 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nanoparticle modification in biological media: implications for oral nanomedicines
Vishnaka Muraleetharan,
Julia Mantaj,
Magda Swedrowska and
Driton Vllasaliu *
*
Institute of Pharmaceutical Science, School of Cancer and Pharmaceutical Science, King's College London, 150 Stamford Street, London, SE1 9NH, UK. E-mail: Driton.vllasaliu@kcl.ac.uk
First published on 6th December 2019
Abstract
Nanomedicine has shown potential in enabling oral administration of poorly absorbable drugs, such as biologics. As part of the process related to optimisation of the safety and efficacy of nanomedicines, it is imperative that the interaction of nanoparticles with the biological systems – including the gut – is fully characterised. In this article, we provide an overview of the major mechanisms by which nanoparticles may transform upon introduction in biological media. Specifically, the phenomena of association, dissolution and biomolecule adsorption are discussed, together with factors which influence the occurrence of each phenomenon. The implications of these phenomena within the context of therapeutic action of nanomedicines, which includes reduced targeting efficiency, are also explored. Finally, we will comment on nanoparticle modification within the gut environment, including the currently available gastrointestinal models for the study of nano-bio interactions, with implications in the area of nanomedicines for oral administration.
1. Introduction
The oral route of drug administration is favoured due to convenient administration (ingestion), which facilitates patient adherence to therapy. However, many drugs, including almost all biologics – the use of which has proliferated in recent years – are predominantly delivered via parenteral formulations because of poor oral bioavailability.1 This can be attributable to a number of factors, including drug degradation in the gastrointestinal (GI) tract and poor intestinal absorption.2 A number of drug delivery approaches are being researched to improve oral bioavailability of drugs such as biologics and poorly soluble drugs. One such approach makes use of drug carriers of nanoscale dimensions, i.e. nanomedicine. These nanosystems are usually based on polymeric nanoparticles that are designed to permeate the intestinal epithelium by targeting epithelial receptors which transport cargo from the luminal side to the basolateral (blood) side.3,4 Targeting of biological transport is thought to be essential as most particulates themselves show poor permeability across the GI mucosa.5Nanoparticle biocorona formation, which arises as a result of interaction of nanoparticles with a range of biomolecules present in biological milieus (e.g. proteins, lipids, nucleic acids, metabolites), is a critical phenomenon that determines the biological fate (therapeutic activity or toxicity) of nanoparticles. The importance of the biomolecular corona formed around nanoparticles in biofluids in determining bioactivity (particle–cell interaction) has been demonstrated, for example following nanoparticle interaction with human plasma (as will be discussed later). However, characterisation of the biomolecular corona of nanoparticles in the gastrointestinal biofluids remains poorly understood.6 This knowledge is crucial since biofluid in the GI environments is complex and is likely to significantly alter the properties and colloidal stability of nanoparticles, and thus also their bioavailability and the biological responses they induce.7 The issue of nanoparticle modification in the gut and resulting implications for orally administered nanomedicines has been highlighted in a recent editorial by Berardi and Bombelli.6 This article aims to further increase the awareness of this (somewhat understudied) issue by providing a broader overview and discussion of nanoparticle behaviour in biological media and a focus on nanoparticle behaviours in the GI environment and consequences for oral nanomedicines.
2. The fate of nanoparticles in biological media and physiological implications
There are multiple mechanisms by which nanoparticles reduce their excess surface energy (to attain a more energetically stable state) on introduction to complex biological media.8 Following the classical Derjaguin–Landau–Verwey–Overbeek (DLVO) theory, nanoparticles may cluster into irregularly-shaped larger entities, namely soft agglomerates or dense aggregates.9 Alternatively, nanoparticles may dissolve, following the Noyes–Whitney relationships.10 Nanoparticles can additionally gain a corona of variable thickness around them, either by adsorbing biomolecules from the surroundings or by reacting with the surroundings (e.g. oxidation), which changes their initial identity.9,112.1. Association
