Open Access Article
Open Access ArticleCollective absorption of 2,4,6-trinitrotoluene into lipid membranes and its effects on bilayer properties. A computational study†
Hong Yangab,
Mi Zhoub,
Huarong Lib,
Liu Liub,
Yang Zhou *b and
Xinping Longb
*b and
Xinping Longb
aSchool of Material Science and Engineering, Tsinghua University, Beijing 100084, China
bInstitute of Chemical Materials, China Academy of Engineering and Physics, Mianyang 621900, China. E-mail: zhouy@caep.cn
First published on 28th November 2019
Abstract
The widely used explosive, 2,4,6-trinitrotoluene (TNT), is a highly toxic chemical, which can cause hepatitis, cataracts, jaundice and so on, in humans. The interaction between TNT and biological membranes is crucial for understanding its toxic effects. Here, we mainly focused on molecular-level mechanisms for the collective adsorption of TNT into lipid membranes and the corresponding effects on bilayer properties by all-atom molecular dynamics simulations. We revealed that TNT can readily form an aggregate in the aqueous phase and quickly approach the surface of the membrane. At low concentrations of TNT (7 mol%), the aggregate is unstable and breaks up after several nanoseconds, and then the dispersed TNT molecules enter the membrane alone. At high concentrations (14 mol%), the aggregate is adsorbed as a whole and remains stable inside the membrane. After some of the TNT is absorbed by the membrane, the remaining TNT across the membrane would have greater permeability, i.e., the calculated permeability coefficient (P) is increased from 1.7 × 10−2 to 18.3 cm s−1. Correspondingly, a higher bioconcentration factor (BCF) was also observed. The increased level is more pronounced in the presence of TNT aggregates (i.e., high concentrations). This phenomenon is closely related to the strong interaction between TNT molecules. The results suggested that TNT molecules that have entered into the membrane can facilitate the membrane uptake, permeation and bioaccumulation of subsequent TNT molecules, exhibiting a synergistic effect. This work has a certain significance for understanding the toxicity of TNT.
Introduction
One of the widely used explosives, 2,4,6-trinitrotoluene (TNT), is a class C carcinogen rated by the Environmental Protection Agency, which can cause cataracts, hemolytic crisis, urinary tract infection, reproductive toxicity, and kidney and liver tumors.1,2 The toxicity of TNT was observed as early as the 1920s,3 and gradually attracted widespread attention.4–6 TNT has toxic effects on all living organisms, including mammals, fish, insects, bacteria and plants.7,8 More importantly, contact with TNT during manufacture, transportation, storage and disposal always threatens the health of humans (e.g. soldiers). Therefore, a thorough understanding of the toxic mechanisms of TNT is essential for the diagnosis and treatment of related diseases.In the past decades, lots of studies explored toxicity effects of TNT,9–12 which is related to the progress of adsorption, distribution, metabolism and excretion (ADME) in organisms. From the perspective of toxicity mechanism, existing works have provided a deep understanding for the metabolism of TNT. In detail, nitro groups of TNT can be metabolized to form nitroso and/or hydroxylamine groups during the two-electron reduction, which further interact with proteins and DNA, causing toxic and mutagenic effects.9,11 Additionally, the one electron reduction of nitro groups can induce oxidative stress, which is involved to several TNT-induced diseases such as cataracts.11,13 As for other processes (adsorption, distribution, and excretion), although several works have also focused on the bioaccumulation of TNT and its metabolites,7,12,14 the deep understanding on molecular-level mechanisms of TNT into related tissues and organs through plasma membranes are also absent, which is a crucial step in the ADME process of TNT.
To the best of our knowledge, there are still few studies on the transmembrane transport of TNT, in spite of its vital importance for better understanding the ADME in organisms. Golius et al.15 calculated the partition coefficient for several explosives (such as TNT, dinitrotoluene, dinitroanisole). Our recent work16 studied the thermodynamics of TNT and its metabolites passively across lipid membranes. We found the accumulation of TNT and metabolites in the membrane results in the increase of water penetration, which is concentration-dependent. In addition, the experiment revealed that the percutaneous permeability of TNT in guinea pig skin was proportional to the amount of TNT.2 Under real physiological conditions, the concentration of drugs and toxicants plays a significant role in their membrane partitioning and permeation.17 For example, Kang and Loverde18 showed that the increasing concentration of paclitaxel would facilitate the drug partitioning into the membrane and affect its distribution in membranes. Casalegno et al.19 also found that the membrane adsorption of dioxins at high concentrations were significantly different from that of single molecules. These results suggested that the concentration of xenobiotics may strongly affect their adsorption, distribution, accumulation and permeation in membranes. Thus, the detailed understanding on the collective adsorption of TNT into membranes at high concentrations can provide insights into ADME (or bioaccumulation) profiles of TNT and further the subsequent toxicity effect.
Although experimental techniques are very good means to estimate the partition and penetration properties of chemicals and their dependence on concentration, temperature and pressure.20 However, for pure experiments, obtaining a detailed molecular level understanding of chemical–membrane interactions remains a challenge. Computer simulation techniques such as molecular dynamics (MD) simulation are powerful complementary tools, which has been increasingly popular in recent years and investigated the permeation and distribution of small molecules,21,22 polar and charged molecules,23 anesthetic,24 carbon nanoparticles.25 Therefore, we have utilized all-atom MD simulations to investigate the collective adsorption of TNT into membranes at various concentrations and the effect of solute concentrations on the passive diffusion of TNT through a model cellular membrane.
