSRu·1.3H2O (M = 528.5 g mol−1) C 45.45(45.92), H 4.73 (4.93), N 7.95(7.93), S 6.06(5.34).1H NMR (600 MHz, DMSO-d6) δ (ppm): 0.93, 1.00 (2d, 3JHH = 6.0 Hz, CH(CH3)2), 2.76 (s, CH3 p-cymene), 2.43 (septet, 3JHH = 6.0 Hz, CH(CH3)2), 4.71, 4.80, 5.13, 5.63 (4d, 3JHH = 6.0 Hz, 4Ar CH), 5.20 (s, CH pyrazole), 7.65 (m, 3H Ar CH), 8.02 (m, 2H Ar CH), 10.35 (s, 1H, –OH pyrazole), 10.80 (s, 1H, –NH pyrazole). 13C NMR (600 MHz, DMSO-d6) δ (ppm): 18.64, 21.24, 21.91, 22.97 (4CH3), 30.54 (CH p-cymene), 80.46, 83.36, 85.64, 85.86, 86.37, 87.93 (12CHAr), 100.47 (CH), 66.94 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 165.83(C–NH), 175.88 (C
O), 165.83(C–NH), 175.88 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). ESI-MS (m/z): 451.1(42%) 452.2(39%) 453.1(63%) 454.1(100%) 456.0(60%).
S). ESI-MS (m/z): 451.1(42%) 452.2(39%) 453.1(63%) 454.1(100%) 456.0(60%).
2.2.2.4. Synthesis of [(η6-p-cymene)Ru(1-[amino(thioxo)methyl]-5-hydroxy-3-methyl-1H-pyrazole) Cl2 (2d). The solution of 1-[amino(thioxo)methyl]-5-hydroxy-3-methyl-1H-pyrazole (48.14 mg, 0.3 mmol) in 8 ml of dry anhydrous dichloromethane was dropped slowly to the solution of dichloro(p-cymene)ruthenium(II)dimer (92.03 mg, 0.15 mmol) in 6 ml of dry anhydrous dichloromethane. Obtained mixture was stirred out under atmospheric argon at room temperature in Schlenk tube. After 24 h of heating, the mixture was concentrated under reduced pressure and then diethyl ether was added. The precipitation was filtered off as brown solid. Single crystals of 2d for X-ray analysis were obtained from recrystallization of precipitated product (method in solvents system in methanol). Yield: (146.98 mg, 92.71%), brown crystals, mp 144.3–145.8 °C. FTIR (KBr cm−1): ν (NH2) 3459, 3367; ν (OH) 3240; ν (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) 1603; ν (C
O) 1603; ν (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1492, 1477; ν (C–N) 1397, ν (N–N) 1021; ν (C
C) 1492, 1477; ν (C–N) 1397, ν (N–N) 1021; ν (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 891. Anal. calcd for C15H22ClN3O2SRu·H2O (M = 491.39 g mol−1) C 36.66(36.09), H 4.51(4.84), N 8.55(8.26), S 6.53(6.22). 1H NMR (600 MHz, DMSO-d6) δ (ppm): 0.98, 1.03 (2d, 3JHH = 6.0 Hz, 6H, CH(CH3)2), 2.07 (s, CH3), 2.37 (s, CH3 pyrazole), 2.55 (septet, 3JHH = 6.0 Hz, CH(CH3)2), 5.50, 5.83, 5.89, 5.92 (4d, 3JHH = 6.0 Hz, 4H Ar CH), 10.13 (s, 1H, OH), 10.56 (s, 1H, NH). 13C NMR (600 MHz, DMSO-d6) δ (ppm): 15.59, 16.69, 17.67, 18.62 (4CH3), 25.54 (C pyrazole), 31.02 (CH p-cymene), 65.82 (CH pyrazole), 80.35, 82.92, 84.61, 85.89, 86.10, 87.49 (6CHAr), 161.68 (C
S) 891. Anal. calcd for C15H22ClN3O2SRu·H2O (M = 491.39 g mol−1) C 36.66(36.09), H 4.51(4.84), N 8.55(8.26), S 6.53(6.22). 1H NMR (600 MHz, DMSO-d6) δ (ppm): 0.98, 1.03 (2d, 3JHH = 6.0 Hz, 6H, CH(CH3)2), 2.07 (s, CH3), 2.37 (s, CH3 pyrazole), 2.55 (septet, 3JHH = 6.0 Hz, CH(CH3)2), 5.50, 5.83, 5.89, 5.92 (4d, 3JHH = 6.0 Hz, 4H Ar CH), 10.13 (s, 1H, OH), 10.56 (s, 1H, NH). 13C NMR (600 MHz, DMSO-d6) δ (ppm): 15.59, 16.69, 17.67, 18.62 (4CH3), 25.54 (C pyrazole), 31.02 (CH p-cymene), 65.82 (CH pyrazole), 80.35, 82.92, 84.61, 85.89, 86.10, 87.49 (6CHAr), 161.68 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 165.24 (C–NH), 175.18 (C
O), 165.24 (C–NH), 175.18 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). ESI-MS (m/z): 389.1(34%) 390.1(38%) 394.1(52%) 392.1(100%) 394.1(54%).
S). ESI-MS (m/z): 389.1(34%) 390.1(38%) 394.1(52%) 392.1(100%) 394.1(54%).
2.3. Crystal structure determination
Single crystal X-ray diffraction data of compounds 2a, 2b, 2c and 2d were collected on a Stoe IPDS-2T diffractometer with graphite-monochromated Mo-Kα radiation; crystals were cooled using a Cryostream 800 open flow nitrogen cryostat (Oxford Cryosystems). Data collection and image processing were performed with X-Area 1.75.46 Intensity data were scaled with LANA (the part of X-Area) in order to minimize different intensities of symmetry-equivalent reflections (the multi-scan method).Structures were solved by direct methods and all non-hydrogen atoms were refined with anisotropic thermal parameters by full-matrix least squares procedure based on F2 using the SHELX-2014 program package.47 The Olex48 and Wingx49 program suites were used to prepare the final cif files. Figures were prepared with the freeware Mercury 4.0.0.50 Hydrogen atoms were usually refined using isotropic model with Uiso(H) values fixed to be 1.5 times Ueq. of C atoms for –CH3 or 1.2 times Ueq. for –CH2 and –CH groups. Disordered molecule of dichloromethane, present in the crystals of compound 2a, was refined as disordered between two positions with occupation factors 0.59(3)/0.41(3). Crystal parameters and refinement details are collected in Table 1.
| Identification code | 2a | 2b | 2c | 2d |
| Empirical formula | C17H25Cl4N3RuS | C18H27Cl2N3RuS | C20H22ClN3ORuS | C15H22ClN3O2RuS |
| Formula weight [u] | 546.33 | 489.45 | 488.98 | 444.93 |
| Temperature [K] | 120(2) | |||
| Wavelength [Å] | 0.71073 | |||
| Crystal system | Orthorhombic | Orthorhombic | Monoclinic | Orthorhombic |
| Space group | Pbca | Pbca | P21/c | P212121 |
| Unit cell dimensions | ||||
| a [Å] | 13.077(7) | 10.6918(18) | 10.6196(18) | 10.398(2) |
| b [Å] | 14.235(6) | 14.539(2) | 12.7321(17) | 12.551(3) |
| c [Å] | 23.698(10) | 26.597(4) | 14.8584(15) | 13.766(5) |
| α [°] | 90 | 90 | 90 | 90 |
| β [°] | 90 | 90 | 107.980(11) | 90 |
| γ [°] | 90 | 90 | 90 | 90 |
| Volume [Å3] | 4412(4) | 4134.5(11) | 1910.9(5) | 1796.6(8) |
| Z | 8 | 8 | 4 | 4 |
| Density (calcd.) [mg m−3] | 1.645 | 1.573 | 1.700 | 1.645 |
| Absorption coefficient [mm−1] | 1.297 | 1.124 | 1.085 | 1.149 |
| Absorption corr. method | Integration | |||
| F(000) | 2208 | 2000 | 992 | 904 |
| Crystal size [mm] | 0.132 × 0.092 × 0.059 | 0.107 × 0.073 × 0.033 | 0.075 × 0.059 × 0.029 | 0.095 × 0.077 × 0.044 |
| θ range for data collect [°] | 2.283 to 25.998 | 2.444 to 25.999 | 3.200 to 25.999 | 2.943 to 25.995 |
| Reflections collected | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 362 362 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 018 018 |
9994 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 472 472 |
| Independent reflections | 4273 | 4043 | 3737 | 3509 |
| R(int) | 0.1075 | 0.0398 | 0.0531 | 0.0678 |
| Completeness to θ = 25.242° | 0.985 | 0.995 | 0.994 | 0.994 |
| Data/restraints/parameters | 4273/2/268 | 4043/0/239 | 3737/0/247 | 3509/0/216 |
| Goodness-of-fit on F2 | 1.082 | 1.040 | 1.019 | 1.037 |
| Final R indices [I > 2sigma(I)] | R1 = 0.0816 | R1 = 0.0284 | R1 = 0.0400 | R1 = 0.0441 |
| wR2 = 0.1567 | wR2 = 0.0563 | wR2 = 0.0776 | wR2 = 0.0829 | |
| R indices (all data) | R1 = 0.1343 | R1 = 0.0426 | R1 = 0.0672 | R1 = 0.0655 |
| wR2 = 0.1763 | wR2 = 0.0610 | wR2 = 0.0860 | wR2 = 0.0906 | |
| Largest diff. peak/hole [e Å−3] | 0.945 and −1.521 | 0.438 and −0.481 | 0.561 and −0.730 | 0.556 and −0.588 |
Crystallographic data for the structural analysis has been deposited with the Cambridge Crystallographic Data Center, no. CCDC 1896357 2a, CCDC 1896358 2b, CCDC 1896359 2c, CCDC 1896360 2d.