The long-range thermodynamic forces between colloidal nanoparticles, governed by Brownian motion, result in interparticle collisions. As per the DLVO theory, nanoparticles may either associate (ir)reversibly or repel upon such contact, depending on their short-range thermodynamic interactions. Surface interactions are simplified by the theory, and the two forces considered to dominate short-range interparticle interactions are attractive van der Waal's (vdW) forces and repulsive electrostatic double layer (EDL) forces. Thus, the likelihood of nanoparticle association is approximately equal to the summation of these forces, which yields either a net attractive or net repulsive.The colloidal stability of nanoparticles is influenced by both their inherent properties (e.g. shape, size and surface charge) and those of the medium in which they are suspended (e.g. pH and ionic strength). To exemplify, the larger a nanoparticle's net surface charge, the greater its colloidal stability. For nanoparticles bearing an amphoteric surface (e.g. metal hydroxides), surface charge is instead a function of the medium's pH. Under pH values close to their isoelectric point or point of zero charge, nanoparticles will have a near-neutral net surface charge. Interparticle repulsions due to the EDL will consequently be reduced, enhancing the effect of vdW attraction and encouraging nanoparticle association.12 Likewise, when subjected to low ionic strengths, a nanoparticle's EDL extends far from its surface, causing interparticle repulsions. Conversely, under high ionic strengths (as is often present in biofluids), the EDL experiences compression and neutralisation, causing nanoparticle association via a strengthened vdW force.11 By posing steric repulsion forces, surface coatings of synthetic polymer, polyelectrolytes or natural organic matter (NOM) may increase the colloidal stability of nanoparticles. Hydrophilic polymers, including polyethylene glycol (PEG) and polyvinylpyrrolidone (PVP), may enable further stabilisation via repulsive short-range hydration force.12,13
The association of nanoparticles in aqueous biological media, including cell culture media, may hinder their cellular targeting efficiency and reduce the representativity of experimental findings. Moreover, depending on their physicochemical properties and the cell type, nanoparticle association may skew their cellular uptake and toxicity profiles. For example, aggregated transferrin-capped gold nanoparticles experienced 25% reduced uptake by A549 and HeLa cells compared to their single, monodispersed counterparts. However, the largest aggregates underwent two-fold greater uptake by MDA-MB 435 cells. Nevertheless, no unique toxic responses were observed with the aggregation state.14
Conversely, Zook et al. identified a substantial decrease in hemolytic toxicity with an increase in size of silver nanoparticle agglomerates (from 43 nm to 1400 nm).15 Similarly, Tripathy et al. reported that, compared to small aggregates, large aggregates of zinc oxide nanoparticles exhibit drastically reduced potency with regards to evoking mitochondrial dysfunction, producing reactive oxygen species (ROS) and causing cellular apoptosis in RAW 264.7 murine macrophages. Interestingly, concentration-dependent aggregation was also identified here, wherein only small aggregates were formed at low concentrations of zinc oxide nanoparticles in phosphate buffer solution (PBS).16
Readers should note that although the association of pristine nanoparticles is possible in biomolecule-free media (in vitro), once in complex biological fluids, the adhesion of biomolecules on nanoparticles would change the surface characteristics of the latter. As a consequence of the new identity, nanoparticles could aggregate,17 or on the contrary, in some cases become more stable.18
2.2. Dissolution
Nanoparticle dissolution involves the movement of constituent molecules of a dissolving solid from its surface to the bulk solution, via a diffusion layer densely occupied by solvated entities, e.g. biomolecules, solute molecules and ions. Accordingly, the material's solubility in the medium and the surface-bulk concentration gradient are considered the major driving forces for this dynamic mechanism. As supported by the Noyes–Whitney equations, nanoparticles exhibit enhanced dissolution kinetics compared to bulk materials due to an immense surface-area-to-volume ratio, a reduced diffusion layer thickness and partially coordinated surface-residing atoms. Furthermore, with a greater proportion of these surface-residing atoms at edges and corners compared to at planar regions, ions and small clusters part from the surface more easily. Since dissolution can determine in vivo nanoparticle persistence, it should be considered upon the interpretation of their biological responses and toxicological profiles.10,19,20Similar to nanoparticle association, dissolution is influenced by several factors, both intrinsic and extrinsic. Nanoparticles often demonstrate a kinetic size effect wherein the maximum concentration of dissolved species is observed instantaneously, after which the concentration decreases until saturation is attained. As identified by Schmidt and Vogelsberger for titanium dioxide nanoparticles, this phenomenon is most evident at larger nanoparticle concentrations.21 An enhanced kinetic size effect is generally associated with smaller nanoparticle sizes. For example, there is sufficient evidence in the literature illustrating an increase in the cytotoxicity of copper oxide when going from bulk/microscale material to smaller nanoparticles, by reason of increased solubility.22,23 Similarly, accelerated silver ion-release with a reduction in particle size, going from macroscale silver foil to nanoparticles of diameter 60 nm and 4.8 nm, has been reported.24 Conversely, no significant difference was observed between the dissolution rates of bulk and nanoparticulate zinc oxide in Osterhout's medium or a synthetic freshwater algal medium.23,25
