Open Access Article
Open Access ArticlePEGylation of protein-imprinted nanocomposites sandwiching CdTe quantum dots with enhanced fluorescence sensing selectivity†
Xiao Hana,
Wenyan Hanb,
Shiting Zhanga,
Zhiqiang Liua and
Guoqi Fu *a
*a
aKey Laboratory of Functional Polymer Materials of Ministry of Education, Institute of Polymer Chemistry, College of Chemistry, Nankai University, Tianjin 300071, China. E-mail: gqfu@nankai.edu.cn; Tel: +86 22 23501443
bKey Laboratory of Bioactive Materials of Ministry of Education, College of Life Science, Nankai University, Tianjin 300071, China
First published on 21st November 2019
Abstract
Fluorescent sensors combining the selective recognition of protein molecularly imprinted polymers (MIPs) and the fluorescent sensing of quantum dots (QDs) have been studied considerably, but their fluorescence sensing selectivity for the target proteins remains to be increased. Herein, we propose a strategy for increasing the sensing selectivity by post-imprinting PEGylation of surface protein-imprinted nanocomposites with embedded QDs. With bovine hemoglobin (BHb) as a model protein template, protein MIP nanolayers were anchored over the CdTe QD decorated SiO2 nanoparticles by the sol–gel process using aminopropyltriethoxy silane and tetraethoxysilicane. PEG chains were then grafted onto the surface of the imprinted nanostructures via the nucleophilic reaction of the surface amine groups with N-hydroxysuccinimide ester-terminal methoxy-PEG, followed by template removal. The resultant PEGylated sensors showed significantly improved aqueous dispersion stability compared with the non-PEGylated controls. More importantly, such PEGylation greatly increased the fluorescence response selectivity, with the Stern–Volmer equation based imprinting factor increasing from 2.7 to 5.4. The PEGylated sensors were applied to determine BHb in bovine serum samples with satisfactory recoveries at three spiking levels ranging from 94.3 to 103.7%, indicating their potential application in real samples.
Introduction
Molecularly imprinted polymers (MIPs) are man-made polymer receptors, which surpass the natural receptors due to their high stability, ease of obtention and low cost. The molecular imprinting process usually involves copolymerization of functional and crosslinking monomers in the presence of a template molecule. Subsequent removal of the template leaves specific cavities (imprinting sites) in the resultant MIPs that are complementary to the template. MIPs have been widely used in a variety of fields, such as affinity separation media, molecular recognition elements for sensors, and specific target-capturing materials in ligand binding assays.1,2 To date, the imprinting of small molecules is now rather developed, but the imprinting against proteins and other biomacromolecules is still challenging, due to their large size, complex structure, variable conformation, and significantly reduced template–monomer interactions in the required aqueous polymerization media.3–5 Toward these limitations, diverse approaches have been studied, such as epitope imprinting,6 surface imprinting,7–14 and nanoscale imprinting.15–17 Among these, surface imprinting over nanomaterials, such as SiO2 or Fe3O4 nanoparticles, carbon nanotubes and quantum dots (QDs), has proved to be an effective solution to both facilitate macromolecular diffusion and achieve high binding capacity.18 Also, the features of the nanocores are naturally combined with the affinity recognition of the MIP nanoshells.Over the past decade, semiconductor QDs, i.e. semiconductor nanocrystals (e.g. CdTe and CdS), have received increasing attention because of their unique properties, such as bright photoluminescence (PL), good photostability, broad excitation but particle size-dependent narrow emission wavelengths, and high quantum yields. Based on the changes of fluorescence spectra, these QD labels as both sensing and recognizing elements have been widely used to detect various species such as ions, small organic molecules, and biomacromolecules.19,20 To improve the selectivity of the QDs based sensors, these years have witnessed a new type of MIP-based QDs sensors (MIP-QD sensors), which combine the selective recognition of MIPs with the fluorescence sense of QDs.21
Surface protein imprinting over the QDs embedding nanomaterials has also received considerable research interest.22–34 The sol–gel approach is frequently employed to form protein-imprinted shells over the QDs based nanocores, due to its mild polymerization conditions and easy operation. More importantly, the PL properties of the internal QDs can be maintained to a great extent, since the resultant imprinted SiO2 nanoshells are optically transparent and inert.19 The sol–gel process often involves simultaneous hydrolysis and condensation of several siloxanes with different functionalities for enhancing the interaction with the protein, among which amino siloxanes are prerequisite and major components for providing strong hydrogen bonding and electrostatic attraction. Till now, the sensing performance of protein MIP-QD sensors, involving selectivity, sensitivity, responsive time and detection range, remains to be improved owing to some limitations: (i) the inherent difficulty in successful protein imprinting as described above; (ii) intrinsic high background fluorescence; (iii) random location of fluorescent indicators outside the imprinted cavities.21 Among these sensing properties, fluorescence response selectivity is very important for a protein MIP-QD sensor. For achieving high response selectivity, the common approach is tuning the components and molar ratios of siloxanes used for the fabrication of protein MIP-QD sensors based on sol–gel process.22,23,25 However, in any case, the Si–OH, –NH2 and other groups residual on the surface can lead to significant nonspecific binding, thus unavoidably decreasing the fluorescence response selectivity. Another approach is based on ratiometric fluorescence technique using dual-emission fluorescence signals, with one for sensing target and another for eliminating environmental effects. It needs to build two individual materials with different fluorescence emission wavelengths, and hence is more complicated than the single-emission method.31,32