2.4. Minimal inhibitory, bactericidal/fungicidal concentration (MIC, MBC/MFC)
The MIC of arene–ruthenium(II) complexes with carbothioamidopyrazole derivatives 2a–2d, as well as the ligands alone 1a–1d and dimer, tested at a final concentration range of 31.2–1000 μg ml−1, were determined by the broth microdilution method, according to the EUCAST guidelines.51 Stock solutions of the lyophilized compounds (80 mg ml−1) were freshly prepared in 100% DMSO (POCh, Poland) and further diluted in liquid culture medium (Mueller-Hinton Broth (BTL, Poland) for bacteria; RPMI-1640 without phenol red (Sigma, USA) for fungi). The highest concentration of the solvent never exceeded 1.25% in order not to limit viability of the microbes. Briefly, MIC was defined as the lowest concentration of the compounds inhibiting bacterial/fungal growth after 24–48 h of co-incubation at 37 °C compared to the appropriate positive controls. The microorganisms in culture medium containing DMSO at the highest final concentration (1.25% vol/vol) served as positive controls. The lowest concentration of the preparations that prevented growth of the bacteria or yeast after subculturing 10 μl from the wells marked as MIC, 2× MIC and 4× MIC on solid (agar) medium (incubation for 24 h at 37 °C) was interpreted as MBC/MFC. Experiments were carried out in duplicate in each of 2 separate sets of experiments.2.5. Cytotoxic activity on human foreskin fibroblasts HFF-1
Cytotoxicity of the compounds was tested on human foreskin fibroblasts HFF-1 (ATCC-SCRC-1041) (LGC Standards Sp. z o.o., Poland) using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction assay. Briefly, HFF-1 were seeded 1 × 105 per well on 96-well plates (Nunc, Denmark) in DMEM (Sigma, USA) with 15% (v/v) fetal calf serum (FCS; Cytogen, Poland) and 1% (v/v) penicillin/streptomycin (Sigma, USA) and cultured for 48 h at 37 °C, 5% CO2. Stock solutions of the lyophilized compounds (25 mM) were freshly prepared in 100% DMSO (POCh, Poland) and further diluted in cell culture medium to obtain the final concentration range of 3.9–500 μM. The final concentration of DMSO did not exceed 2%, i.e. the value that did not affect viability of eukaryotic cells. The cells in culture medium containing DMSO (2% v/v) served as positive control (100% of cells viability). HFF-1 were exposed to arene–ruthenium(II) complexes, ligands and dimer for 24 h under the above culture conditions. The supernatants were removed, the cells were washed and next, 100 μl fresh culture medium and 50 μl MTT (1.5 mg ml−1 in PBS; Sigma, USA) was added and the incubation was continued for 2 h. After the aspiration of the MTT-containing medium, 100 μl of 20% sodium dodecylsulfate (SDS; Sigma, USA) in the mixture of dimethylforamide (DMF) with water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added to dissolve blue formazan crystals produced by metabolically active cells. After overnight incubation at room temperature, the absorbance was read (λ = 550 nm) using multifunctional plate reader Victor2 (Wallac, Finland) and compared with positive control to calculate the percentage of cell viability and the IC50 (concentration contributing to 50% loss of viability). IC50 were calculated based on curve equations determined for the lines of trends the cell viability dependence on tested compounds concentration. Two independent experiments with two replicates in each were performed.
1) was added to dissolve blue formazan crystals produced by metabolically active cells. After overnight incubation at room temperature, the absorbance was read (λ = 550 nm) using multifunctional plate reader Victor2 (Wallac, Finland) and compared with positive control to calculate the percentage of cell viability and the IC50 (concentration contributing to 50% loss of viability). IC50 were calculated based on curve equations determined for the lines of trends the cell viability dependence on tested compounds concentration. Two independent experiments with two replicates in each were performed.
2.6. Anticancer activity of ligands and their complexes 2a–2d
Cytotoxicity of all ligands and their complexes was tested against human leukemia promyelocytic HL-60, lymphoblastic NALM-6 and human skin melanoma WM-115 cell lines. Carboplatin and cisplatin were used as a reference. The cytotoxic effect was determined using MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, Sigma-Aldrich, St. Louis, MO] assay as described before. All tested compounds were incubated with tested cell lines for 46 h. Next, cells were treated with the MTT reagent and incubation was continued for another 2 h. IC50 values (expressed as the concentration of the tested compound required to reduce the cells survival fraction to 50% of the control) were calculated from concentration–response curves. Stock solutions of the tested compounds (1a–1d, 2a–2d, dimer, carboplatin and cisplatin) were every time freshly prepared in DMSO solvent. All ligands 1a–1d were diluted in complete culture medium to final concentration 10−7 to 10−3 M, while arene Ru(II)complexes 2a–2d were diluted to final concentration 10−7 to 10−4 M. In these assay, DMSO concentration never exceeded 0.2% and had no influence on cell growth. Moreover, cultured cells that grew in the absence of drugs were used as a control.2.7. Cleavage of pUC57 DNA
The DNA cleavage ability of the ruthenium(II) complexes was studied by using agarose gel electrophoresis. Supercoiled plasmid pUC57 DNA (4634bp), dissolved in 10 mM Tris–HCl 50 mM NaCl buffer (pH 7.2), was treated with tested compounds (50, 100, 200, 300 and 400 μM). All mixtures were incubated at 37 °C for 24 h and then the loading buffer (1 μl) containing 25% bromophenol blue, 0.25% xylene cyanol and 30% glycerol, was added.Each sample (10 μl) was loaded into 1% w/v agarose gel. Electrophoresis was performed at 75 V in Tris–acetate–EDTA (TAE) buffer. The gel was stained with Midori green advance and then photographed under UV light. The proportion of DNA in each fraction was quantitatively estimated from the intensity of each band with the Syngen BTX-20.M system using the Scion Image software. All experiments were carried out in triplicate under the same conditions.
2.8. DFT calculations
DFT calculations i.e. geometry optimization, were performed using ADF program (version 2018.01)52–54 with BP86-D potential and TZP basis in SCF model as implemented in ADF.55–57 All calculations were performed in vacuo.3. Results and discussion
3.1. Synthesis of compounds
All ligands 1a–1d can be obtained in reactions of thiosemicarbazide derivatives with appropriate diketones or ketoesters. Cyclic pyrazole-bearing compounds with the carbothioamide group may exist as several tautomeric forms (Scheme 2).21Carbothioamidopyrazoles 1a–1d possess three potential coordination sites: the nitrogen atom of the pyrazole ring, as well as the nitrogen and sulfur atoms of the thioamide group. Each ligand may act as a neutral, bi- or monodentate moiety.22 In our experiments, all ligands bind to the ruthenium(II) ion as bidentate N,S-donors to form the corresponding complexes 2a–2d. All complexes were synthesized in the reaction between a dichloro(p-cymene)ruthenium(II)dimer (3) and appropriate carbothioamidopyrazoles containing various substituents in C-3 and C-5 position of pyrazole ring: dimethyl substituent (for complex 2a), diethyl (for complex 2b), phenyl/methyl and carbonyl or hydroxyl group (for complexes 2c/2d). All reactions were executed with ligand: metal molar ratio (L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M) 2
M) 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and the reactions for complex 2a and 2b were carried out in anhydrous dicholoromethane, used as a solvent (Scheme 3).