In compliance with the Gibbs–Thomson effect, the dissolution tendency and equilibrium solubility of particles and surface structures increase with a reduced radius of positive curvature (i.e. convex); this is due to their energetic instability. In some instances, their equilibrium solubility may even surpass their saturation concentration, resulting in precipitation and/or Ostwald ripening. It can thus be deduced that nanoparticles bearing a reduced radius of negative curvature (i.e. concave) demonstrate better colloidal stability.10
As discussed above, the colloidal stability of nanostructures is often enhanced by the hindrance of interparticle association through surface coatings. However, the ligands of such stabilising agents may instead alter the dissolution kinetics of the nanoparticles they harbour.19 For instance, in water at 25 °C, PVP-stabilised silver nanoparticles have demonstrated a greater degree and rate of dissolution compared to citrate-stabilised silver nanoparticles. It was postulated that this was because the citrate layer served as a chemical barrier by reducing the departing silver ions.26 Contrastingly, Huynh and Chen observed that, over a 30 min duration in water, citrate-stabilised silver nanoparticles experienced greater dissolution than PVP-stabilised silver nanoparticles.27
As well as nanoparticle properties, the nature of the medium (e.g. pH and ionic strength) may influence nanoparticle dissolution through its effects on the above mentioned phenomenon, agglomeration and aggregation.28 The association of individual nanoparticles reduces their exposed surface area, thereby hindering ion release to the bulk medium and lowering the degree of dissolution. Additionally, the association of nanostructures reduces the number of surface sites which may be oxidised.29 For instance, the association of goethite nanorods at reduced pH values and/or increased ionic strengths was observed to quench, either largely or completely, their dissolution.30
Changes in the saturation concentration and dissolution kinetics of nanoparticles due to the availability of organic (e.g. NOM and polysaccharides) and/or inorganic (e.g. simulated biofluid and buffer) entities, are also documented in the literature. Dissolution is reportedly ‘catalysed’ by ionic and organic species capable of forming soluble complexes with the released ions. Conversely, the generation of less soluble complexes impedes dissolution.10 For instance, copper oxide nanoparticles have demonstrated near-complete dissolution in the presence of tryptone and yeast extract, which are rich in amino acids. However, no significant difference in dissolution was measured upon their exposure to sodium chloride.22 For silver nanoparticles, the addition of a small amount of chloride has been associated with a significant reduction in the rate of release of soluble species.31 Contrastingly, cysteine is reported to enhance the dissolution of silver nanoparticles.32
The complex relationship between nanoparticle dissolution and the various intrinsic (e.g. nanoparticle composition, shape, surface area, surface chemistry, etc.) and extrinsic factors (e.g. media) is depicted in Fig. 1. Understanding these factors, facilitates the understanding and prediction of the fate and biological response of nanoparticles.
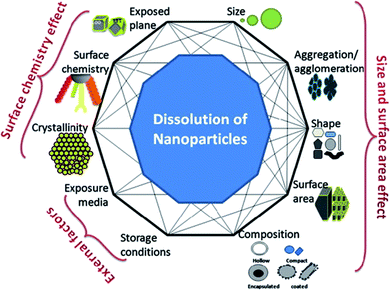 | ||
| Fig. 1 A simplistic representation showing the factors that can affect dissolution of nanoparticles and the possible interconnectivity among the factors, themselves. This figure has been reproduced from ref. 28 with permission from Elsevier. | ||
The immense contribution of dissolution to the cytotoxicity of nanoparticles comprising toxic constituent ions (e.g. zinc oxide), in comparison to that of nanoparticles which are themselves toxic (e.g. cerium dioxide), has been described in the literature.33 To exemplify, divalent and trivalent cations of copper and iron (respectively) may evoke site-specific DNA damage.34 Also, exposure to hexavalent and divalent cations of chromium and cadmium (respectively) has been shown to enhance hepatic lipid peroxidation.35 A greater proportion of copper nanoparticles (relative to copper carbonate salt) were reported to experience intestinal absorption, accumulate in the brain tissue and, at greater dietary doses, lead to hepatic damage in rat models.36 Furthermore, divalent silver and zinc cations are known to damage the bacterial membrane, thereby causing bacterial death.37
The employment of nanostructures (which may be considered a reservoir of cations) for medical applications therefore necessitates careful consideration. The extent of both the bactericidal and hemolytic activity of silver nanoparticles has correlated with the quantity of silver ions released.15,38 Likewise, oxidative stress and cytotoxicity by cadmium-containing quantum dots have been associated directly with the intracellular release of constituent divalent cadmium cations.39 However, while enhanced dissolution kinetics of nanoparticles has been employed in the case of drug nanocrystals to improve drug dissolution, oral nanomedicines are generally based on insoluble nanoparticulate carriers, designed to retain the drug until it reaches the target.