Polyethylene glycol (PEG) is a highly flexible, nonionic, hydrophilic and biocompatible polymer. It is extensively used in biomedical fields as a blocking agent to resist nonspecific adsorption of proteins on implant surfaces and nanoparticles intended for therapy, imaging and biodetection.35,36 The protein-repelling effect of PEG is attributed to the low free energy at PEG–water interface and the absence of hydrogen bonding and electrostatic interactions with proteins.37 Surface modification of the non-cavity regions of the protein MIPs in the presence of the template with a PEG-like protein-resistant polymer, so-called a post-imprinting modification strategy, is a promising method for lowering nonspecific protein adsorption. For the first time, Luan et al.38 grafted PEG chains from the surface of protein-imprinted siloxane copolymers before template removal by reaction of the surface silanol groups with methoxy-PEG-silane for reducing nonspecific binding. Quite recently, also via post-imprinting modification, we grafted nonlinear PEG chains from protein-imprinted nanoparticles fabrication via surface-initiated RAFT polymerization,39 and the PEG-modified protein-imprinted nanoparticles showed remarkable increased binding selectivity. Can these post-imprinting PEGylation strategies be employed to enhance the fluorescence response selectivity of a protein MIP-QD sensor? To the best of our knowledge, till now there is no such study.
Herein, we propose a new approach for post-imprinting PEGylation of protein MIP-QDs sensors for increasing fluorescent sensing selectivity. Protein-imprinted nanoshells were anchored over the SiO2 nanoparticles decorated with lots of CdTe quantum dots by sol–gel process. PEG chains were then grafted from the imprinted nanoshells via a rather active nucleophilic reaction of the surface amine groups with N-hydroxysuccinimide ester-terminal methoxy-PEG. This PEGylation can be more effective than that based on the modification of the surface silanol groups reported previously.38 With bovine hemoglobin (BHb) as a model template, the PEGylated protein MIP-QD sensors were thoroughly characterized by transmission electronic microscopy (TEM), dynamic light scattering (DLS), thermogravimetric analysis (TGA) and Fourier-transformation infrared (FT-IR), and their fluorescent sensing properties were studied with both single-protein solutions and simulated biosamples. After PEGylation, the protein MIP-QD sensors showed significantly enhanced fluorescent detection selectivity, in addition to increased aqueous dispersion stability.
Experimental
Materials
Cadmium chloride hydrate (CdCl2·2.5H2O), trisodium citrate dehydrate, sodium borohydride (NaBH4) were obtained from Tianjin Chemical Reagents Co., Tianjin. Tetraethyl orthosilicate (TEOS), sodium tellurite (Na2TeO3), mercaptosuccinic acid (MSA), 3-aminopropyltriethoxysilane (APTES), (2-N-morpholino)ethanesulfonic acid (MES), 4-(dimethylamino)pyridine (DMAP) and N,N′-disuccinimidyl carbonate (DSC) were purchased from Aladdin, Shanghai. Polyethylene glycol monomethylether (mPEG) with an average molecular weight (Mw) of 1900 was purchased from Alfa Aesar. Cytochrome c (Cyt c), bovine serum albumin (BSA) and lysozyme (Lyz) were purchased from Sangon, Shanghai. Bovine hemoglobin (BHb) was obtained from Sigma, Shanghai. Bovine blood was purchased by Xiaochuan Biotech. Co. Ltd. (Tianjin, China). The NHS-terminal mPEG (denoted as PEG-NHS) was synthesized by the reaction of mPEG with DSC according to the literature,40 as described in ESI.†Characterization
Fluorescence (FL) measurements were performed on an F-4600 fluorospectrophotometer (Hitachi). The morphologies and structures of the nanoparticles were observed by a TEM (Tecnai G2 F20, FEI). Zeta potentials and hydrodynamic diameters (Dh) of the particles were measured by DLS with a NanoBrook Omni (Brookhaven) laser light scattering spectrometer at the wavelength of 659 nm at 90° angle. FT-IR spectra were determined on a Bio-Rad FTS 135 FT-IR spectrometer over KBr pellets. TGA was performed by a NETZSCH TG 209 thermogravimetric analyzer under nitrogen atmosphere with a heating rate of 10 °C min−1 up to 800 °C. For investigation of the dispersion stability of the MIP and MIP-PEG nanoparticles, the transmittance of their dispersion (4 mg mL−1) in phosphate buffer (pH 7.0, 10 mM) at a wavelength of 600 nm was monitored as a function of time using an UV-vis spectrophotometer (TU-1900, Purkinje General Instrument Co., Beijing) equipped with a temperature controller.Synthesis of SiO2/CdTe QDs assemblies
Carboxyl-coated CdTe QDs were obtained according to the previous literature.41 Briefly, CdCl2·2.5H2O (71.1 mg) and trisodium citrate dehydrate (800 mg) were dissolved to 50 mL of water in a three-necked flask and stirred for 20 min, followed by adding Na2TeO3 (17.72 mg), MSA (200 mg) and NaBH4 (100 mg). The mixture was refluxed at 100 °C for about 5 hours, affording green fluorescent CdTe QDs.The amine-functionalized SiO2 nanoparticles (denoted as SiO2–NH2) were synthesized by a modified Stöber method.42 Ammonium hydroxide (30 mL) and ethanol (500 mL) were mixed to a three-necked flask and at 40 °C for 20 min with mechanical stirring, then TEOS (5 mL) was added and the mixture was stirred overnight. APTES (0.5 mL) was added into the flask drop by drop and the reaction was continued for 24 h. The resultant SiO2–NH2 particles were collected by centrifugation, and then washed with ethanol and water several times.