1 and the reactions for complex 2a and 2b were carried out in anhydrous dicholoromethane, used as a solvent (Scheme 3).
In other reactions, the anhydrous dichloromethane and isopropyl alcohol were used as a solvent for complexes 2c and 2d. All reactions were carried out at room temperature. Complexes 2a–2d were obtained as colored powders or solids. Compounds 2a–2d were structurally characterized with the use of the single crystal X-ray diffraction analysis.
3.2. Characterization of ligand 1b and half-sandwich complexes 2a–2d
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) group were observed shifted to 1594 cm−1 and 1571 cm−1 (for ligands) and to higher regions stretching from 1606 to 1651 cm−1 (for all complexes). Complexes of amino group protons in NMR spectra gave signals from 9.15 ppm to 11.40 ppm. The characteristic bands in IR spectra were assigned a range from 3273 cm−1 to 3459 cm−1 of the amino group of complexes 2a–2d. The 13C NMR resonance of the C
N) group were observed shifted to 1594 cm−1 and 1571 cm−1 (for ligands) and to higher regions stretching from 1606 to 1651 cm−1 (for all complexes). Complexes of amino group protons in NMR spectra gave signals from 9.15 ppm to 11.40 ppm. The characteristic bands in IR spectra were assigned a range from 3273 cm−1 to 3459 cm−1 of the amino group of complexes 2a–2d. The 13C NMR resonance of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S group gave signals for complexes 2a–2d 145.70, 152.01, 175.88 and 175.18 ppm, respectively. Important signal v(C
S group gave signals for complexes 2a–2d 145.70, 152.01, 175.88 and 175.18 ppm, respectively. Important signal v(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) for complexes showed stretching vibrations to 872 cm−1 (for compounds 2a–2c), 891 cm−1 (for compound 2d), while for ligand they were shifted to 886 cm−1. It may suggest that sulfur atom in substituents C
S) for complexes showed stretching vibrations to 872 cm−1 (for compounds 2a–2c), 891 cm−1 (for compound 2d), while for ligand they were shifted to 886 cm−1. It may suggest that sulfur atom in substituents C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S can be a possible site of metal coordination.
S can be a possible site of metal coordination.| Complex | Type ions [XRuL]+ | Type ions [XRuLCl]+ | Sandwich ions | Ions in negative mode |
|---|---|---|---|---|
| 2a | 388.1(34%), 389.1(42%), 390.1(72%), 392.0(40%) | 423.1(42%), 425(69%), 426.1(100%), 427.1(36%), 428.0(76%) | 811.9(11%), 812.9(13%), 813.6(10%), 814.8(19%), 815.6(13%), 816.9(10%) | Not observed |
| 2b | 415.1(45%), 416.1(49%), 417.1(59%), 418.1(100%), 419.1(33%), 420(74%) | 452.1(20%), 453.1(43%), 454.1(63%), 456.0(54%) | Not observed | |
| 2c | 451.1(42%), 452.2(39%), 453.1(63%), 454.1(100%), 456.0(60%) | Not observed | 486.4(29%), 487(64%), 488.3(100%), 489.3(35%), 490(64%) | |
| 2d | 389.1(34%), 390.1(38%), 394.1(52%), 392.1(100%), 394.1(54%) | 426.1(9%), 426.9(8%), 428.1(15%), 430.1(11%) | 780.0(6%), 781.0(5%), 782.0(12%), 782.4(6%), 783.0(11%), 783.8(4%) | Not observed |
Various types of ions were observed for all tested compounds. The most intense signals were obtained for ions, created after losing one or two chlorine atoms. Simulated isotope patterns for 2a with the chemical formula [C16H23Cl2N3SRu]+ cations exactly matched the mass spectra of complexes 2b, which means that ionization of these compounds took place via the loss of chloride ions. Relevant charts and scan of their mass spectrum are provided in ESI (Fig. 1S†). Very interesting signals were observed at around 815 Da for complex 2a and around 783 Da for complex 2d. It was found that these compounds can exist as a complex of two ligands or two complexes. According to literature, this is a common feature for half-sandwich Ru(II) complexes, where ruthenium is connected to the electronegative substituents. Examples are shown in Fig. 2S in ESI.†25
The tandem analysis was performed for ions with the highest abundance. All ions [XRuLCl]+, during the first step, eliminate fragments of hydrogen chloride. Further fragments of ms3 led to leaving HN = C = S (59 Da). Similar effects were observed for ligand (1b). The molecule of thiocyanate molecule was easily eliminated. The presented analysis of ESI-MS spectra shows that used compounds are stable in the solutions and it is difficult to break ruthenium–sulfur coordination bonds or permanent nitrogen in tandem reactions even at 200 eV voltage on the capillary and 5000 eV on the needle.
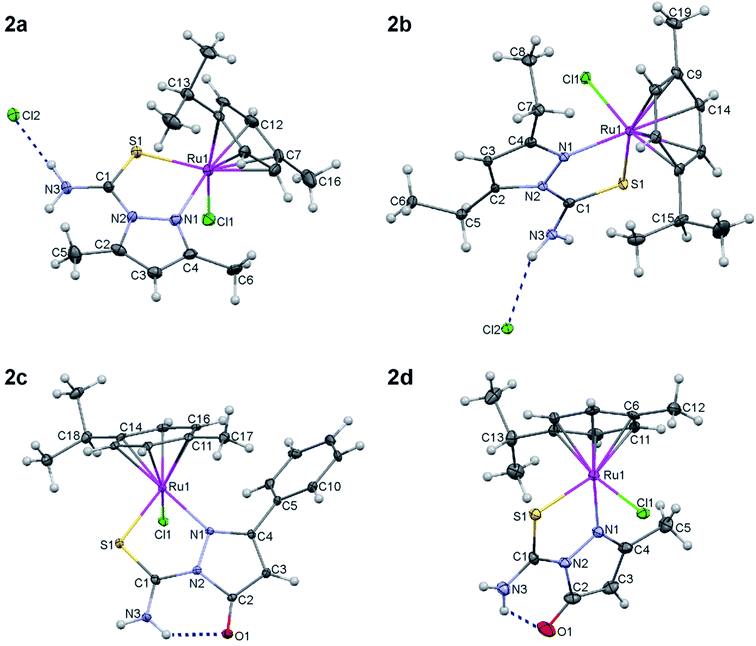
3.3. Crystal structures
The molecular structures of the complexes are illustrated in Fig. 1. Important bond lengths and angles are collected in Table 3. As it was shown in Scheme 3, two types of ruthenium complexes can be distinguished among the compounds – two ionic 2a, 2b and two molecular species 2c, 2d. The Ru(II) complex ions in 2a and 2b are accompanied by chloride counter ions. Interestingly, the changes in the molecular geometry, triggered by the change in the character of the heteroorganic ligand (neutral vs. anionic), are not conspicuous. They are rather visible within the ligands than in the ligand – metal interaction.| 2a | 2b | 2c | 2d | |
|---|---|---|---|---|
| Bond lengths [Å] | ||||
| Ru1–N1 | 2.101(8) | 2.104(2) | 2.109(4) | 2.087(6) |
| Ru1-centroid of p-cymene (CPC) | 1.704 | 1.691 | 1.683 | 1.688 |
| Ru1–S1 | 2.357(3) | 2.3260(7) | 2.3635(12) | 2.369(2) |
| Ru1–Cl1 | 2.411(3) | 2.4144(7) | 2.4393(11) | 2.429(2) |
| S1–C1 | 1.693(10) | 1.686(3) | 1.688(4) | 1.703(10) |
| N2–N1 | 1.385(10) | 1.401(3) | 1.410(5) | 1.404(9) |
| N2–C1 | 1.401(12) | 1.393(3) | 1.368(5) | 1.353(11) |
| N2–C2 | 1.416(12) | 1.397(3) | 1.438(5) | 1.434(11) |
| N1–C4 | 1.333(13) | 1.326(3) | 1.346(5) | 1.326(10) |
| C1–N3 | 1.309(12) | 1.313(4) | 1.312(6) | 1.320(11) |
| C4–C3 | 1.419(14) | 1.416(4) | 1.395(6) | 1.403(12) |
| C2–C3 | 1.351(14) | 1.359(4) | 1.406(6) | 1.375(14) |
| C2–O1 | — | — | 1.250(5) | 1.254(11) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Angles [°] | ||||
| N1–Ru1–CPC | 132.44 | 135.14 | 134.08 | 132.16 |