2.3. Biomolecule adsorption
On exposure to physiological milieus, nanoparticles may interact with a range of biomolecules including lipids, nucleic acids and metabolites. The adsorption of proteins to nanoparticle surface establish a protein corona has been given particular attention in the recent years, particularly within the context of nanomedicine targeting. Since many proteins are amphipathic, they have the potential to interact with almost any nanoparticle surface in their vicinity. Consequently, the interfacial interactions of nanoparticles with proteins are inevitable in such environments. In concordance, as expressed thermodynamically by ΔadsG < 0 (where ΔadsG symbolises the Gibbs free energy of adsorption), this mechanism of lowering the excess surface energy of nanoparticles is spontaneous.40The surface adsorption of serum proteins by particles (opsonisation) was first postulated by Wright and Douglas in 1903.41 Later in 1962, whilst assessing the surface adsorption of fibrinogen in blood plasma, Vroman noticed that the maximum surface concentration of fibrinogen was attained at an intermediate time point, suggestive of its substitution by other proteins with time.42 Described by the Vroman effect, corona formation is a dynamic and competitive process. The initial stages of adsorption are dominated by small (high mobility), abundant proteins. However, with time, some of these proteins are substituted by those which are larger (low mobility), less abundant and have greater affinity for the surface.43 An equilibrium is gradually attained, after which each protein has equal rates of association and dissociation, and further exchange causes no change in the corona's composition.44 As illustrated in Fig. 2, the protein corona branches into a ‘hard corona’ and ‘soft corona’; these denote a firmly bound, equilibrium state layer (wherein proteins exhibit high affinity for the nanoparticle surface), and a loosely bound layer (comprised of low-affinity proteins in rapid exchange with those in the medium), respectively.45,46
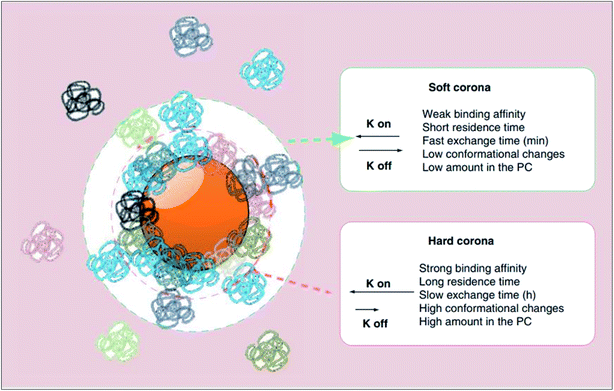 | ||
| Fig. 2 A schematic representation of the protein corona (PC) which branches into two layers, namely the hard and soft coronas. As listed, proteins which establish the hard corona exhibit higher affinity for the nanoparticle surface and thus have a longer surface residence time and slower exchange rate with proteins in the bulk medium than do proteins of the soft corona (as represented by a smaller rate constant (K off) in the order of hours as oppose to minutes). Hard corona proteins, of which there are more, also experience greater conformational changes than do soft corona proteins. This figure has been reproduced from ref. 11 with permission from Future Medicine. | ||
An intrinsic influencer of protein corona composition is nanoparticle size. For both citrate-stabilised and PVP-stabilised silver nanoparticles, proteomic analysis of the acquired corona revealed that 110 nm nanoparticles associated with almost 1.8 times more serum proteins than did 20 nm nanoparticles.47 The increased curvature of the smaller nanoparticles likely suppressed the adsorption of proteins, particularly those more bulky and rigid.48 Contrastingly, due to a size much smaller and bigger (respectively) than the most abundant serum proteins, neither 4 nm nor 40 nm gold nanoparticles acquired a complete protein corona.49 Concordantly, 30 nm gold nanoparticles were reported to adsorb 2.3 times more plasma proteins than 50 nm gold nanoparticles.50 This discrepancy suggests the effect of nanoparticle composition. Concerning protein conformation within the corona, 6 nm silica nanoparticles evoked a six-fold reduced modification in the secondary structure of human carbonic anhydrase 1 than did 15 nm silica nanoparticles.51 Similarly, increasing the size of silica nanoparticles from 4 nm to 15 nm substantially decreased the stability of adsorbed ribonuclease A due to increased protein unfolding.52
The association of nanoparticles in solution is known to both lower their exposed surface area for protein adsorption (which reduces the density of the corona) and induce surface rugosity (which evokes conformational changes in the adsorbed proteins).53 Regarding nanoparticle morphology, gold nanorods have been observed to undergo greater protein adsorption than gold nanospheres of equivalent size.54 Likewise, neutral nanoparticles have been reported to bind fewer proteins than their charged counterparts.55 Between positively-charged and negatively-charged nanoparticles, the former adsorb the greatest number of proteins.56 Moreover, employing gold nanoparticles bearing self-assembled monolayers of functionalised alkyl chains, it was proved that increasing surface hydrophobicity increases the corona's density. This is since protein adsorption is mediated (in part) by hydrophobic interactions.57