Methods
Simulation systems
In this work, two sizes of lipid bilayers were constructed by CHARMM-GUI.26 The large-size membrane system contained 512 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine (POPC) lipids and around 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 water molecules, and the small-size system consisted of 128 POPC lipids and around 13
900 water molecules, and the small-size system consisted of 128 POPC lipids and around 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 water molecules. Both systems have been equilibrated for 100 ns. The final simulation box was about 13.0 × 13.0 × 10.0 nm3 for the large-size system, and 6.5 × 6.5 × 13.0 nm3 for the small-size system, respectively. Based on the large-size membrane, we introduced various number of TNT molecules into the aqueous phase by the Packmol software package,27 and built two simulation systems containing 7 mol% and 14 mol% TNT (also called as 7%-system and 14%-system), respectively. The distance between inserted TNT molecules and bilayer center in the z direction (the normal direction of membrane) ranged from 4.0 to 4.4 nm. Water molecules within 1.5 nm of TNT were subsequently deleted to remove bad contacts in the simulation box. Finally, the 7%-system was composed of 512 lipids, 30
000 water molecules. Both systems have been equilibrated for 100 ns. The final simulation box was about 13.0 × 13.0 × 10.0 nm3 for the large-size system, and 6.5 × 6.5 × 13.0 nm3 for the small-size system, respectively. Based on the large-size membrane, we introduced various number of TNT molecules into the aqueous phase by the Packmol software package,27 and built two simulation systems containing 7 mol% and 14 mol% TNT (also called as 7%-system and 14%-system), respectively. The distance between inserted TNT molecules and bilayer center in the z direction (the normal direction of membrane) ranged from 4.0 to 4.4 nm. Water molecules within 1.5 nm of TNT were subsequently deleted to remove bad contacts in the simulation box. Finally, the 7%-system was composed of 512 lipids, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 water molecules and 40 TNT molecules, while the 14%-system consisted of 512 lipids, 30
500 water molecules and 40 TNT molecules, while the 14%-system consisted of 512 lipids, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 091 water molecules and 80 TNT molecules.
091 water molecules and 80 TNT molecules.
MD simulations
CHARMM36 force field28 and TIP3P model29 were chosen for lipids and water molecules, respectively. The parameters of TNT were obtained from the standard CGenFF Force Field.30 The NPT ensemble was chosen for all simulations. After 1 ns pre-equilibrium, the 7%-system and 14%-system were run for 200 ns and 300 ns, respectively. The temperature was fixed at 310 K using a velocity rescale thermostat31 with a time constant of 0.2 ps. The Parrinello–Rahman algorithm32 was used to maintain semi-isotropic pressure at 1 bar with a time constant of 1 ps and a compressibility of 4.5 × 10−5 bar−1. The LINCS algorithm33 was chosen to constrain the bonds involving hydrogen atoms, while bond lengths and angles of water were constrained using the SETTLE algorithm.34 The integration time step was set to 2 fs and the neighbor list was updated every 10 steps. The electrostatic interaction was computed by particle mesh Ewald (PME) algorithm35 with the real space cutoff of 1.2 nm, a grid spacing of 0.16 nm and fourth-order interpolation. The van der Waals (VDW) interaction was switched smoothly to zero over the region between 1.0 and 1.2 nm. The steepest descent algorithm was used for energy minimizations. The simulations were performed by the GROMACS v5.1.2 (ref. 36) and the snapshots were produced by the VMD v1.9.2.37Potential of mean force (PMF)
Gibbs frees energy (ΔG) profiles (or potential of mean force, PMF) for TNT crossing lipid bilayers along the z direction were calculated by the umbrella sampling method.38 Considering the high computational cost, the small-size system (containing 128 lipids) was selected for the calculations of PMF. 10 and 40 TNT molecules were randomly inserted in the region with 1.5 nm from the bilayer center along the z direction, resulting in the initial structure of TNT-10 and TNT-40 systems, respectively. These two systems were also simulated for 120 ns, and the final structures were used as the initial input of steered molecular dynamics (SMD).39 Considering the symmetry of bilayer, we only simulated the translocation across one side, from aqueous phase to the bilayer centre. Along the reaction coordinate from bulk water (z = 4 nm) to the centre of the bilayer (z = 0 nm), all 32 separate windows were defined by a given z position, and the starting structure of each window was selected from the trajectories obtained from the SMD simulation. In the bulk water, the spacing was 0.2 nm; while in the bilayer the spacing was reduced to 0.1 nm. Each window was performed for 50 ns, totalling 1.6 μs. The first 30 ns of each window were used for the system equilibration, and the later 20 ns were used to calculate PMF profiles by the weighted histogram analysis method (WHAM).40 The force constant of the harmonic spring was 1000 kJ mol−1 nm−2 for all windows. For the PMF calculation of multi-TNT system, we only constrain the z-position of incoming TNT molecule, and the remaining TNT molecules are allowed to move freely in all directions.Permeability coefficient
The permeability coefficient (P) was estimated according to the inhomogeneous solubility diffusion model,41 in which P was expressed as a function of ΔG(z) and diffusion coefficient D(z), as presented in eqn (1)
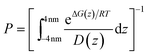 | (1) |
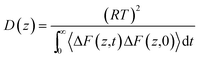 | (2) |
| ΔF(z,t) = F(z,t) − 〈F(z,t)〉 | (3) |
Results and discussion
Membrane adsorption of TNT
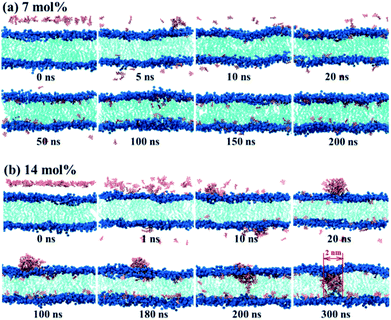