| N1–Ru1–S1 | 79.7(2) | 80.93(6) | 81.72(10) | 80.9(2) |
| S1–Ru1–CPC | 127.42 | 125.21 | 126.44 | 126.34 |
| N1–Ru1–Cl1 | 84.2(2) | 81.90(6) | 84.55(10) | 83.7(2) |
| Cl1–Ru1–CPC | 127.43 | 128.84 | 126.46 | 128.22 |
| S1–Ru1–Cl1 | 89.50(10) | 89.14(8) | 87.62(4) | 89.64(8) |
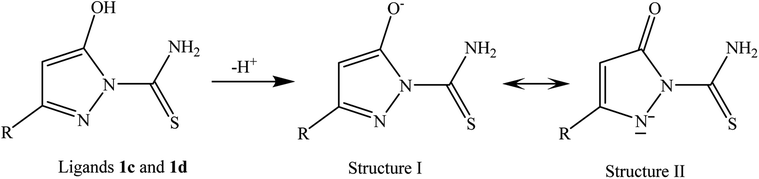
Carbon–oxygen bond in 2c and 2d is short – 1.250 Å – indicating the double bond character of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group; in carbonyl compounds, it is approximately 1.21–1.23 Å, single C–O is usually around 1.43 Å. The carbonyl bond of the ligand in 2c and 2d is slightly elongated in comparison with the typical range but this may be explained by the hydrogen bonding interaction between O1 and the adjacent amino group. Crystal structures of 2c and 2d prove that binding to the metal ion forces the mesomeric structure II of the deprotonated ligands 1c and 1d as illustrated in Scheme 4.
O group; in carbonyl compounds, it is approximately 1.21–1.23 Å, single C–O is usually around 1.43 Å. The carbonyl bond of the ligand in 2c and 2d is slightly elongated in comparison with the typical range but this may be explained by the hydrogen bonding interaction between O1 and the adjacent amino group. Crystal structures of 2c and 2d prove that binding to the metal ion forces the mesomeric structure II of the deprotonated ligands 1c and 1d as illustrated in Scheme 4.
Deprotonation of the hydroxyl ligands 2c and 2d influences C3–C4 within the pyrazole ring; the bond is shortened in comparison to the analogous bond in 2a and 2b because of larger double bond character, whereas N1–N2 become slightly longer because of the electrostatic repulsion (see Table 3 and Scheme 3). However, the bonds to Ru remain almost the same for nearly all surrounding atoms in all complexes with the exception of Ru–Cl1 which is elongated by ca. 0.021 Å in the neutral 2c and 2d. It may be also noticed that Ru–CPC (see Table 1 for explanation) is slightly shortened in 2c/2d.
The exchange of neutral ligands for anionic ones does not change the “stiff” piano-stool geometry typical for the Ru(II)–arene complexes e.g.;26–30 the angles around Ru atom remain alike in all the studied compounds (Table 3). The overlays of the structures are presented in ESI as Fig. 3S.† As it was shown in Fig. 3S,† 2a and 2d adopt the most similar geometry; thus, it can be concluded that the mutual position of the ligands is mainly guided by the sterical hindrance, exerted by the substituents and not by the charge on the ligand.
Pseudo-tetrahedral coordination of four different ligands makes all complexes chiral but three out of four compounds crystallize in centrosymmetric groups as racemic mixtures. Only 2d crystallized as one of two enantiomers in a chiral P212121 space group.
The crystal packings and intermolecular interactions in solid 2a–2d are presented in ESI (Fig. 4S–8S†). Except for 2d, the complexes usually form hydrogen bonded dimers in the solid state. In the crystals of 2d, additional molecule of water, inserted between the molecules of Ru(II) complex, creates more complicated pattern of hydrogen bonds leading to the formation of 1D chain that is aligned approximately along the a-axis. Parameters of intra- and intermolecular hydrogen bonds are collected in Table 1S of the ESI.†
3.4. Antimicrobial activity
Direct antimicrobial activity of arene–ruthenium(II) complexes with a carbothioamidopyrazole derivatives 2a–2d, as well as the ligands alone 1a–1d and dimer, was tested on representatives of pathogenic, opportunistic and commensal microorganisms, including staphylococci, enterococci, Pseudomonas, Proteus and Candida. The results of minimal inhibitory concentration (MIC) and minimal bactericidal/fungicidal concentration (MBC/MFC) are presented in Table 4.| Compound | MIC [μg ml−1] MBC/MFC [μg ml−1] | |||||
|---|---|---|---|---|---|---|
| S. aureus ATCC 29213 | S. epidermidis ATCC 12228 | E. faecalis ATCC 29212 | P. aeruginosa ATCC 25619 | P. vulgaris ATCC 8427 | C. albicans ATCC 10231 | |
| 1a | >1000 | >1000 | >1000 | >1000 | >1000 | >1000 |
| >1000 | >1000 | >1000 | >1000 | >1000 | >1000 | |
| 1b | 1000 | >1000 | >1000 | >1000 | 1000 | 1000 |
| >1000 | >1000 | >1000 | >1000 | 1000 | 1000 | |
| 1c | 1000 | 62.5 | 500 | >1000 | >1000 | >1000 |
| >1000 | 1000 | >1000 | >1000 | >1000 | >1000 | |
| 1d | 1000 | 250 | 250 | >1000 | >1000 | >1000 |
| >1000 | 250 | >1000 | >1000 | >1000 | >1000 | |
| 2a | 125 | 62.5 | 62.5 | >1000 | >1000 | 250 |
| 125 | 125 | 125 | >1000 | >1000 | >1000 | |
| 2b | 62.5 | 31.2 | 62.5 | >1000 | >1000 | 250 |
| 125 | 62.5 | 62.5 | >1000 | >1000 | >1000 | |
| 2c | 31.2 | <31.2 | <31.2 | >1000 | >1000 | >1000 |
| 62.5 | <31.2 | <31.2 | >1000 | >1000 | >1000 | |
| 2d | 500 | 500 | 500 | >1000 | 1000 | 1000 |
| 500 | 500 | 1000 | >1000 | 1000 | >1000 | |
| Dimer | >1000 | >1000 | >1000 | >1000 | >1000 | 1000 |
| >1000 | >1000 | >1000 | >1000 | >1000 | >1000 | |
Antimicrobial effect of the compounds tested was observed against Gram-positive bacteria: Staphylococcus aureus, Staphylococcus epidermidis and Enterococcus faecalis, while all tested compounds had influence on Gram-negative bacteria (Pseudomonas aeruginosa, Proteus vulgaris) growth over the concentration range that was tested. The only exception was weak antimicrobial activity of 1b and 2d against P. vulgaris ATCC 8427 with MIC/MBC level of 1000 μg ml−1. It has to be noted that antimicrobial effect was observed for arene–ruthenium(II) complexes, while dimer alone and most of the used ligands remained inactive. Even if selected ligand possessed antimicrobial activity against defined microbial strain, such as 1c against S. epidermidis ATCC 12228 and E. faecalis ATCC 29212 (MIC at 62.5 μg ml−1 and 500 μg ml−1, respectively), the activity of corresponding complex 2c in the same configurations was more than 2-fold and 16-fold, respectively, stronger (MIC < 31.2 μg ml−1). Only compounds 2a and 2b were active against Candida albicans with MIC at 250 μg ml−1. However, both complexes indicated only fungistatic, but not fungicidal activity (MFC above the tested concentration range).