For self-assembly of the SiO2–NH2 nanoparticles with the CdTe QDs, the SiO2–NH2 nanoparticles (100 mg) were dispersed in 5 mL of MES buffer (50 mM, pH 5.5). To this dispersion, 20 mL of the CdTe QDs solution were added, and then the mixture was rapidly stirred for 2 h in the dark. Thus obtained assemblies (denoted by SiO2/CdTe) were collected by centrifugation and then washed three times with water.
Synthesis of core–shell BHb-imprinted nanocomposites and their PEGylation
Core–shell BHb-imprinted nanocomposites were synthesized according to the literature.25 The above obtained SiO2/CdTe assemblies (∼20 mg) were re-dispersed in 40 mL of ultrapure water, and then BHb (20 mg) and APTES (40 μL) were added to this dispersion. After stirring for 0.5 h, TEOS (80 μL) and NH3·H2O (80 μL) were added and the stirring was continued for 4 h at 25 °C. Thus obtained BHb-imprinted nanoparticles were collected by centrifugation and then washed repeatedly with methanol/0.1 M NaOH (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) solution for template removal until no template was detected by UV-vis spectrophotometer, followed by washing with ethanol and ultrapure water two times, respectively, and finally lyophilized for future use. The non-imprinted nanoparticles were prepared the same way but without the template. The as-synthesized fluorescent BHb-imprinted and non-imprinted nanoparticles were denoted as SiO2/CdTe/MIP and SiO2/CdTe/NIP, respectively.
1, v/v) solution for template removal until no template was detected by UV-vis spectrophotometer, followed by washing with ethanol and ultrapure water two times, respectively, and finally lyophilized for future use. The non-imprinted nanoparticles were prepared the same way but without the template. The as-synthesized fluorescent BHb-imprinted and non-imprinted nanoparticles were denoted as SiO2/CdTe/MIP and SiO2/CdTe/NIP, respectively.
For PEGylation, the core–shell BHb-imprinted nanoparticles without template removal or the non-imprinted nanoparticles synthesized the same way described above were dispersed in 2 mL of ethanol containing PEG-NHS (2 mg) and TEA (0.6 μL dissolved in 10 μL of ethanol), and this suspension was stirred at 25 °C for 15 h. After that, template removal and subsequent washing were carried out by the same way as detailed above. The resultant PEGylated BHb-imprinted and non-imprinted nanoparticles were denoted as SiO2/CdTe/MIP-PEG and SiO2/CdTe/NIP-PEG, respectively. It was proved that the PEGylation did not affect removal of the template by measuring the BHb amounts removed from the SiO2/CdTe/MIP-PEG and SiO2/CdTe/MIP nanoparticles using a UV-vis spectrophotometer, respectively.