As demonstrated by Maiorano et al. utilising Dulbecco's Modified Eagle medium (DMEM) and Roswell Park Memorial Institute medium (RPMI), supplemented with Fetal Bovine Serum (FBS), biofluid composition is a prominent extrinsic influencer of corona formation. The two media (which vary in their salt, glucose and amino acid content) induced dissimilar adsorption patterns, with citrate-stabilised gold nanoparticles gaining a more abundant and stable corona in DMEM at all tested sizes.58 Consistently, an increase in plasma concentration (from 3% to 80%) corresponded to a notable increase in protein adsorption for polystyrene nanoparticles. For silica nanoparticles, increasing plasma concentration instead induced considerable changes in the protein pattern, illustrative of competitive adsorption by less-abundant, high-affinity proteins due to enhanced adhesion at higher plasma concentrations.59 Finally, nanoparticles which interact with proteins electrostatically have been reported to experience pH-dependent corona formation by reason of an altered protein binding affinity.60
The instantaneous adsorption of proteins by nanoparticles may alter their physicochemical attributes (including size and surface charge), thereby providing them with a new biological identity. Thus, it is not pristine nanoparticles, but those which are biomolecule-coated that largely govern physiological responses such as systemic circulation time, bioavailability, cellular uptake and cytotoxicity.61 For instance, the formation of a protein corona around transferrin-functionalised silicon dioxide nanoparticles was shown to eliminate their transferrin-mediated targeting capability due to masked receptor-binding sites (Fig. 3). In another study, the coating of gold nanoparticles with polyacrylic acid was shown to cause the unfolding of adsorbed fibrinogen which, consequently, evoked an inflammatory cascade.11
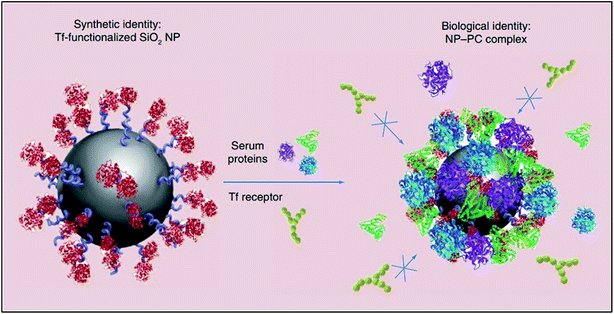 | ||
| Fig. 3 A schematic representation of the loss of transferrin (Tf)-mediated specificity by Tf-functionalised silicon dioxide nanoparticles due to the formation of a protein corona which impedes receptor-binding site interactions. This figure has been reproduced from ref. 11 with permission from Future Medicine. | ||
By contrast, serum proteins adsorbed to amine-functionalised polystyrene nanoparticles were reported to counterweight the nanoparticle surface charge, be retained upon their uptake (by both A549 and 1321N1 cells), and only experience degradation once trafficked to lysosomes. The harbouring of these positively charged nanoparticles averted their direct encounter with cell membranes, which would have otherwise induced cell death.62 Similarly, the generation of ROS (a measure of cytotoxicity) in human acute monocytic leukaemia (THP-1) cells by cobalt oxide nanoparticles was observed to be lower following incubation in complete cell culture medium for 48 h, than in PBS for the same duration.63
3. Nanoparticle modification in the gastrointestinal environment
3.1. The changing gastrointestinal environment
Luminal fluid within the upper small intestine uniquely comprises chyme from the stomach, as well as secretions from the following compartments: liver (bile; bicarbonate, bile pigments, bile salts, cholesterol, phospholipids, and organic wastes), pancreas (bicarbonate, amylases, lipases and proteases including trypsin and chymotrypsin) and small intestinal wall (mineral ions; bicarbonate, chloride and sodium, and water).66 The digestion of silver nanoparticles with food compounds (carbohydrates, fatty acids and proteins) was reported to have a negligible impact on nanoparticle uptake by intestinal Caco-2 cells, whilst the digestion of silver nanoparticles in the absence of food compounds reduced cellular uptake by 60%.67 In another study, coatings of bovine serum albumin and casein were found to limit the adhesion of carboxylate-functionalised polystyrene nanoparticles (of 20 nm, 100 nm and 200 nm diameter) to Caco-2 cells. Conversely, exposure to meat extract caused no obvious impact on nanoparticle–cell interaction. Likewise, incubation in murine intestinal fluid increased the adherence of 20 nm and 100 nm nanoparticles to Caco-2 cells.68 Furthermore, treatment with in vitro digestive solution reportedly decreased the ROS generation potential of silicon dioxide and zinc oxide nanoparticles, albeit with no detectable effects on cytotoxicity.69
In addition to compartmentalisation, the composition of GI fluid is governed by mixing patterns, fluid absorption into the intestinal wall and transit through the intestinal tract. Several secretions of the small intestine are induced by food consumption, which gives rise to notable variation in GI fluid composition with prandial state. To exemplify, the consumption of a protein-rich meal (within the usual range of dietary intake) is followed by a surge in free and peptide amino acids within the intraluminal contents, originating from the exogeneous protein.70 Accordingly, the influence of protein-, fat- and carbohydrate-rich meals, as well as high-calorie meals, on drug absorption have been postulated.71 Thus ultimately, the prediction of nanoparticle transformation within the GI environment necessitates the consideration of regional and prandial effects on GI fluid composition, and thereby biomolecule corona formation.