Initially, arene–ruthenium(II) complexes were not going to be used as antimicrobial agents. However, considering the fact that human microbiome plays an important role in preservation of healthy skin balance and mucous membrane barriers, as well as taking into account the etiology of many pathological changes (e.g. wound infections, cancer ulcerations), it is important to test antimicrobial activity of potential therapeutics. Epidemiological data indicate that wound infections frequently occur in patients with cancer, particularly with breast, head and neck cancers, as well as melanoma. The wound results from the tumor cells that infiltrate the skin and underlying tissues (malignant wounds) or is a consequence of complications of medical procedures (surgery, radiotherapy, chemotherapy).33–35 Since patients with cancers are mostly affected by opportunistic infections with a predominance of staphylococci as their etiological agents, the results in which we demonstrated biostatic and biocidal effect of tested complexes (mainly 2a–2c) against Gram-positive bacteria, such as S. aureus, S. epidermidis, and E. faecalis, seem to be favorable. Potential local application of tested arene–ruthenium(II)complexes on the skin could simultaneously reduce the development of melanoma cells and protect against wounds infections. Nevertheless, the range of antimicrobial activity of such preparations strongly depends on their structure and composition. For example, we did not observe biocidal activity of arene–ruthenium(II)complexes against Gram-negative bacteria and Candida yeast, while Kulkarni et al.36 demonstrated good activity of the pyrazole-based copper complexes with thiosemicarbazide arms against E. coli and P. aeruginosa, comparable with gentamycin activity, when they used 500 μg of the compounds in the disc-diffusion method. At the same time, other transition metal e.g. Co(II), Ni(II), and Zn(II) complexes did not show such significant antimicrobial activity.37 Similar discrepancies in the antimicrobial activity were observed by Mandal et al.37 who described a strong biostatic effect of cadmium and mercury complexes of 5-methyl pyrazole-3yl-N-(20-methylthiophenyl) methyleneimine, (MPzOATA) ligand, against some Gram-positive and Gram-negative bacteria, while nickel complexes with the same ligands ([Ni(MPzOATA)2](Cl)(PF6); [Ni(MPzOATA)2](ClO4)2CH3CN; [Ni(MPzOATA)2](BF4)2H2O) did not express such activity. Interestingly, the MICs of cadmium and mercury complexes against S. aureus reached similar values (35 μg ml−1 and 20 μg ml−1, respectively) as our 2b and 2c ruthenium complexes.38 Therefore, we postulate that every single new anticancer drug should be assessed for its antimicrobial effect specific test microorganisms, selected on the basis of its potential site of application and formulation.
3.5. Cytotoxic activity
To test cytotoxicity of arene–ruthenium(II) complexes with carbothioamidopyrazole derivatives 2a–2d, as well as the ligands alone 1a–1d and dimer against regular eukaryotic cells, human foreskin fibroblasts HFF-1 (ATCC-SCRC-1041) were chosen. The influence of tested compounds on the cell viability is shown in Fig. 2.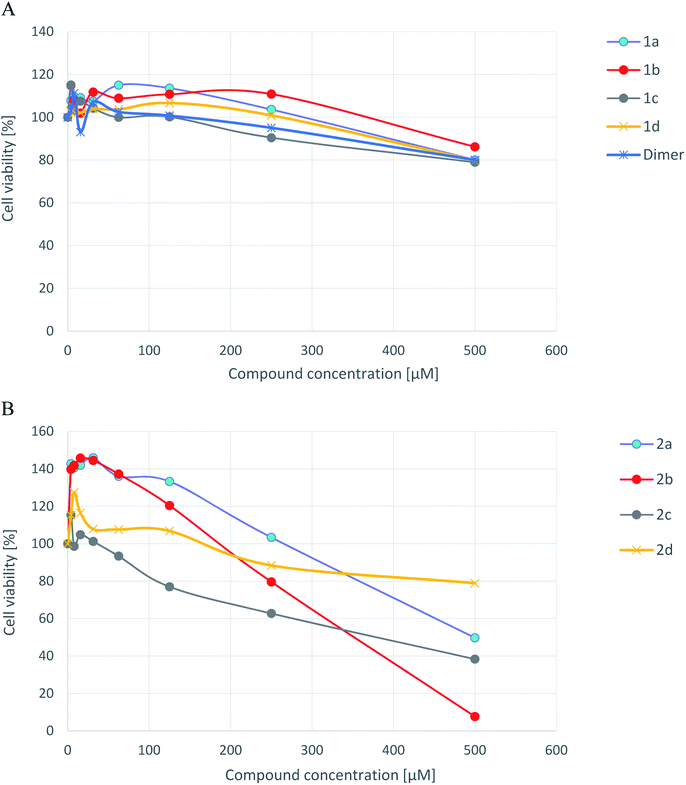 | ||
| Fig. 2 Cytotoxicity of ligands 1a–1d and dimer [A], and arene–ruthenium(II) complexes 2a–2d [B] for human foreskin fibroblasts line HFF-1 measured by the MTT method. | ||
Neither ligands nor dimer alone were cytotoxic against human fibroblasts over the tested concentration range. IC50 values were as high as 1220.5; 1740.2; 1001.1; 1384.0 μM for 1a–1d, respectively, and 1211.2 μM for dimer. Most of arene–ruthenium(II) complexes with pyrazole derivatives indicated cytotoxicity against tested cell lines. The strongest cytotoxicity showed complexes 2b and 2c with IC50 values 358.3 μM and 381.4 μM respectively. Slightly weaker cytotoxic effect indicated compound 2a with IC50 value 545.1 μM. The viability of HFF-1 cells did not drop below 80% in the presence of compound 2d (no cytotoxic effect), for which IC50 value was 850.5 μM. Interestingly, the complexes 2a and 2b used at low concentrations (3.9–31.2 μM) seems to stimulate human fibroblast divisions (metabolic activity of those cells was about 39–45% higher than untreated control cells). However, the possible mitogenic activity of 2a and 2b arene–ruthenium(II) complexes requires confirmation in the future studies.
3.6. Anticancer activity of novel ruthenium complexes
The in vitro anticancer activity was tested for all obtained arene–ruthenium(II) compounds 2a–2d and their ligands 1a–1d against HL-60 (leukemia promyelocytic), NALM-6 (leukemia lymphoblastic) and WM-115 (human melanoma) cell lines. Also, dichloro(p-cymene)ruthenium(II) dimer was tested against the above cell lines. Cisplatin and carboplatin were used as a ref. 31. The colorimetric MTT assay was used to determine the influence of broad range of compounds concentrations (from 10−7 to 10−5 M) on cells lines (exposure time was 48 h). Obtained results, expressed as IC50 values for all tested compounds, are presented in Table 5.| Compounds | Cell culture IC50a [μM] | ||
|---|---|---|---|
| HL-60 | NALM-6 | WM-115 | |
| a IC50 values [μM] were calculated at concentration of a tested compound required to reduce the fraction of surviving cell to 50% of that observed in comparison to the control probe, non treated cell. Mean values are presented of parameter IC50 ± SD from 4 experiments.b The values for referential compounds can be found in literature.23 | |||
| 1a | 583.9 ± 84.3 | 569.3 ± 21.9 | >1000 |
| 1b | 506.37 ± 47.08 | 468.52 ± 44.36 | 481.29 ± 39.43 |
| 1c | 618.0 ± 32.0 | 152.3 ± 26.7 | 751.5 ± 118.1 |
| 1d | 778.8 ± 125.3 | 614.7 ± 60.9 | 90.91 ± 10.2 |
| 2a | 88.86 ± 6.06 | 51.55 ± 5.71 | 60.24 ± 6.30 |
| 2b | 80.83 ± 3.9 | 40.03 ± 5.59 | 54.88 ± 5.94 |
| 2c | 86.51 ± 8.02 | 11.71 ± 1.62 | 26.66 ± 3.28 |
| 2d | 594.0 ± 52.0 | 491.4 ± 31.0 | 7.99 ± 0.87 |
| Dimer | 400.86 ± 46.22 | 373.89 ± 40.78 | >1000 |
| Cisplatinb | 0.8 ± 0.1 | 0.7 ± 0.3 | 18.2 ± 4.3 |
| Carboplatinb | 4.3 ± 1.3 | 0.7 ± 0.2 | 422.2 ± 50.2 |
The highest antitumor activity was observed for complex 2d against WM-115 cell line (IC50 = 7.99 ± 0.87 μM), while complex 2c was the most active against NALM-6 cell line (IC50 = 11.71 ± 1.62 μM). It went out that compound 2d is more cytotoxic than cisplatin and carboplatin. IC50 values for complexes 2a–2c were similar to each other against HL-60, NALM-6 and WM-115 cell lines and at the same time lower than for ligands. Complex 2d was inactive against acute leukemia (HL-60) as well as lymphoblastic (NALM-6) cell lines. All ligands 1a–1d and dichloro(p-cymene)ruthenium(II) dimer were inactive against all the tested cell lines: HL-60, NALM-6 and WM-115.