Fluorescence measurements
All fluorescence detections were performed with the slit widths of 5 nm for excitation and 10 nm for emission, respectively. The excitation wavelength was set to 420 nm, and the emission wavelengths were scanned from 450 nm to 650 nm. The freeze-dried nanoparticles could be well re-dispersed in phosphate buffer (10 mM, pH 7.0), and the concentration was set at 0.1 mg mL−1. The pH value was chosen according to its effect on the quenching efficiency of the nanoparticles (see ESI Fig. S1†). The protein concentrations employed for all fluorescence measurements were set at 1.8 μM unless otherwise stated.Analysis of real samples
The practicability and accuracy of the SiO2/CdTe/MIP-PEG nanoparticles were assessed by detecting BHb in bovine serum. Briefly, the bovine serum was diluted 2000-fold with phosphate buffer (pH 7.0, 10 mM). The SiO2/CdTe/MIP-PEG nanoparticles (0.1 mg) were added into 1 mL of the diluted bovine serum solution, which was spiked with BHb at three concentration levels of 0.6, 1.2 and 1.8 μM. The fluorescence spectra of the suspension were recorded and all the tests were performed in triplicates.Results and discussion
Synthesis and characterization of PEGylated BHb-imprinted nanocomposites
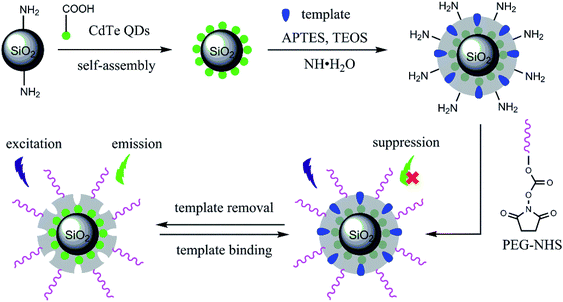
Scheme 1 illustrates the procedure for the synthesis of BHb-imprinted nanocomposites sandwiching CdTe quantum dots and bearing PEG chains. Carboxyl-capped CdTe QDs were self-assembled on the surface of amine-modified SiO2 nanoparticles by electrostatic interaction. Over this core–satellite structures, BHb-imprinted nanoshells were anchored via sol–gel process based on simultaneous hydrolysis and condensation of APTES and TEOS, which were used as functional monomer and crosslinker, respectively. Finally, PEG chains were grafted from the imprinted nanoshells' surface via the nucleophilic reaction of the surface residual amine groups with PEG-NHS. The reaction mechanism for the PEGylation is shown in ESI Scheme S1.† Fluorescence emission upon template removal and fluorescence suppression upon template binding are also demonstrated owing to the embedded CdTe QDs.The different nanostructures were observed by TEM (see ESI Fig. S2†). As shown from Fig. S2a,† the SiO2–NH2 nanoparticles are narrowly size-dispersed, with an average diameter of about 60 nm. In Fig. S2b,† a number of tiny CdTe QDs can be seen from the surface of the SiO2/QDs assemblies. As seen from Fig. S2c and d,† the imprinted and non-imprinted nanoshells can be discriminated due to their some lower contrast in comparison with the internal SiO2/QDs. As seen from Fig. S2e and f,† the PEG chains grafted cannot be distinguished from the SiO2/QDs/MIP-PEG and SiO2/QDs/NIP-PEG nanocomposites probably due to rather low grafting amounts.
The zeta potentials of the nanoparticles were monitored by DLS. As shown in Fig. 1A, the SiO2–NH2 nanoparticles are positive, and hence the carboxyl capped CdTe QDs (oppositely charged) could be self-assembled on the surface. The SiO2/QDs assemblies even became negative, indicating large amount of CdTe QDs adsorbed on the surface. The SiO2/CdTe/MIP and SiO2/CdTe/NIP nanoparticles turned to positive charges because of using amino silane as the functional monomer. After PEGylation, the final SiO2/CdTe/MIP-PEG and SiO2/CdTe/NIP-PEG nanoparticles are significantly less positive than corresponding SiO2/CdTe/MIP and SiO2/CdTe/NIP controls, respectively, thanks to the charge screening effect of the nonionic PEG chains grafted. The SiO2/CdTe/MIP-PEG dispersion in water shows much better stability than the un-PEGylated controls (see Fig. 1B and ESI Fig. S3†), also proving successful surface grafting of PEG chains.
Fig. 1C shows the FT-IR spectra of the different nanoparticles. The characteristic signals of the Si–O–Si stretching vibration at 1064 cm−1, Si–O vibration at 788 cm−1 and –NH2 vibration at 3414 cm−1 were observed in the spectrum of the SiO2–NH2 particles. In the spectrum of the SiO2/CdTe particles, a new peak appears at 1377 cm−1 due to the O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–O– from the carboxyl groups capped on the surface of CdTe QDs. The SiO2/CdTe/MIP and SiO2/CdTe/NIP nanoparticles show no distinct peak because of no new functional groups introduced during the sol–gel process. In the spectra of the SiO2/CdTe/MIP-PEG and SiO2/CdTe/NIP-PEG nanoparticles, new peaks occur at 2932 and 1377 cm−1, corresponding to –CH2 and O
C–O– from the carboxyl groups capped on the surface of CdTe QDs. The SiO2/CdTe/MIP and SiO2/CdTe/NIP nanoparticles show no distinct peak because of no new functional groups introduced during the sol–gel process. In the spectra of the SiO2/CdTe/MIP-PEG and SiO2/CdTe/NIP-PEG nanoparticles, new peaks occur at 2932 and 1377 cm−1, corresponding to –CH2 and O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–O– stretching vibrations, respectively, and the peak 1537 cm−1 can be attributed to O
C–O– stretching vibrations, respectively, and the peak 1537 cm−1 can be attributed to O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–NH–, which was formed by the reaction of PEG-NHS with the –NH2 on the surface of the SiO2/CdTe/MIP nanoparticles. Therefore, the PEGylation was successful. This was further confirmed by TGA. As shown in Fig. 1D, the final weight loss of the SiO2/CdTe/MIP-PEG composites is significantly larger than the SiO2/CdTe/MIP controls, and the PEG grafting percentage relative to the whole SiO2/CdTe/MIP-PEG composites was estimated to be 2.8 wt%.