The preprandial stomach is characterised by acidic pH values of 1.0 to 2.5, whilst average pH values of 6.6 and 7.5 are found in the proximal small intestine and terminal ileum, respectively.74 By contrast, the ingestion of food or liquid may cause a steep rise in gastric pH, up to values of 6.0 or higher depending on the composition and volume of the ingested matter.75 Similarly, the pH of the gastric mucus layer increases from approximately 1.0 in the lumen, to near-neutral values at the epithelial cell surface.76 A rise in pH towards the point-of-zero-charge of titanium dioxide nanoparticles has shown to increase the diameter and decrease the mobility of nanoparticle aggregates. As supported by the DLVO theory, this is due to the suppression of their surface ionisation which weakens interparticle EDL repulsions, thereby promoting nanoparticle aggregation.77,78 Thus, the inconsistency of GI pH must also be considered as a potential influencer of the therapeutic efficacy of nanoparticles and nano-scale drug delivery systems, through its modification of nanoparticle surface properties.
3.2. Gastrointestinal models for the study of nanoparticle behaviour
Major impediments to the characterisation of interactions between nanoparticles and the biological environment within the gut, which may explain the knowledge gap in this area to date, stem from the unavailability of reliable models of the GIT, in addition to analytical difficulties presented by the complex mixture of biomolecules (e.g. lipids, proteins and surfactants) within GI fluids. In vivo studies obviously enable a more reliable evaluation of nanomedicines, including establishing their toxicological profile, therapeutic response, drug–drug and drug–food interactions (and subsequent adverse effects), which are otherwise unidentifiable in vitro.79 However, it is imperative to select an animal model which best simulates the relevant human system. For example, rats are commonly employed to predict the human intestinal permeation of conventional chemicals. However, as a prerequisite, they must undergo surgery and anaesthesia, which may cause abnormal GI transit times. As a result of ethical considerations, costs, and inter-species variability, there is considerable interest in using representative in vitro models of the GI biofluids and GIT tissue for the study of nanoparticle behaviour post-ingestion/administration.Fasted and fed state SIF (FaSSIF-V2 and FeSSIF-V2, respectively) are highly simplistic models of intraluminal composition. As such, they are deprived of vital intraluminal components which may render in vitro findings unreliable. For example, whilst HIF harbour a variety of bile salts, FaSSIF-V2 and FeSSIF-V2 comprise pure sodium taurocholate only.82 FeSSIF-V2 additionally fails to replicate the intricate ultrastructure of postprandial HIF (which includes mixed micelles and vesicles), likely due to the absence of lipids and lipolysis products. Furthermore, the movement and enzymatic degradation of food (which impact colloid formation) within the GIT are unaccounted for.64,83
The addition of pancreatin (and calcium chloride) is one approach to improve the complexity of FeSSIF-V2.84 The substitution of sodium taurocholate with crude bile salt extracts (in both FaSSIF-V2 and FeSSIF-V2) has also proven to better predict in vivo conditions. However, to ensure the reproducibility of experimental results, it is often necessary to standardise the quantity of bile salt added to crude extract batches.85
However, the Caco-2 transwell system accounts for neither the constant fluid flow nor the fluid shear stress tolerated by epithelial cells in vivo, thereby skewing the predicted bioavailability of chemicals and/or nanomedicines.91,92 Caco-2 cell monolayers additionally lack a mucus coating, which serves to (i) control the intestinal absorption of matter and (ii) shield the epithelium from harmful intraluminal contents in vivo.89 Expanding on the latter, bile salts are considerably toxic (irritant) to epithelial cells despite being a major constituent of intestinal media. Thus, the potentially detrimental impact of SIF on the Caco-2 intestinal barrier model (which may otherwise be a useful strategy to model both the intestinal luminal fluid and epithelium) may cause the overestimation of transepithelial permeation.93 One strategy to omit this is to instead employ co-culture models (i.e. Caco-2 cells with mucus-secreting HT29-MTX cells).94
4. Conclusions
Nanoscale drug delivery systems have been proposed and are being developed to improve oral drug bioavailability. However, detailed characterisation of the interactions between these nanosystems and the biological environment within the gut is currently lacking, with existing literature on the interactions and behaviour changes of nanoparticles in physiological environments focusing on the blood. Only limited studies have assessed the modification of nanoparticles on exposure to the GI tract conditions (e.g. digestion) and these point to significant modification of nanosystems in the GI environment, impacting their bioactivity (including cell uptake and toxicity). More research is needed in this area, particularly considering the complexity of the GI tract and the different factors that could influence nanoparticle behaviour, including the changing gastrointestinal fluid composition and mechanical forces. Importantly, study of nanoparticle behaviours in the GI environment has to precede and inform research into the design and development of oral nanomedicines. Otherwise, current studies focusing on formulation of nanoparticles for oral delivery, particularly those conducted in vitro and utilising simple and non-representative models (e.g. media), may be of limited value.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was partially funded by the Engineering and Physical Sciences Research Council (EPSRC); grant reference number EP/P002544/2.References
- Y. Yun, Y. W. Cho and K. Park, Nanoparticles for oral delivery: targeted nanoparticles with peptidic ligands for oral protein delivery, Adv. Drug Delivery Rev., 2013, 65(6), 822–832 CrossRef CAS PubMed.