IC50 values for complexes 2a–2c against all tested cancer cell lines (HL-60, NALM-6, WM-115) were achieved at the concentration range from 11.7 to 88.9 μM, while against normal fibroblasts HFF-1 the same effect was observed at a concentration above 358 μM. The anticancer activity of 2d complex was strongly dependent on the used cell line. The best results were observed against melanoma cell line WM-115 with the activity above 2-fold stronger than cisplatin (7.9 μM versus 18.2 μM of IC50 values, respectively). Based on IC50 values arene–ruthenium(II) complexes with a pyrazole might be good alternative to cisplatin. A similar cytotoxic effect of 2d against HL-60 and NALM-6 was achieved only for concentration above 491 μM. The application of complex 2d as anticancer agent seems to be safe because as it was already mentioned before 50% cytotoxicity against HFF-1 cells was obtained at the concentration 850.5 μM.
3.7. Cleavage of pUC57 DNA
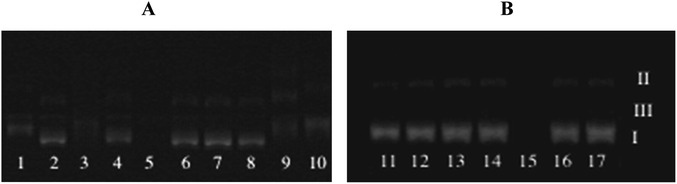
The cleavage of plasmid pUC57 DNA by tested compounds was tested with gel electrophoresis.The supercoiled form of DNA (Form I) that occurs naturally, when nicked, gives an open circular relaxed form (Form II) and further cleaves to a linear form (Form III). During electrophoresis, Form I shows the fastest migration compared to Forms II and III. Form II migrates slowly prior to its relaxed structure, while Form III migrates between the positions of Forms I and II.32 Pure DNA diluted in the mixture of DMSO (the same amount as for tested compounds) + TrisHCl/NaCl buffer without treatment with tested compound was used as control.
Lanes 1–17 (Fig. 3) represent DNA incubated with increasing amounts of the complexes, with concentrations of 50, 100 and 150 μM, respectively (results of the usage of 150 μM of compound are presented only for compound 2d and 2a). In most cases, complex concentration above 150 μM produced DNA sedimentation (except dimer compound that caused DNA sedimentation even in concentration 50 μM).
The results indicated that chosen ruthenium(II) complexes 2b, 2d can play a similar role as nucleases by cleavage the DNA Form I into Form II and Form III. It seems that both complexes are able to cut the DNA strand at two positions. The complexes cause an increase in the band intensity of Form III, while the intensity of Form I progressively decreased and these effects are definitely dependent on the concentration of the tested compound.
Compound 2a and 2c have no influence on the supercoiled plasmid DNA in any of tested concentrations.
All tested ligands 1a–1d as well as dimer 3 have no effect on the DNA strands, which means that the effect caused by compounds 2b, 2d is a result of their complex structures, not the activity of the ligands alone. However, it has to be noticed that only small concentrations of the dimer were used to the studies as it caused strong sedimentation of the DNA in higher concentrations.
The cytotoxicity of ruthenium complexes against cancer cell lines might be related to their ability to interactions with DNA.38,39 Many metal complexes can change the structure and stability of DNA by hydrogen bonding and π stacking between its strands.40 It has been well documented that transition–metal complexes are not only able to interact with DNA but also to damage its structure as well as other cellular structures. This might be a promising strategy for anticancer drug design.41 It has been proved that DNA is the main biological target of cisplatin – the successful anticancer drug.42 Ruthenium(II)–arene complexes similarly to cisplatin are less reactive at high chloride concentrations (e.g. in blood plasma), while at low chloride concentrations inside a cell, they are activated by aquation and then are able to interact with DNA and proteins.43,44
The cytotoxic effect of compounds 2b, 2d might be related to their ability to cleave the DNA as they cut the supercoiled DNA in two positions leading to the linear form of the DNA. It was also proved that the influence of complexes 2b, 2d on the DNA structure is associated with their own activity as ligands 1a, 1b, 1d did not affect the DNA. However it has to be noted, that while although DNA binding could be the mechanism of action, other factors e.g. cell uptake will have an influence on the cytotoxicity and can depend on the nature of the compound.
3.8. DFT calculations
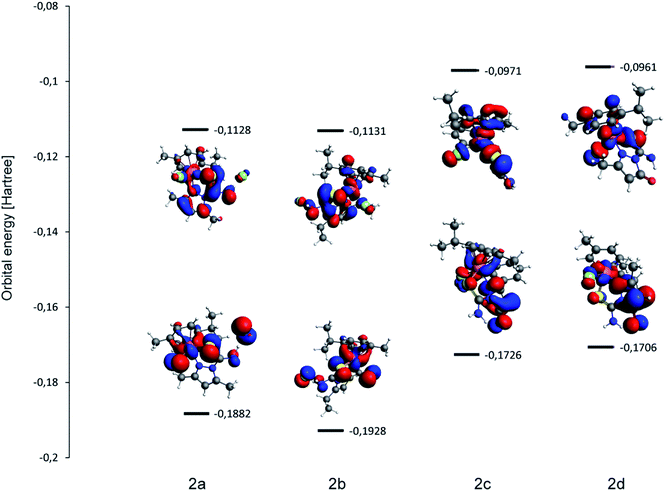
The results of DFT calculations confirm the correctness of the measured structures and diamagnetic character of Ru(II) in the studied complexes. The basic pseudotetrahedral structures were preserved during the geometry optimizations (energy minimization procedure) as shown in Fig. 6S.† Important bond lengths and bond angles of coordinated ligands in the X-ray experimental and calculated structures are compared in Table 2S.† Major shifts during the optimization procedure were connected with the positions of chloride anions, which were deprived of the nearest neighbors. Though the molecule of water was not included in the calculated structure of 2d, the geometry of this complex molecule remained approximately constant during the optimization procedure. Calculations confirm the effects observed in the experimental structures for example they show the strengthening of Ru-p-cymene centroid bonds within the molecular complexes in comparison to cationic ones.The electronic structures are obviously different for the pair of cationic complexes 2a and 2b and molecular 2c and 2d. Orbitals HOMO of the cationic species are built from localized p orbitals of sulfur and nitrogen atoms of N,S-organic ligands, whereas the corresponding HOMO orbitals of molecular complexes have a large contribution of delocalized π orbitals of pyrazole ligands (Fig. 4).
Molecular electrostatic potential (MEP) maps of 2a–2d illustrated in Fig. 5 show the distribution of charge on the isodensity surface 0.03. Though the relatively large differences between the minimum and maximum values of MEP are expected for the ionic complexes 2a and 2b, they are even larger for molecular species 2c and 2d, which was not obvious before the analysis. The minimum values of MEP for 2c and 2d are located at the oxygen O1 atom of the pyrazole ligand and the lowest value of −0.063 eV considering all the studied complexes is reached for 2d. Interestingly, the crystal structure of 2d features a molecule of water hydrogen bonded to O1, which is a nice experimental proof of the relatively large negative MEP value connected with O1 within this complex.