At first, we investigated the processes of TNT entering the lipid bilayer from the aqueous phase at two concentrations (i.e., 7 mol% and 14 mol%, which correspond to 40 and 80 TNT molecules, respectively) by unconstrained MD simulations. It is worth noting that the choice of these two concentrations is only to explore the different mechanisms of TNT membrane uptake. The basis for selection is not the actual physiological concentration, but our previous simulation results for judging the formation of aggregates.16 As we all know, a small piece of phospholipid membrane and its surrounding waters can be only considered in simulations, so TNT concentrations here cannot be directly compared with those in real physiological conditions especially the different size scales. In addition, to minimize the influence of box size on the result under the high TNT concentration, the large-size membrane (consisted of 512 POPC lipids) was used. Initially, TNT molecules were inserted in the region with 4.0–4.5 nm from the bilayer center (i.e., 2.0–2.5 nm from the membrane surface) along the z direction and evenly distributed in the x–y plane.For the system with the lower concentration (7%-system, Fig. 1a), we found that several TNT molecules could rapidly attract together in the aqueous phase and form a small aggregate in the first 5 ns, although the other molecules still existed as a single molecule. It should be noted that, due to the use of periodic boundary conditions, individual TNT molecules would enter the membrane from the other side of box, as shown in the snapshots at 5 ns of Fig. 1a. At the higher concentration (14%-system), Fig. 1b shows that TNT molecules could also form two small aggregates rapidly. However, unlike the 7%-system, these two aggregates would come into contact due to the motion and eventually merge into a larger aggregate at t = 20 ns. The larger aggregate has been stable during the simulation time and gradually adsorbed as a whole by the bilayer surface in the next 280 ns. In detail, several TNT molecules on the aggregate will first enter the cell membrane, and stay at the head–tail interface. Then, the remaining molecules on the aggregate would quickly enter into the cell membrane. The TNT molecules making up the aggregate would readily move toward the central region of membrane and interact with TNT molecules that diffuse into the opposite leaflet (t = 200 ns), resulting in the formation of a membrane-spanning aggregate (t = 300 ns).
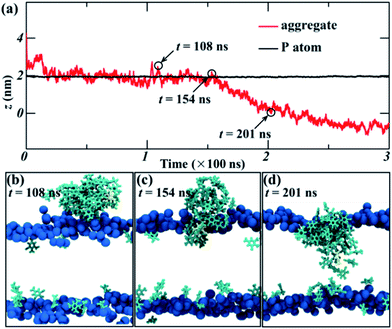
In order to further explore the collective adsorption of TNT, Fig. 2 presented more details about the adsorption process of TNT aggregates from the surface into the interior of lipid bilayer in the 14%-system. We noted that although the aggregate entered into the membrane as a whole, the relative position of individual TNT molecules in the aggregate is not fixed, and the entire aggregate is creeping forward. As an example, TNT25 (a TNT molecule with atomic number 25) was observed to reside in the middle of the aggregate when the simulation time t = 108 ns (Fig. 2a and b). However, at the next moment (t = 154 ns), it left the original position and move closer to the surface of membrane (Fig. 2a and c). At the same time, other TNT molecules also move to the inside of the membrane in the similar way. Therefore, the whole aggregate appears to creep forward. After the remaining molecules were adsorbed into the membrane, TNT25 has moved to the central region of the membrane, and then interacts with TNT molecules in the opposite leaflet (Fig. 2c). As shown in Fig. 1 and 2, the above results clearly show that the adsorption of TNT molecules into membranes is concentration-dependent, which is qualitatively consistent with experimental results.42 At low concentrations, the permeation of single molecule is the predominant mode of absorption; at high concentrations, the collective adsorption as an aggregate plays a dominant role (Fig. 1). The relative motion of single TNT molecule along the aggregate (Fig. 2) may be an important factor for collective adsorptions and further the final formation of the membrane-spanning aggregate.
Aggregate size
To quantitatively describe the aggregate (or cluster) during the partitioning of TNT into lipid bilayer, we characterized the aggregate size by counting the number of TNT molecules in the largest aggregate in two systems. The results are shown in Fig. 3. The criterion for judging whether the molecule belongs to the same aggregate is that the distance between two molecules is less than 0.35 nm, which ensures that the main peaks of the radial distribution functions (RDF) between TNT molecules in our simulations are taken into account (data not shown). For the 7%-system, an unstable aggregate formed in the initial stage consists of about 10 TNT molecules. Then, the number of TNT molecules in this aggregate gradually decreases to 2–3 within 20 ns (Fig. 3a), indicating the breakup of the aggregate. For the 14%-system, there are two aggregates formed in the initial stage and the larger one contains about 25 TNT molecules. After 20 ns, the two aggregates merged into one, which is composed of about 40–50 TNT molecules (Fig. 3b), corresponding to the aggregate size of about 2 nm (Fig. 1).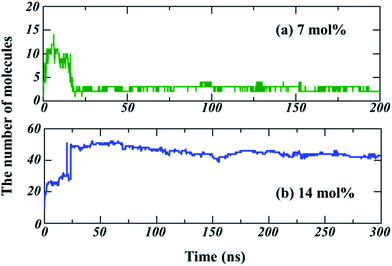 | ||
| Fig. 3 The number of TNT molecules contained in the largest aggregate with the simulation time for (a) 7%-system, and (b) 14%-system. | ||
These results revealed that hydrophobic TNT molecules first aggregate in the aqueous phase. The size of the initial aggregate determines its stability. Small aggregates (e.g., ∼10 molecules) are unstable, while larger aggregates (e.g., more than 20 molecules) would remain stable for at least several hundred nanoseconds and could enter into the membrane as a whole. We noted that the aggregate size was dependent on the initial TNT concentration, suggesting its crucial role in the adsorption behaviors of TNT.