- G. P. Carino and E. Mathiowitz, Oral insulin delivery, Adv. Drug Delivery Rev., 1999, 35(2–3), 249–257 CrossRef CAS PubMed.
- E. M. Pridgen, F. Alexis, T. T. Kuo, E. Levy-Nissenbaum, R. Karnik, R. S. Blumberg, R. Langer and O. C. Farokhzad, Transepithelial transport of Fc-targeted nanoparticles by the neonatal Fc receptor for oral delivery, Sci. Transl. Med., 2013, 5(213), 213ra167 CrossRef PubMed.
- Y. Shi, X. Sun, L. Zhang, K. Sun, K. Li, Y. Li and Q. Zhang, Fc-modified exenatide-loaded nanoparticles for oral delivery to improve hypoglycemic effects in mice, Sci. Rep., 2018, 8(1), 726 CrossRef PubMed.
- S. J. Cao, S. Xu, H. M. Wang, Y. Ling, J. Dong, R. D. Xia and X. H. Sun, Nanoparticles: Oral Delivery for Protein and Peptide Drugs, AAPS PharmSciTech, 2019, 20(5), 190 CrossRef PubMed.
- A. Berardi and F. Baldelli Bombelli, Oral delivery of nanoparticles – let's not forget about the protein corona, Expert Opin. Drug Delivery, 2019, 16(6), 563–566 CrossRef PubMed.
- A. Murphy, K. Sheehy, A. Casey and G. Chambers, Potential of biofluid components to modify silver nanoparticle toxicity, J. Appl. Toxicol., 2015, 35(6), 665–680 CrossRef CAS.
- A. Tsuda and N. K. Venkata, The role of natural processes and surface energy of inhaled engineered nanoparticles on aggregation and corona formation, NanoImpact, 2016, 2, 38–44 CrossRef PubMed.
- D. R. Baer, M. H. Engelhard, G. E. Johnson, J. Laskin, J. Lai, K. Mueller, P. Munusamy, S. Thevuthasan, H. Wang, N. Washton, A. Elder, B. L. Baisch, A. Karakoti, S. V. Kuchibhatla and D. Moon, Surface characterization of nanomaterials and nanoparticles: important needs and challenging opportunities, J. Vac. Sci. Technol., A, 2013, 31(5), 50820 CrossRef.
- P. Borm, F. C. Klaessig, T. D. Landry, B. Moudgil, J. Pauluhn, K. Thomas, R. Trottier and S. Wood, Research strategies for safety evaluation of nanomaterials, part V: role of dissolution in biological fate and effects of nanoscale particles, Toxicol. Sci., 2006, 90(1), 23–32 CrossRef CAS PubMed.
- C. Corbo, R. Molinaro, A. Parodi, N. E. Toledano Furman, F. Salvatore and E. Tasciotti, The impact of nanoparticle protein corona on cytotoxicity, immunotoxicity and target drug delivery, Nanomedicine, 2016, 11(1), 81–100 CrossRef CAS.
- T. L. Moore, L. Rodriguez-Lorenzo, V. Hirsch, S. Balog, D. Urban, C. Jud, B. Rothen-Rutishauser, M. Lattuada and A. Petri-Fink, Nanoparticle colloidal stability in cell culture media and impact on cellular interactions, Chem. Soc. Rev., 2015, 44(17), 6287–6305 RSC.
- L. Guerrini, R. A. Alvarez-Puebla and N. Pazos-Perez, Surface Modifications of Nanoparticles for Stability in Biological Fluids, Materials, 2018, 11(7), 1154 CrossRef PubMed.
- A. Albanese and W. C. Chan, Effect of gold nanoparticle aggregation on cell uptake and toxicity, ACS Nano, 2011, 5(7), 5478–5489 CrossRef CAS PubMed.
- J. M. Zook, R. I. Maccuspie, L. E. Locascio, M. D. Halter and J. T. Elliott, Stable nanoparticle aggregates/agglomerates of different sizes and the effect of their size on hemolytic cytotoxicity, Nanotoxicology, 2011, 5(4), 517–530 CrossRef CAS PubMed.