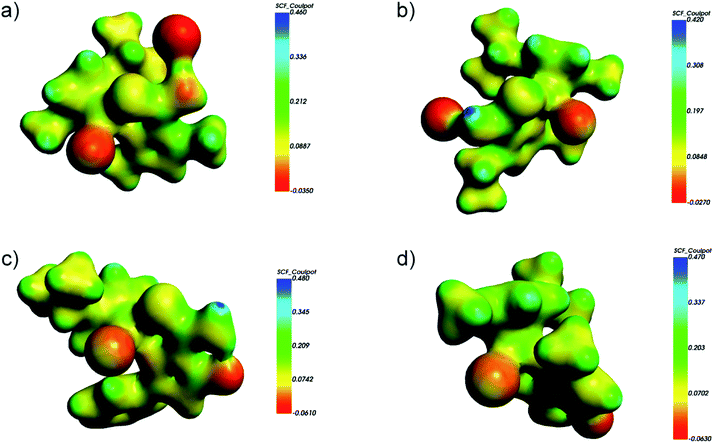 | ||
| Fig. 5 The calculated (DFT BP86-D/TZP) electrostatic potential mapped at the isodensity surface 0.03 for: (a) 2a; (b) 2b; (c) 2c; (d) 2d. | ||
The charge distribution within the complexes may be further described with the partial charges and dipole moments collected in Table 6. Despite the different character of the complex species the partial charge of the Ru, which is often connected with the cytotoxicity of ruthenium complexes 45 remains within relatively narrow range. The major difference is noticed for 2a and here the partial positive charge of Ru is lowest, which is in agreement with the lowest cytotoxicity in most studied concentrations as discussed in the previous chapters. The large negative partial charge is connected with O1 and according to the charges arising from the Mulliken population analysis it is even higher than negative charge ascribed to the chloride anion Cl2. The dipole moments are comparable for 2c and 2d. The largest value of the dipole moment of 2a seems to be more connected with the spatial arrangement of atoms than the charge separation.
| Atom | Complex | |||||||
|---|---|---|---|---|---|---|---|---|
| 2a | 2b | 2c | 2d | |||||
| Mulliken | Hirshfeld | Mulliken | Hirshfeld | Mulliken | Hirshfeld | Mulliken | Hirshfeld | |
| Ru1 | +0.381 | +0.276 | +0.420 | +0.284 | +0.449 | +0.286 | +0.461 | +0.284 |
| Cl1 | −0.400 | −0.292 | −0.386 | −0.278 | −0.398 | −0.274 | −0.397 | −0.300 |
| Cl2 | −0.520 | −0.389 | −0.528 | −0.383 | — | — | ||
| S1 | +0.041 | −0.005 | −0.036 | −0.043 | −0.069 | −0.074 | −0.076 | −0.068 |
| O1 | — | — | −0.618 | −0.273 | −0.615 | −0.271 | ||
| N1 | −0.304 | −0.059 | −0.326 | −0.062 | −0.339 | −0.091 | −0.345 | −0.096 |
| N2 | +0.084 | +0.032 | −0.076 | +0.040 | −0.089 | +0.008 | −0.116 | +0.007 |
| N3 | −0.099 | −0.143 | −0.139 | −0.143 | +0.052 | −0.134 | +0.010 | −0.138 |
| Dipole moments [D] | ||||||||
| 9.945 | 7.418 | 6.929 | 7.004 | |||||
Another calculated feature that may be compared with the cytotoxicity results is HOMO–LUMO gap illustrated in Fig. 4 (together with the shape of frontier orbitals). Low energy separation of frontier orbitals is correlated with complex reactivity and geometrical flexibility (the ease of excitation). As listed in Fig. 4 caption there are no major differences between the studied complexes, however complex 2b features the largest HOMO–LUMO gap of 2.169 eV and in 2d the gap is smallest (2.027 eV) thus the value is not related to the ionic/molecular character of the complex.
4. Conclusions
In response to the demand for effective anticancer drugs, the new arene–ruthenium(II) complexes were synthesized. All compounds were obtained with high yields. X-ray experiment indicated that all complexes have nitrogen–sulfur coordination create piano stool complexes. Electrospray ionization mass spectrometry confirmed complexes structure 2a–2d in solid and solution. An analysis of ESI-MS spectra shows that all used compounds are stable in solutions and it is difficult to break ruthenium–sulfur coordination bonds or permanent nitrogen in tandem reactions. Coordination bonds in all complexes 2a–2d are stable and not fragmented, whereas other bonds decompose in these molecules. It was demonstrated, that the various structure of these complexes (presence of carbonyl, hydroxyl, phenyl groups in pyrazole ring) influenced their biological activity, including anticancer effect. It has to be noted that compound 2d showed higher cytotoxicity against melanoma cell line WM-115 than cisplatin. The compound 2c demonstrated good activity against lymphoblastic cell line NALM-6. Although the new arene–ruthenium(II) complexes indicated moderated cytotoxicity also against normal eukaryotic cells, as well (see the effect against human fibroblasts line HFF-1) the application of tested complexes, based on IC50 values (stronger cytotoxic effect against cancer than regular cells), seems to be still safe. The cytotoxic effect of compounds 2b, 2d might be a result of their ability to cleave the DNA. Assuming that new preparations will appear to have a more universal effect, a huge part of this research was focused not only on their anticancer, but also on antimicrobial activity. A potential application of tested complexes on skin or mucous membranes can affect opportunistic activity of natural microbiota and well as pathogens, which often cause wound infections in cancer patients. As new arene–ruthenium(II) compounds indicated biostatic and biocidal activity against Gram-positive bacteria, including S. aureus, S. epidermidis and E. faecalis, they could not only inhibit proliferation of cancer cells, but also protect the patients against malignant wound infections. Nevertheless, further studies, including in vivo tests, are necessary in this regard.The DFT analysis proved that there is no simple correlation between the ionic/molecular character of the complex and its features such as partial charges, dipole moments or HOMO–LUMO separations.
Conflicts of interest
The authors declare no conflict of interests.Acknowledgements
Financial support from Medical University of Lodz (grant no. 503/3-066-02/503-31-001 to E. Budzisz) and (grant no. 502-03/3-066-02/502-34-093 to E. Namiecinska) is gratefully acknowledged.References
- A. K. Singh, D. S. Pandey, Q. Xu and P. Braunstein, Coord. Chem. Rev., 2014, 270–271, 31–56 CrossRef CAS.
- S. B. Fricker, Dalton Trans., 2007, 43, 4903–4917 RSC.
- Z. Ude, I. Romero-Canelón, B. Twamley, D. Fitzgerald Hughes, P. J. Sadler and C. J. Marmion, J. Inorg. Biochem., 2016, 160, 210–217 CrossRef CAS PubMed.
- T. S. Morais, T. J. L. Silva, F. Marques, M. P. Robalo, F. Avecilla, P. J. A. Madeira, P. J. G. Mendes, I. Santos and M. H. Garcia, J. Inorg. Biochem., 2012, 114, 65–74 CrossRef CAS PubMed.
- A. Weiss, R. H. Berndsen, M. Dubois, C. Müller, R. Schibli, A. W. Griffioen, P. J. Dyson and P. Nowak-Sliwinska, Chem. Sci., 2014, 5, 4742–4748 RSC.
- A. Srishailam, Y. P. Kumar, N. M. D. Gabra, P. V. Reddy, N. Deepika, N. Veerababu and S. Satyanarayana, J. Fluoresc., 2013, 23, 897–908 CrossRef CAS PubMed.
- G. Sűss-Fink, Dalton Trans., 2010, 39, 1673–1688 RSC.
- F. V. Rocha, C. V. Barra, A. V. G. Netto, A. E. Mauro, I. Z. Carlos, R. C. G. Frem, S. R. Ananias, M. B. Quilles, A. Stevanato and M. C. da Rocha, Eur. J. Med. Chem., 2010, 45, 1698–1702 CrossRef CAS PubMed.
- F. Wang, J. Xu, K. Wu, S. K. Weidt, C. L. MacKay, P. R. R. Langridge-Smith and P. J. Sadler, Dalton Trans., 2013, 42, 3188–3195 RSC.
- G. S. Smith and B. Therrien, Dalton Trans., 2011, 40, 10793–10800 RSC.
- S. Karabasannavar, P. Allolli, I. N. Shaikh and B. M. Kalshetty, Indian J. Pharm. Educ., 2017, 51, 490–501 CrossRef.
- Z. K. Jaćimović, M. Kosović, S. B. Novaković, G. Giester and A. Radović, J. Serb. Chem. Soc., 2015, 80, 867–875 CrossRef.