In our simulations, we have tried our best to build a simulation system with the initial concentration close to the real physiological conditions. Lotufo et al.43 found that the concentration of TNT in viscera of sheepshead minnows could reach 300 μmol kg−1, corresponding to the concentration of 0.3 mM if the density was assumed to be 1 g mL−1. In addition, Lachance et al.7 found that the concentration of amine metabolite of TNT (2A, 2-amino-4,6-dinitrotoluene) in earthworm tissues is 3.0 μmol g−1, corresponding to about 3.0 mM. The higher concentration of TNT was observed in the tissues with higher lipid content (the difference can be up to 50 times).44 In our work, the simulation box only represented a small piece of lipid-rich tissue (the volume of lipids exceeds 1/3 of the total volume) and so TNT likely accumulates in this domain to a higher concentration. Even so, the commonly used 128-POPC system is still difficult to describe such a low concentration system. Thus, we used a large-size model membrane with 512 POPC lipids, in which 7 and 14 mol% correspond to the TNT concentration of about 39.3–78.6 mM. In fact, the concentration is still higher than that in real conditions. However, we believe that the TNT concentration in our simulation systems is achievable in the typical nanoscale physiological volume, for example in the local region of body-part rich in lipids.
Effects of aggregates on bilayer structures
The partitioning of solutes into the membrane might generally result in the significant change for bilayer structures. The profiles of the density of lipids and water molecules along the bilayer normal (z direction) are given in Fig. 4. The results demonstrate that the thickness of the lipid bilayer (the distance between the peak positions of lipid density) is not significantly influenced by the partition of TNT in membranes. In addition, no large change was found for the density profile of water, suggesting that the partitioning of TNT molecules into the membrane (even in the presence of membrane-spanning aggregate) does not destroy the integrity of the bilayer or lead to the formation of water pores. For TNT density profiles, we found that a part of TNT molecules had entered the membrane in the first 30 ns of simulations with the density peak at z = ±1.5 nm, well corresponding to the results from our previous work.16 After a few hundred nanoseconds of simulations, most of TNT molecules are absorbed into the membrane from the bulk water.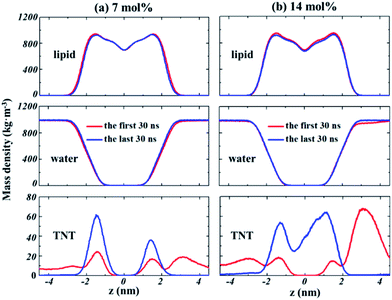 | ||
| Fig. 4 The evolution of the density profiles along the bilayer normal (z direction) for lipids, water and TNT molecules in different systems. (a) 7%-system, (b) 14%-system. | ||
At high concentration (14%-system, Fig. 4b), the non-zero value of TNT density in the central region indicated that TNT molecules could be stably distributed at the hydrophobic core of bilayers. Furthermore, it should be emphasized that in the first 30 ns of the 14%-system, a very high peak of TNT density was found at the position of z = 3 nm, which corresponds to the aggregate located on the membrane surface. This aggregate occupies the space of water molecules, causing the water density to drop slightly in this region (Fig. 4b). The behaviour of lipid tails is evaluated by the deuterium order parameter (SCD), which can be calculated from the last 30 ns of trajectory as
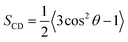 | (4) |
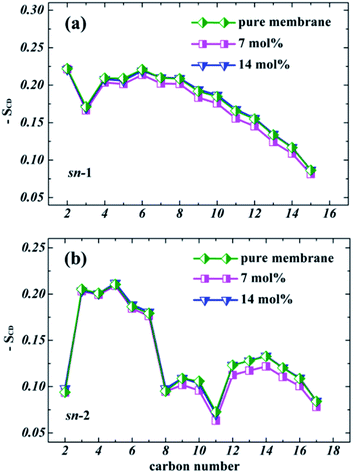 | ||
| Fig. 5 Deuterium order parameters (−SCD) obtained for sn-1 (a) and sn-2 (b) chains of POPC lipids with various TNT concentrations. | ||
Effects of aggregates on bilayer permeability
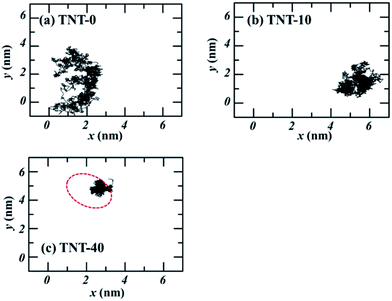
Kang et al.18 observed the water finger along the aggregate of paclitaxel inside the POPC membrane. Our previous work16 also found that more water molecules can access the core of the membrane due to the partition of TNT and its metabolites in bilayers. Thus, to investigate the effect of TNT distributions in the membrane (random or aggregate) on the permeation properties of lipid bilayer, we calculated the PMF profile of a TNT molecule transferring across lipid bilayers with different TNT concentrations. It should be noted that the 128-POPC bilayer model was used due to the high computational cost of umbrella sampling calculations, in which we directly inserted 10 and 40 TNT molecules into the hydrophobic region of membrane, respectively. The final equilibrium structures (also named as TNT-10 and TNT-40 system, respectively) were chosen to represent the random distribution and aggregate of TNT molecules in membrane formed by the large 512-POPC system. As shown in Fig. 6a, in the TNT-10 system, TNT molecules were randomly distributed at the head–tail interface and no stable cluster or aggregate was observed, which is very similar to the large 7%-system in Fig. 1. Similarly, a membrane-spanning aggregate was formed in the TNT-40 system, which contains about 15–20 TNT molecules. The aggregate size in the TNT-40 system (∼1 nm) is smaller than that in the large-size 14%-system, which is attributed to the different number of molecules initially placed in the systems.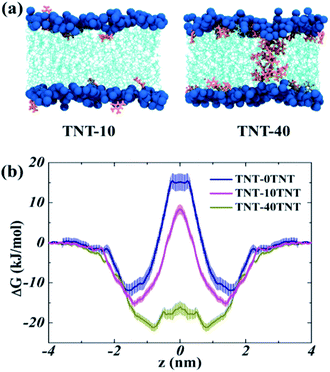 | ||