C–NH–, which was formed by the reaction of PEG-NHS with the –NH2 on the surface of the SiO2/CdTe/MIP nanoparticles. Therefore, the PEGylation was successful. This was further confirmed by TGA. As shown in Fig. 1D, the final weight loss of the SiO2/CdTe/MIP-PEG composites is significantly larger than the SiO2/CdTe/MIP controls, and the PEG grafting percentage relative to the whole SiO2/CdTe/MIP-PEG composites was estimated to be 2.8 wt%.
Fluorescence properties
The excitation and emission spectra of the synthesized CdTe QDs were measured (see ESI Fig. S4†), which show a wide excitation spectrum but a very narrow emission spectrum. The fluorescence emission spectra of QDs, SiO2/QDs/MIP and SiO2/QDs/MIP-PEG were measured with an excited light of 420 nm. As shown in Fig. 2, the SiO2/QDs/MIP and SiO2/QDs/MIP-PEG nanoparticles shared fluorescence spectra similar to that of the CdTe QDs, despite a slight red shift of the emission peaks. This red shift is consistent with that reported previously.30,43 It could be ascribed to the formation of SiO2/QDs/MIP hybrid structure, which could increase the effective size of the QDs and thereby reduce the quantum size effect, hence causing a red shift of the photoluminescence maximum.30 Also, the SiO2/QDs/MIP-PEG nanoparticles showed only slight decrease in the fluorescence intensity compared with the SiO2/QDs/MIP controls without PEGylation, and their dispersion turned to green under UV light. Moreover, the fluorescence intensity of SiO2/QDs/MIP-PEG nanoparticles was quenched by the added template protein BHb, mainly due to the specific interaction between the template and imprinted cavities. The fluorescence emission spectrum of the SiO2/QDs/MIP-PEG nanoparticles showed no spectral overlap with the absorption spectrum of BHb (see ESI Fig. S5†), therefore the fluorescence quenching of CdTe QDs could be ascribed to the electron transfer between QDs and BHb.32Since the emission should be stable during fluorescence detection, the fluorescence stability of the imprinted nanoparticles was examined by repeatedly detecting the fluorescence intensity every 5 min at the maximum emission peak. As shown in Fig. 3a, the emission of the SiO2/QDs/MIP and SiO2/QDs/MIP-PEG particles remained stable within 40 min, which was enough for the following fluorescence detection. The response time of the fluorescence sensors was also tested in order to assess BHb binding kinetics to the imprinted cavities. As seen from Fig. 3b, within about 10 min, the fluorescence intensities of both SiO2/QDs/MIP and SiO2/QDs/MIP-PEG nanoparticles decreased to a constant level, indicative of achieving binding equilibriums. It also suggested that the surface grafted PEG chains did not show significant impact on the protein accessibility to the imprinting sites in the imprinted nanoshells. Therefore, protein binding to the fluorescence sensors was performed for 15 min before fluorescence detection in the following study.
Optimization of PEGylation
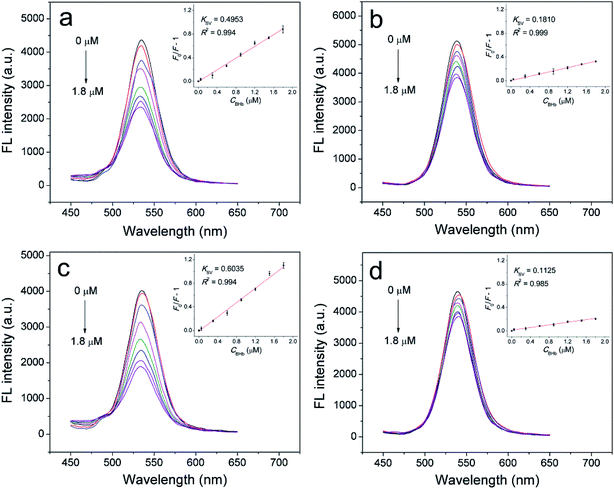
The previous study by Luan et al.38 showed that the PEG grafting amounts affected the recognition performance of the resultant PEGylated protein-imprinted materials. Therefore, we synthesized SiO2/QDs/MIP-PEG nanoparticles with varied amounts of PEG-NHS added to the reaction mixture and then examined their fluorescence quenching by BHb. mPEG (Mw 1900) was chosen for the synthesis of PEG-NHS, since it facilitated purification of the product via precipitation, and also overlong mPEG would decrease the reaction activity of the resultant PEG-NHS. The fluorescence quenching is quantified by F0/F − 1, where F0 and F are the fluorescent intensities in the absence and presence of quencher, respectively. The ratio of F0/F − 1 for the SiO2/QDs/MIP-PEG relative to that for the SiO2/QDs/NIP-PEG, denoted as (F0/F − 1)rel, was employed to evaluate the relative fluorescence quenching. As seen from Fig. 4, with increasing PEG-NHS concentrations, the BHb-induced fluorescence quenching for the SiO2/QDs/NIP-PEG nanoparticles decreased gradually, probably due to the decreased nonspecific binding. For the SiO2/QDs/MIP-PEG nanoparticles, however, the