- A. Ansari, A. Ali, M. Asif and S. Shamsuzzaman, New J. Chem., 2017, 41, 16–41 RSC.
- A. D. Kumar, S. Bharath, R. N. Dharmappa, S. Naveen, N. K. Lokanath and K. A. Kumar, Res. Chem. Intermed., 2018, 44, 5635–5652 CrossRef.
- K. Karrouchi, S. Radi, Y. Ramli, J. Taoufik, Y. N. Mabkhot, F. A. Al-aizari and M. Ansar, Molecules, 2018, 23, 1–86 CrossRef PubMed.
- Y.-J. Cui, L.-Q. Tang, Ch-M. Zhang and Z.-P. Liu, Molecules, 2018, 23, 1–9 Search PubMed.
- E. Budzisz, U. Krajewska, M. Różalski, A. Szuławska, M. Czyż and B. Nawrot, Eur. J. Pharmacol., 2004, 502, 59–65 CrossRef CAS PubMed.
- E. Budzisz, M. Malecka, I.-P. Lorenz, P. Mayer, R. Kwiecień, P. Paneth, U. Krajewska and M. Rozalski, Inorg. Chem., 2006, 45, 9688–9695 CrossRef CAS PubMed.
- M. Sobiesiak, I.-P. Lorenz, P. Mayer, M. Wozniczka, A. Kufelnicki, U. Krajewska, M. Rozalski and E. Budzisz, Eur. J. Med. Chem., 2011, 46, 5917–5926 CrossRef CAS.
- M. Grazul, E. Besic-Gyenge, C. Maake, M. Ciolkowski, M. Czyz, R. K. O. Sigel and E. Budzisz, J. Inorg. Biochem., 2014, 135, 68–76 CrossRef CAS PubMed.
- E. Namiecinska, M. Sobiesiak, M. Malecka, P. Guga, B. Rozalska and E. Budzisz, Curr. Med. Chem., 2019, 26, 664–693 CrossRef CAS PubMed.
- I. Radosavljević Evans, J. A. K. Howard, L. E. M. Howard, J. S. O. Evans, Z. K. Jacimović, V. S. Jevtović and V. M. Lovac, Inorg. Chim. Acta, 2004, 357, 4528–4536 CrossRef.
- M. Sobiesiak, T. Muzioł, M. Rozalski, U. Krajewska and E. Budzisz, New J. Chem., 2014, 38, 5349–5361 RSC.
- M. Sobiesiak, M. Cieślak, K. Królewska, J. Kaźmierczak-Barańska, B. Pasternak and E. Budzisz, New J. Chem., 2016, 40, 9761–9767 RSC.
- P. Pelagatti, M. Carcelli, F. Calbiani, C. Cassi, L. Elviri, C. Pelizzi, U. Rizzotti and D. Rogolino, Organometallics, 2005, 24, 5836–5844 CrossRef CAS.
- J. Grau, V. Noe, C. Ciudad, M. J. Prieto, M. Font-Bardia, T. Calvet and V. Moreno, J. Inorg. Biochem., 2012, 109, 72–81 CrossRef CAS PubMed.
- A. A. Nazarov, C. G. Hartinger and P. J. Dyson, J. Organomet. Chem., 2014, 751, 251–260 CrossRef CAS.
- P. Kumar, R. K. Gupta and D. S. Pandey, Chem. Soc. Rev., 2014, 43, 707–733 RSC.
- P. Rogala, A. Jabłońska-Wawrzycka, K. Kazimierczuk, A. Borek, A. Błażejczyk, J. Wietrzyk and B. Barszcz, J. Mol. Struct., 2016, 1126, 74–82 CrossRef CAS.
- J. M. Gichumbi, H. B. Friedrich and B. Omondi, J. Organomet. Chem., 2016, 808, 87–96 CrossRef CAS.
- E. Budzisz, U. Krajewska and M. Rozalski, Pol. J. Pharmacol., 2004, 56, 473–478 CAS.
- R. R. Sinden, DNA Structure and Function, Academic Press Limited, London, 1994, pp. 131–133 Search PubMed.
- I. Fromantin, S. Watson, A. Baffie, A. Rivat, M. C. Falcou, I. Kriegel de Rycke and Y. Ingenior, Ostomy/Wound Manag., 2014, 60, 38–48 Search PubMed.
- W. G. Payne, D. K. Naidu, C. K. Wheeler, D. Barkoe, M. Mentis, R. E. Salas, D. J. Ir. Smith and M. C. Robson, Eplasty, 2008, 8, e9 Search PubMed.
- K. Rolston, L. Nesher and J. T. Tarrand, Infect. Dis. Ther., 2014, 3, 245–256 CrossRef PubMed.
- N. V. Kulkarni, A. Kamath, S. Budagumpi and V. K Revankar, J. Mol. Struct., 2011, 1006, 580–588 CrossRef CAS.
- S. Mandal, M. Mondal, J. K. Biswas, D. B Cordes, A. M. Z. Slawin, R. J. Butcher, M. Saha and N. Ch. Saha, J. Mol. Struct., 2018, 1152, 189–198 CrossRef CAS.
- C. X. Zhang and S. J. Lippard, Curr. Opin. Chem. Biol., 2003, 7, 481–489 CrossRef CAS.
- L. R. Ferguson and W. A. Denny, Mutat. Res., Fundam. Mol. Mech. Mutagen., 2007, 623, 14–23 CrossRef CAS PubMed.
- J. R. Aldrich-Wright, S. R. Vagg and P. A. Williams, Coord. Chem. Rev., 1997, 166, 361–389 CrossRef CAS.
- Y. Jung and S. J. Lippard, Chem. Rev., 2007, 107, 1387–1407 CrossRef CAS PubMed.
- K. Gkionis, J. A. Platts and J. Grant Hill, Inorg. Chem., 2008, 47, 3893–3902 CrossRef CAS PubMed.
- F. Wang, J. Bella, J. A. Parkinson and P. J. Sadler, J. Biol. Inorg Chem., 2005, 10, 147–155 CrossRef CAS PubMed.
- F. Wang, H. Chen, S. Parsons, I. D. H. Oswald, J. E. Davidson and P. J. Sadler, Chem. - Eur. J., 2003, 9, 5810–5820 CrossRef CAS PubMed.
- Z. Sochorová Vokáčová, I. Turel and J. V. Burda, J. Mol. Model., 2018, 24, 98 CrossRef PubMed.
- STOE and C. GmbH, X-area - software package for collecting single-crystal data on STOE area-detector diffractometers, for image processing, scaling reflection intensities and for outlier rejection, Darmstadt, Germany, 2015 Search PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 2015, 71, 3–8 CrossRef PubMed.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, Appl. Crystallogr., 2009, 42, 339–341 CrossRef CAS.
- L. J. Farrugia, J. Appl. Crystallogr., 2012, 45, 849–854 CrossRef CAS.
- C. F. Macrae, I. J. Bruno, J. A. Chisholm, P. R. Edgington, P. McCabe, E. Pidcock, L. Rodriguez-Monge, R. Taylor, J. van de Streek and P. A. Wood, J. Appl. Crystallogr., 2008, 41, 466–470 CrossRef CAS.
- European Committee on Antimicrobial Susceptibility Testing – EUCAST, http://www.eucast.org, 2012.
- G. te Velde, F. M. Bickelhaupt, E. J. Baerends, C. Fonseca Guerra, S. J. A. van Gisbergen, J. G. Snijders and T. Ziegler, J. Comput. Chem., 2001, 22, 931–967 CrossRef CAS.
- C. Fonseca Guerra, J. G. Snijders, G. te Velde and E. J. Baerends, Theor. Chem. Acc., 1998, 99, 391–403 Search PubMed.
- ADF2018, SCM, Theoretical Chemistry, Vrije Universiteit, Amsterdam, The Netherlands, 2018, http://www.scm.com Search PubMed.
- D. Becke, Phys. Rev. A, 1988, 38, 3098–3100 CrossRef PubMed.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter, 1988, 37, 785–789 CrossRef CAS PubMed.
- E. van Lenthe and E. J. Baerends, J. Comput. Chem., 2003, 24, 1142–1156 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1896357–1896360. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ra08736b |
| This journal is © The Royal Society of Chemistry 2019 |