| Fig. 6 (a) The snapshots of the membrane systems used in the umbrella sampling. (b) The PMF profiles of TNT transferring across different systems. | ||
All PMF profiles are shown in Fig. 6b, and the PMF profile of TNT though the pure POPC membrane was taken from our previous work.16 In the pure POPC membrane, the free energy of an incoming TNT molecule (ΔGwater) decreases by −12.0 kJ mol−1 from the aqueous phase into the interior of lipid bilayers, without an obvious barrier in the membrane–water interface. The profile had a minimum (zmin) at z = 1.51 nm, just below the head groups of lipids. Then, TNT overcomes an energy barrier of 27.9 kJ mol−1 from one leaflet to another (ΔGbarrier). We found that the incorporation of 10 TNT molecules (with several molecules moving into the bulk water) in lipid bilayers have a great effect on the transmembrane process of subsequent TNT molecules. Specifically, in the TNT-10 system, the position of zmin moves to z = 1.39 nm, indicating that the subsequent TNT tend to reside in the region closer to the center of membrane. Moreover, the value of ΔGwater increases to −15.2 kJ mol−1, which suggests the promoted ability of TNT partitioning into the membrane. ΔGbarrier declines by ∼4.3 kJ mol−1, from 27.9 to 23.6 kJ mol−1, which indicates that the presence of TNT inside the membrane promotes the capability of subsequent TNT molecules to transfer through the membrane. An analogous or more significant tendency was also found in the TNT-40 system (with a membrane-spanning aggregate), in which zmin moves further from z = 1.39 nm to z = 0.77 nm, and ΔGwater also increases to −21 kJ mol−1. Importantly, there is only a small barrier (ΔGbarrier = 4.8 kJ mol−1) in the central region of the membrane, indicating the great enhanced ability of TNT molecules to pass through the hydrophobic core. In the recent MD study reported by Gupta and co-workers,47 it was also obtained that C60 could enter the membrane more easily in the presence of other C60 molecules inside the bilayer at relatively low concentrations.
The quantitative results describing the membrane affinity and permeability of TNT in different systems are shown in Table 1. Liposome–water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KLW) and bioconcentration factor (BCF) were calculated by utilizing the method provided in our previous work.16 The results of log
KLW) and bioconcentration factor (BCF) were calculated by utilizing the method provided in our previous work.16 The results of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KLW demonstrate that the presence of TNT molecules in the head–tail interface of membrane would increase the value of log
KLW demonstrate that the presence of TNT molecules in the head–tail interface of membrane would increase the value of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KLW from 1.61 to 2.11, and the higher concentration (40 TNT) resulting in the formation of a membrane-spanning aggregate would lead to the further increase of log
KLW from 1.61 to 2.11, and the higher concentration (40 TNT) resulting in the formation of a membrane-spanning aggregate would lead to the further increase of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KLW to 3.19. Experimentally, Golius et al.48 found that the liposome–water partition coefficient of TNT was 2.18, which was slightly larger than the calculated result from the TNT-0 system, and very close to the one obtained from the TNT-10 system. It indicates that the concentration of solutes may be an important factor in evaluating the partition coefficient by MD simulations. BCF is defined as the ratio of a chemical concentration in the organism to that in the surrounding water, which could describe the ability of solutes to accumulate in the membrane. Our results reveal that the initial adsorption of a portion of TNT into the membrane would enhance the adsorption (or accumulation) of subsequent incoming TNT molecules in the membrane, especially in the formation of aggregates. In experiments, Conder et al.42 also demonstrated that the initial concentration of TNT and its metabolites significantly affected the measured BCFs, e.g., the value of BCF of metabolites at high concentration (240 to 620 nmol g−1) was approximately 1.1–2.2 times higher than that at lower concentration (35–260 nmol g−1), in good agreement with our results. Additionally, we calculated the permeability coefficients (P) to quantitatively estimate the changes of the ability of TNT to pass through the membrane. The results (Table 1) also showed that the permeability of TNT was significantly enhanced, with an increase in permeability coefficient (P) by 1–3 orders of magnitude (from 0.017 to 18.3 cm s−1).