fluorescence quenching increased firstly to a peak, and then dropped down. The former increase may be attributed to the enhancement of the already formed imprinting sites by the grafted PEG chains in the presence of the template, which is coincided with our previous study about post-imprinting modification of surface protein-imprinted nanoparticles with nonlinear PEG chains via graft polymerization.39 The latter decrease may be ascribed to the steric hindrance due to the excessively grafted PEG chains. Compared with the SiO2/QDs/MIP nanoparticles with an (F0/F − 1)rel of 2.6, the SiO2/QDs/MIP-PEG nanoparticles prepared with a PEG-NHS concentration of 1 mg mL−1 showed the highest (F0/F − 1)rel, and thus employed in this study.Selectivity and sensitivity of the sensors
To study the selectivity as well as sensitivity of the fluorescent sensors, the fluorescent spectra of both kinds of imprinted and non-imprinted nanocomposites were recorded in the presence of BHb with increasing concentrations. The fluorescence quenching (F0/F − 1), as defined above, followed the Stern–Volmer equation| F0/F − 1 = KSVCq | (1) |
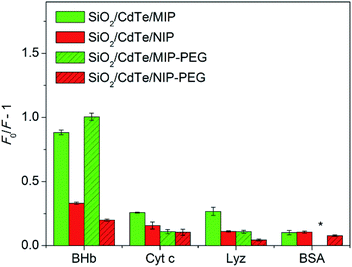
The selectivity of the MIP sensors for the template was further examined by comparing their fluorescent response toward BHb with that toward other reference proteins having different Mw and isoelectric point (pI) at a same protein concentration (1.8 μM). The Mw and pI of all the proteins used are BHb (Mw 64.5k, pI 6.8), BSA (Mw 66k, pI 4.8), Lyz (Mw 14k, pI 11.1), Cyt c (Mw 12.4k, pI 10.2). As shown in Fig. 6, both the SiO2/QDs/MIP and SiO2/QDs/MIP-PEG nanoparticles showed much stronger fluorescence quenching to BHb than to other reference proteins, while the non-imprinted nanoparticles show approximately comparable fluorescence quenching to all the proteins studied. This is because the specific imprinted cavities with the memory of the shape, size, and functionality of BHb were generated during the synthesis of BHb-imprinted nanocomposites, therefore BHb preferentially entered into recognition sites to quench CdTe QDs, although BSA has a comparable size with BHb, and Lyz and Cyt c have smaller sizes which seem to make them easier to approach the cavities.
The PEGylation also affected fluorescent sensing sensitivity of the imprinted nanoparticles. The limit of detection (LOD) of the imprinted nanocomposites was measured on the basis of 3σ/s, where σ means the standard deviation of the blank measurements, and s means the slope of calibration curve (herein, s = KSV).32 The LOD of the SiO2/QDs/MIP nanoparticles was determined to be 36.2 nM, while the LOD of the SiO2/QDs/MIP-PEG nanoparticles decreased to 27.8 nM mainly due to their higher KSV.
Practical applicability and performance comparison
The practical applicability of the SiO2/QDs/MIP-PEG sensor was further studied in bovine serum samples spiked with different BHb concentrations. As shown in Table 1, the recoveries ranged from 96.2–103.7% with relative standard deviations (RSDs) of 1.7–3.6%. Therefore, the SiO2/QDs/MIP-PEG sensor may be practically employed for sensitive and accurate determination of low BHb contents in real samples.| Spiked (μM) | Found (μM) | Recovery ± RSD (%) |
|---|---|---|
| 0 | 0 | — |
| 0.60 | 0.58 ± 0.01 | 96.2 ± 1.7 |
| 1.20 | 1.14 ± 0.04 | 94.9 ± 3.6 |
| 1.80 | 1.87 ± 0.06 | 103.7 ± 3.2 |
The fluorescent sensing performance for BHb detection was compared with other BHb-imprinted nanoparticles embedding semiconductor QDs reported previously. As shown in Table 2, the SiO2-NPs/CdTe/MIP-PEG as well as SiO2-NPs/CdTe/MIP nanoparticles prepared in this work showed LOD comparable with those of the others listed. However, the SiO2-NPs/CdTe/MIP-PEG particles synthesized in this work exhibited remarkable higher IF value than all the single-fluorescence counterparts, and even approached the ratiometric sensors. Although ratiometric sensors usually demonstrated higher sensing selectivity than single-fluorescence sensors,21 the presented post-imprinting PEGylation approach may be applied to the construction of ratiometric sensors for further improve their sensing performance.
| Sensors | Fluorescence mode | Linear range | LOD | IF | Ref. |
|---|---|---|---|---|---|
| UCNPs/MOFs/MIP | Single | 1.55–9.3 μM | 960 nM | 1.8 | 28 |
| Mn-doped ZnS QDs/MIP | Single | 0.1–5 μM | 38 nM | 3.1 | 26 |
| Mn-doped ZnS QDs/MIP | Single | 0.1–10 μM | 32 nM | 3.1 | 27 |
| CdTe/thermosensitive MIP | Single | 0.28–4 μM | 160 nM | 2.8 | 24 |
| SiO2-NPs/CdTe/MIP | Single | 0.02–2.1 μM | 9.4 nM | 3.8 | 25 |
| CdTe/SiO2 + CdTe/MIP | Ratiometric | 0.05–3 μM | 9.6 nM | 6.8 | 32 |
| SiO2-NPs/CdTe/MIP | Single | 0.05–1.8 μM | 36.2 nM | 2.7 | This work |
| SiO2-NPs/CdTe/MIP-PEG | Single | 0.05–1.8 μM | 27.8 nM | 5.4 | This work |
Conclusions
In summary, we have demonstrated an approach for post-imprinting PEGylation of surface protein-imprinted nanocomposites containing CdTe QDs with BHb as a model protein template for increasing the fluorescence sensing selectivity. The protein-imprinted nanocomposites were fabricated via sol–gel process, and then the PEGylation was successfully achieved before template removal by the nucleophilic reaction of the surface residual amine groups with N-hydroxysuccinimide ester-terminal methoxy-PEG. The PEGylation could not only improve the aqueous dispersion stability of the imprinted nanocomposites, but also greatly increase their fluorescence sensing selectivity. Thus obtained PEGylated sensors may be potentially applied for protein detection in real samples.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 21544006 and 21674051), the Natural Science Foundation of Tianjin City (no. 15JCYBJC47400), and the National Key R&D Program of China (2017YFC11044001).Notes and references
- L. X. Chen, S. F. Xu and J. H. Li, Chem. Soc. Rev., 2011, 40, 2922–2942 RSC.
- L. X. Chen, X. Y. Wang, W. H. Lu, X. Q. Wu and J. H. Li, Chem. Soc. Rev., 2016, 45, 2137–2211 RSC.
- D. R. Kryscio and N. A. Peppas, Acta Biomater., 2012, 8, 461–473 CrossRef CAS PubMed.
- S. J. Li, S. S. Cao, M. J. Whitcombe and S. A. Piletsky, Prog. Polym. Sci., 2013, 39, 145–163 CrossRef.
- M. Dabrowski, P. Lach, M. Cieplak and W. Kutner, Biosens. Bioelectron., 2018, 102, 17–26 CrossRef CAS PubMed.
- K. G. Yang, S. W. Li, L. K. Liu, Y. W. Chen, W. Zhou, J. Q. Pei, Z. Liang, L. H. Zhang and Y. K. Zhang, Adv. Mater. DOI:10.1002/adma.201902048.
- X. T. Shen, T. C. Zhou and L. Ye, Chem. Commun., 2012, 48, 8198–8200 RSC.
- A. Nematollahzadeh, W. Sun, C. S. A. Aureliano, D. Lutkemeyer, J. Stute, M. J. Abdekhodaie, A. Shojaei and B. Sellergren, Angew. Chem., Int. Ed., 2011, 50, 495–498 CrossRef CAS PubMed.
- T. Shiomi, M. Matsui, F. Mizukami and K. Sakaguchi, Biomaterials, 2005, 26, 5564–5571 CrossRef CAS.
- G. Q. Fu, H. Y. He, Z. H. Chai, H. C. Chen, J. Kong, Y. Wang and Y. Z. Jiang, Anal. Chem., 2011, 83, 1431–1436 CrossRef CAS PubMed.
- X. J. Li, B. L. Zhang, W. Li, X. F. Lei, X. L. Fan, L. Tian, H. P. Zhang and Q. Y. Zhang, Biosens. Bioelectron., 2014, 51, 261–267 CrossRef CAS PubMed.
- A. Abbas, L. M. Tian, J. J. Morrissey, E. D. Kharasch and S. Singamaneni, Adv. Funct. Mater., 2013, 23, 1789–1797 CrossRef CAS PubMed.
- R. Hu, J. Y. Luan, E. D. Kharasch, S. Singamaneni and J. J. Morrissey, ACS Appl. Mater. Interfaces, 2017, 9, 245–251 Search PubMed.
- S. Ambrosini, S. Beyazit, K. Haupt and B. T. S. Bui, Chem. Commun., 2013, 49, 6746–6748 RSC.
- Y. Hoshino, T. Kodama, Y. Okahata and K. J. Shea, J. Am. Chem. Soc., 2008, 130, 15242–15243 CrossRef CAS PubMed.
- A. Cutivet, C. Schembri, J. Kovensky and K. Haupt, J. Am. Chem. Soc., 2009, 131, 14699–14702 CrossRef CAS PubMed.
- G. Q. Pan, Q. P. Guo, C. B. Cao, H. L. Yang and B. Li, Soft Matter, 2013, 9, 3840–3850 RSC.
- Y. Q. Lv, T. W. Tan and F. Svec, Biotechnol. Adv., 2013, 31, 1172–1186 CrossRef CAS PubMed.