KLW to 3.19. Experimentally, Golius et al.48 found that the liposome–water partition coefficient of TNT was 2.18, which was slightly larger than the calculated result from the TNT-0 system, and very close to the one obtained from the TNT-10 system. It indicates that the concentration of solutes may be an important factor in evaluating the partition coefficient by MD simulations. BCF is defined as the ratio of a chemical concentration in the organism to that in the surrounding water, which could describe the ability of solutes to accumulate in the membrane. Our results reveal that the initial adsorption of a portion of TNT into the membrane would enhance the adsorption (or accumulation) of subsequent incoming TNT molecules in the membrane, especially in the formation of aggregates. In experiments, Conder et al.42 also demonstrated that the initial concentration of TNT and its metabolites significantly affected the measured BCFs, e.g., the value of BCF of metabolites at high concentration (240 to 620 nmol g−1) was approximately 1.1–2.2 times higher than that at lower concentration (35–260 nmol g−1), in good agreement with our results. Additionally, we calculated the permeability coefficients (P) to quantitatively estimate the changes of the ability of TNT to pass through the membrane. The results (Table 1) also showed that the permeability of TNT was significantly enhanced, with an increase in permeability coefficient (P) by 1–3 orders of magnitude (from 0.017 to 18.3 cm s−1).
These results suggest a synergistic effect that TNT molecules previously entering the membrane will markedly promote the permeability and accumulation of latter molecules in the membrane. Furthermore, in the TNT-40 system, the more pronounced changes in the free energy profile might be related to the formation of the membrane-spanning aggregate, in which some TNT molecules could stably reside in the hydrophobic core, and further affect the local barrier of the bilayer center. In the recent simulation, Wang et al.49 reported the effect of concentration on the adsorption and transmembrane process of cimetidine. In detail, 8 mol% of cimetidine would lead to a decrease in the ability of subsequent cimetidine molecules across the membrane and a decline of passive permeability, which was attributed to the insignificant interactions between cimetidine molecules. Therefore, we would infer that the solute–solute interaction might play a crucial role in the membrane adsorption, bioaccumulation and the passive permeation of TNT molecules.
Interactions between TNT molecules
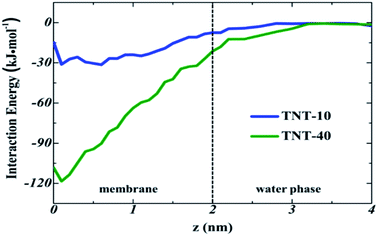
To confirm this inference, we investigated the interaction between TNT molecules in different systems. Fig. 7 depicts the trajectories of incoming TNT molecules in the x–y plane of the bilayer center. The results clearly shows that the lateral diffusion of incoming TNT molecules would be limited by several TNT molecules inside the membrane (random and aggregate). For the pure bilayer without TNT molecules, the incoming TNT molecule has a wide range of lateral diffusion, as shown in Fig. 7a. When the bilayer was introduced 10 TNT molecules, the lateral diffusion region of the subsequent incoming TNT would significantly contract (Fig. 7b). This phenomenon is more pronounced in the system with the TNT aggregate (Fig. 7c). This result indicated the strong interactions between TNT molecules inside the membrane.To quantitatively describe the interaction between TNT molecules, we calculated the interaction energy of the incoming TNT with other TNT molecules in membranes as a function of the distance from the bilayer center, as shown in Fig. 8. During the adsorption process of the incoming TNT molecule from the aqueous phase into the membrane, the interaction energy is gradually decreasing, indicating that the solute–solute interaction is thermodynamically favorable for this process. In the TNT-10 system, the value of interaction energy decreases by −30.9 kJ mol−1, and a greater reduction (−118.3 kJ mol−1) occurs in the TNT-40 system with the higher concentration of TNT (i.e., in the presence of TNT aggregate). Furthermore, the component of interaction energy (Fig. S2†) suggests that the VDW interaction between TNT molecules plays a more important role in their attraction and clustering, although both VDW and electrostatic interactions contribute to the change of interaction energy.
From the interaction energy point of view, we can also clarify the different aggregation behavior of TNT at different concentrations as presented in Fig. 1. In the aqueous phase, the single TNT is fully hydrated and the process of aggregation will lead to the dehydration of TNT. TNT–water interaction energy profiles (Fig. S3†) show that this dehydration process is energetically unfavorable and will compete with the favorable TNT–TNT interactions during the adsorption of single TNT into aggregates. At the high concentration, TNT is likely to form the larger aggregate which will further result in a more favorable interaction between the aggregate and a single TNT (Fig. 8). Therefore, the aggregates observed at higher concentrations are more stable (Fig. 3).