- R. Freeman and I. Willner, Chem. Soc. Rev., 2012, 41, 4067–4085 RSC.
- S. S. M. Rodrigues, D. S. M. Ribeiro, J. X. Soares, M. L. C. Passos, M. L. M. F. S. Saraiva and J. L. M. Santos, Coord. Chem. Rev., 2017, 330, 127–143 CrossRef CAS.
- Q. Yang, J. H. Li, X. Y. Wang, H. L. Peng, H. Xiong and L. X. Chen, Biosens. Bioelectron., 2018, 112, 54–71 CrossRef CAS PubMed.
- W. Zhang, X. W. He, Y. Chen, W. Y. Li and Y. K. Zhang, Biosens. Bioelectron., 2011, 26, 2553–2558 CrossRef CAS PubMed.
- W. Zhang, X. W. He, Y. Chen, W. Y. Li and Y. K. Zhang, Biosens. Bioelectron., 2012, 31, 84–89 CrossRef CAS PubMed.
- W. Zhang, X. W. He, W. Y. Li and Y. K. Zhang, Chem. Commun., 2012, 48, 1757–1759 RSC.
- D. Y. Li, X. W. He, Y. Chen, W. Y. Li and Y. K. Zhang, ACS Appl. Mater. Interfaces, 2013, 5, 12609–12616 CrossRef CAS PubMed.
- L. Tan, C. C. Kang, S. Y. Xu and Y. W. Tang, Biosens. Bioelectron., 2013, 48, 216–223 CrossRef CAS PubMed.
- L. Tan, C. Huang, R. F. Peng, Y. W. Tang and W. Li, Biosens. Bioelectron., 2014, 61, 506–511 CrossRef CAS PubMed.
- T. Guo, Q. L. Deng, G. Z. Fang, D. H. Gu, Y. K. Yang and S. Wang, Biosens. Bioelectron., 2016, 79, 341–346 CrossRef CAS PubMed.
- W. Zhang, W. Liu, P. Li, H. B. Xiao, H. Wang and B. Tang, Angew. Chem., Int. Ed., 2014, 53, 12489–12493 CAS.
- Z. Zhang, J. H. Li, X. Y. Wang, D. Z. Shen and L. X. Chen, ACS Appl. Mater. Interfaces, 2015, 7, 9118–9127 CrossRef CAS PubMed.
- H. Z. Lu and S. F. Xu, Talanta, 2017, 165, 482–488 CrossRef CAS PubMed.
- X. Y. Wang, S. M. Yu, W. Liu, L. W. Fu, Y. Q. Wang, J. H. Li and L. X. Chen, ACS Sens., 2018, 3, 378–385 CrossRef CAS PubMed.
- X. Y. Wang, J. L. Yu, J. H. Li, Q. Kang, D. Z. Shen and L. X. Chen, Sens. Actuators, B, 2018, 255, 268–274 CrossRef CAS.
- X. Zhang, S. Yang, R. Jiang, L. Q. Sun, S. P. Pang and A. Q. Luo, Sens. Actuators, B, 2018, 254, 1078–1086 CrossRef CAS.
- X. Qian, X. H. Peng, D. O. Ansari, Q. Yin-Goen, G. Z. Chen, D. M. Shin, L. Yang, A. N. Young, M. D. Wang and S. Nie, Nat. Biotechnol., 2007, 26, 83–90 CrossRef PubMed.
- L. Cheng, C. Wang, L. Feng, K. Yang and Z. Liu, Chem. Rev., 2014, 114, 10869–10939 CrossRef CAS PubMed.
- T. Pochechueva, A. Chinarev, N. Bovin, A. Fedier, F. Jacob and V. Heinzelmann-Schwarz, J. Immunol. Methods, 2014, 412, 42–52 CrossRef CAS PubMed.
- J. Y. Luan, K. K. Liu, S. Tadepalli, Q. S. Jiang, J. J. Morrissey, E. D. Kharasch and S. Singamaneni, ACS Appl. Mater. Interfaces, 2016, 8, 23509–23516 CrossRef CAS PubMed.
- X. Yang, Y. Sun, Y. Xiang, F. T. Qiu and G. Q. Fu, Analyst, 2019, 144, 5439–5448 RSC.
- T. Miron and M. Wilchek, Bioconjugate Chem., 1993, 4, 568–569 CrossRef CAS PubMed.
- Y. Zhu, Z. Li, M. Chen, H. M. Cooper, G. Q. Lu and Z. P. Xu, Chem. Mater., 2012, 24, 421–423 CrossRef CAS.
- Z. P. Ran and W. L. Yang, RSC Adv., 2014, 4, 37921–37927 RSC.
- S. F. Xu, H. Z. Lu, J. H. Li, X. L. Song, A. X. Wang, L. X. Chen and S. B. Han, ACS Appl. Mater. Interfaces, 2013, 5, 8146–8154 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra08556d |
| This journal is © The Royal Society of Chemistry 2019 |