Conclusions
In summary, we investigated the concentration dependence of TNT membrane adsorption and the effect of TNT concentration on the structures and properties of cellular membranes by all-atom MD simulations. In order to get closer to the real physiological concentration, two large bilayer model of 512 POPC lipids were utilized to obtain the rational concentration of TNT molecules (7 and 14 mol%) in membranes. At the lower concentration of 7 mol%, the adsorption of TNT molecules into the membrane is carried out in a single molecule, although unstable TNT aggregates are also formed in the aqueous phase. At the higher concentration (14 mol%), the collective adsorption of TNT molecules into the membrane as a stable aggregate is firstly observed.The absorbed TNT molecules into membranes have no significant effects on the membrane structure. However, they have a strong influence on the membrane performance such as permeability. TNT molecules that have already entered the membrane can affect the transport of subsequent incoming TNT molecules across the membrane, e.g., the greater ability of incoming TNT molecules to be adsorbed into the bilayer, a shift of free-energy minimum toward the bilayer center and a decreased barrier in the central region. Based on PMF profiles, the calculated liposome–water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KLW) of subsequent incoming TNT molecules increase from 1.61 to 2.11 (TNT-10 system) and 3.19 (TNT-40 system) with the increase of TNT concentrations. Accordingly, the predicted BCF values also increase from 9.9 mL g−1 to 26.6 and 225.6 mL g−1, in good agreement with experimental results.42 In addition, the permeability coefficient of TNT also shows the remarkable enhancement, with the increase from 0.017 cm s−1 to 0.69 and 18.3 cm s−1 for TNT-10 and TNT-40 system, respectively. These results suggest a synergistic effect in the transmembrane transport of TNT molecules, i.e., the presence of TNT inside the membrane can facilitate the membrane adsorption, accumulation and permeation of subsequent TNT molecules. Furthermore, the analysis of interaction energy indicates that solute–solute interactions, especially VDW interactions, played a crucial role in this process.
KLW) of subsequent incoming TNT molecules increase from 1.61 to 2.11 (TNT-10 system) and 3.19 (TNT-40 system) with the increase of TNT concentrations. Accordingly, the predicted BCF values also increase from 9.9 mL g−1 to 26.6 and 225.6 mL g−1, in good agreement with experimental results.42 In addition, the permeability coefficient of TNT also shows the remarkable enhancement, with the increase from 0.017 cm s−1 to 0.69 and 18.3 cm s−1 for TNT-10 and TNT-40 system, respectively. These results suggest a synergistic effect in the transmembrane transport of TNT molecules, i.e., the presence of TNT inside the membrane can facilitate the membrane adsorption, accumulation and permeation of subsequent TNT molecules. Furthermore, the analysis of interaction energy indicates that solute–solute interactions, especially VDW interactions, played a crucial role in this process.
We note that the analogous synergistic effect has been also observed for the hydrophobic paclitaxel.18 In addition, several studies have reported the collective adsorption as aggregate into the membrane for dioxins,19 thioridazine50 and C60 molecules.47 Therefore, our observations and results in this study may be also applicable to some other toxicants or drug molecules. The molecular-level understanding of their passive transport at different concentrations across lipid bilayers allows us to clarify their distribution, metabolism and toxic or therapeutic effects in organisms.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the financial support from the Foundation of State Key Laboratory of NBC Protection for Civilian (No. SKLNBC2018-18), National Natural Sciences Foundation of China (No. 21905260), and CAEP (No. 2014-1-075).Notes and references
- U.S. Agency for Toxic Substances and Disease Registry, Toxicological profile for 2,4,6-trinitrotoluene (TNT), ATSDR publication CAS 118-96-7, ATSDR, Atlanta, 1995, www.atsdr.cdc.gov/toxprofiles/tp.asp?id=677%26tid=125 Search PubMed.
- J. Yinon, Toxicity and Metabolism of Explosives, CRC Press, Boca Raton, FL, 1990 Search PubMed.
- C. Voegtlin and J. M. Johnson, U. S. Pub. Health Rep., 1919, 34, 1307–1313 CrossRef CAS.
- L. Berthe-Corti, H. Jacobi, S. Kleihauer and I. Witte, Chemosphere, 1998, 37, 209–218 CrossRef CAS PubMed.
- A. Prasath, L. Panneerselvan, A. Provatas, R. Naidu and M. Megharaj, Ecotoxicology, 2016, 25, 1873–1879 CrossRef CAS PubMed.
- S. H. Liang, D. W. Hsu, C. Y. Lin, C. M. Kao, D. J. Huang, C. C. Chien, S. C. Chen, I. J. Tsai and C. C. Chen, Ecotoxicol. Environ. Saf., 2017, 138, 39–46 CrossRef CAS PubMed.
- B. Lachance, A. Y. Renoux, M. Sarrazin, J. Hawari and G. I. Sunahara, Chemosphere, 2004, 55, 1339–1348 CrossRef CAS PubMed.
- E. J. Johnston, E. L. Rylott, E. Beynon, A. Lorenz, V. Chechik and N. C. Bruce, Science, 2015, 349, 1072–1075 CrossRef CAS PubMed.
- R. María Dolores, P. Eva, C. Francisco and M. Conrado, FEMS Microbiol. Rev., 2010, 32, 474–500 Search PubMed.
- K. Tzafestas, M. M. Razalan, I. Gyulev, A. M. Mazari, B. Mannervik, E. L. Rylott and N. C. Bruce, New Phytol., 2017, 214, 294–303 CrossRef CAS PubMed.
- P. Kovacic and R. Somanathan, J. Appl. Toxicol., 2014, 34, 810–824 CrossRef CAS PubMed.
- G. R. Lotufo, J. B. Belden, J. C. Fisher, S. F. Chen, R. A. Mowery, C. K. Chambliss and G. Rosen, Environ. Pollut., 2016, 210, 129–136 CrossRef CAS PubMed.
- Y. Kumagaia, T. Wakayama, S. Li, A. Shinohara, A. Iwamatsu, G. Sun and N. Shimojo, FEBS Lett., 2000, 478, 295–298 CrossRef.
- D. R. Ownby, J. B. Belden, G. R. Lotufo and M. J. Lydy, Chemosphere, 2005, 58, 1153–1159 CrossRef CAS PubMed.
- A. Golius, L. Gorb, O. Isayev and J. Leszczynski, J. Biomol. Struct. Dyn., 2018, 1–19 Search PubMed.
- H. Yang, H. Li, L. Liu, Y. Zhou and X. Long, J. Phys. Chem. B, 2019, 123, 6481–6491 CrossRef CAS PubMed.
- S. M. Loverde, J. Phys. Chem. Lett., 2014, 5, 1659–1665 CrossRef CAS PubMed.
- M. Kang and S. M. Loverde, J. Phys. Chem. B, 2014, 118, 11965–11972 CrossRef CAS PubMed.
- M. Casalegno, G. Raos and G. Sello, Phys. Chem. Chem. Phys., 2015, 17, 2344–2348 RSC.
- M. Paloncýová, R. Devane, B. Murch, K. Berka and M. Otyepka, J. Phys. Chem. B, 2014, 118, 1030–1039 CrossRef PubMed.
- D. Bemporad, J. W. Essex and C. Luttmann, J. Phys. Chem. B, 2004, 108, 4875–4884 CrossRef CAS.
- M. Casalegno, G. Raos and G. Sello, Phys. Chem. Chem. Phys., 2016, 18, 17731–17739 RSC.
- I. Vorobyov, T. Olson, J. Kim, R. E. Koeppe II, O. Andersen and T. Allen, Biophys. J., 2014, 106, 586–597 CrossRef CAS PubMed.
- S. Vemparala, C. Domene and M. L. Klein, Acc. Chem. Res., 2010, 43, 103–110 CrossRef CAS PubMed.
- J. Wong-Ekkabut, S. Baoukina, W. Triampo, I. M. Tang, D. P. Tieleman and L. Monticelli, Nat. Nanotechnol., 2008, 3, 363–368 CrossRef CAS PubMed.
- S. Jo, T. Kim, V. G. Iyer and W. Im, J. Comput. Chem., 2008, 29, 1859–1865 CrossRef CAS PubMed.
- L. Martínez, R. Andrade, E. G. Birgin and J. M. Martínez, J. Comput. Chem., 2009, 30, 2157–2164 CrossRef PubMed.
- S. E. Feller and A. D. MacKerell, J. Phys. Chem. B, 2000, 104, 7510–7515 CrossRef CAS.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- K. Vanommeslaeghe, E. Hatcher, C. Acharya, S. Kundu, S. Zhong, J. Shim, E. Darian, O. Guvench, P. Lopes and I. Vorobyov, J. Comput. Chem., 2010, 31, 671–690 CAS.
- G. Bussi, D. Donadio and M. Parrinello, J. Chem. Phys., 2007, 126, 14101 CrossRef PubMed.
- M. Parrinello and A. Rahman, J. Appl. Phys., 1998, 52, 7182–7190 CrossRef.
- B. Hess, J. Chem. Theory Comput., 2008, 4, 116–122 CrossRef CAS PubMed.
- S. Miyamoto and P. A. Kollman, J. Comput. Chem., 1992, 13, 952–962 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1998, 98, 10089–10092 CrossRef.
- D. S. D. Van, E. Lindahl, B. Hess, G. Groenhof, A. E. Mark and H. J. Berendsen, J. Comput. Chem., 2005, 26, 1701–1718 CrossRef PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS.
- G. M. Torrie and J. P. Valleau, J. Comput. Phys., 1977, 23, 187–199 CrossRef.
- B. Isralewitz, M. Gao and K. Schulten, Curr. Opin. Struct. Biol., 2001, 11, 224–230 CrossRef CAS.
- S. Kumar, D. Bouzida, R. H. Swendsen, P. A. Kollman and J. M. Rosenberg, J. Comput. Chem., 1992, 13, 1011–1021 CrossRef CAS.
- S. J. Marrink and H. J. C. Berendsen, J. Phys. Chem., 1994, 98, 4155–4168 CrossRef CAS.
- J. M. Conder, T. W. L. Point and A. T. Bowen, Aquat. Toxicol., 2004, 69, 199–213 CrossRef CAS PubMed.
- G. R. Lotufo and M. J. Lydy, Arch. Environ. Contam. Toxicol., 2005, 49, 206–214 CrossRef CAS PubMed.
- G. R. Lotufo, Ecotoxicol. Environ. Saf., 2011, 74, 301–306 CrossRef CAS PubMed.
- L. Janosi and A. A. Gorfe, J. Chem. Theory Comput., 2010, 6, 3267–3273 CrossRef CAS PubMed.
- J. P. M. Jämbeck and A. P. Lyubartsev, J. Chem. Theory Comput., 2012, 8, 2938–2948 CrossRef PubMed.
- R. Gupta and B. Rai, Nanoscale, 2017, 9, 4114–4127 RSC.
- A. Golius, L. Gorb, S. A. Michalkova, F. C. Hill, M. Shukla, A. B. Goins, D. R. Johnson and J. Leszczynski, Chemosphere, 2016, 148, 322–327 CrossRef CAS PubMed.
- H. Wang and F. Meng, Mol. Simul., 2016, 42, 1292–1297 CrossRef CAS.
- W. Kopec and H. Khandelia, J. Comput.-Aided Mol. Des., 2014, 28, 123–134 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: The number of TNT molecules contained in the largest aggregate with the simulation time for TNT-10 system and TNT-40 system; the structural properties of membrane in different systems; interaction energy decomposition. See DOI: 10.1039/c9ra08408h |
| This journal is © The Royal Society of Chemistry 2019 |