- N. Tripathy, T. K. Hong, K. T. Ha, H. S. Jeong and Y. B. Hahn, Effect of ZnO nanoparticles aggregation on the toxicity in RAW 264.7 murine macrophage, J. Hazard. Mater., 2014, 270, 110–117 CrossRef CAS PubMed.
- P. Zhang, X. Y. Xu, Y. P. Chen, M. Q. Xiao, B. Feng, K. X. Tian, Y. H. Chen and Y. Z. Dai, Protein corona between nanoparticles and bacterial proteins in activated sludge: characterization and effect on nanoparticle aggregation, Bioresour. Technol., 2018, 250, 10–16 CrossRef CAS PubMed.
- S. Dominguez-Medina, J. Blankenburg, J. Olson, C. F. Landes and S. Link, Adsorption of a Protein Monolayer via Hydrophobic Interactions Prevents Nanoparticle Aggregation under Harsh Environmental Conditions, ACS Sustainable Chem. Eng., 2013, 1(7), 833–842 CrossRef CAS PubMed.
- W. Utembe, K. Potgieter, A. B. Stefaniak and M. Gulumian, Dissolution and biodurability: important parameters needed for risk assessment of nanomaterials, Part. Fibre Toxicol., 2015, 12, 11 CrossRef.
- S. W. Bian, I. A. Mudunkotuwa, T. Rupasinghe and V. H. Grassian, Aggregation and dissolution of 4 nm ZnO nanoparticles in aqueous environments: influence of pH, ionic strength, size, and adsorption of humic acid, Langmuir, 2011, 27(10), 6059–6068 CrossRef CAS PubMed.
- J. Schmidt and W. Vogelsberger, Dissolution kinetics of titanium dioxide nanoparticles: the observation of an unusual kinetic size effect, J. Phys. Chem. B, 2006, 110(9), 3955–3963 CrossRef CAS PubMed.
- C. Gunawan, W. Y. Teoh, C. P. Marquis and R. Amal, Cytotoxic origin of copper(II) oxide nanoparticles: comparative studies with micron-sized particles, leachate, and metal salts, ACS Nano, 2011, 5(9), 7214–7225 CrossRef CAS PubMed.
- M. Mortimer, K. Kasemets and A. Kahru, Toxicity of ZnO and CuO nanoparticles to ciliated protozoa Tetrahymena thermophila, Toxicology, 2010, 269(2–3), 182–189 CrossRef CAS PubMed.
- J. Liu, D. A. Sonshine, S. Shervani and R. H. Hurt, Controlled release of biologically active silver from nanosilver surfaces, ACS Nano, 2010, 4(11), 6903–6913 CrossRef CAS.
- N. M. Franklin, N. J. Rogers, S. C. Apte, G. E. Batley, G. E. Gadd and P. S. Casey, Comparative toxicity of nanoparticulate ZnO, bulk ZnO, and ZnCl2 to a freshwater microalga (Pseudokirchneriella subcapitata): the importance of particle solubility, Environ. Sci. Technol., 2007, 41(24), 8484–8490 CrossRef CAS PubMed.
- S. Kittler, C. Greulich, J. Diendorf, M. Koller and M. Epple, Toxicity of silver nanoparticles increases during storage because of slow dissolution under release of silver ions, Chem. Mater., 2010, 22(16), 4548–4554 CrossRef CAS.
- K. A. Huynh and K. L. Chen, Aggregation kinetics of citrate and polyvinylpyrrolidone coated silver nanoparticles in monovalent and divalent electrolyte solutions, Environ. Sci. Technol., 2011, 45(13), 5564–5571 CrossRef CAS PubMed.
- S. K. Misra, A. Dybowska, D. Berhanu, S. N. Luoma and E. Valsami-Jones, The complexity of nanoparticle dissolution and its importance in nanotoxicological studies, Sci. Total Environ., 2012, 438, 225–232 CrossRef CAS PubMed.
- J. Liu and R. H. Hurt, Ion release kinetics and particle persistence in aqueous nano-silver colloids, Environ. Sci. Technol., 2010, 44(6), 2169–2175 CrossRef CAS PubMed.
- G. Rubasinghege, R. W. Lentz, H. Park, M. M. Scherer and V. H. Grassian, Nanorod dissolution quenched in the aggregated state, Langmuir, 2010, 26(3), 1524–1527 CrossRef CAS PubMed.
- C. Levard, S. Mitra, T. Yang, A. D. Jew, A. R. Badireddy, G. V. Lowry and G. E. Brown Jr, Effect of chloride on the dissolution rate of silver nanoparticles and toxicity to E. coli, Environ. Sci. Technol., 2013, 47(11), 5738–5745 CrossRef CAS PubMed.
- A. P. Gondikas, A. Morris, B. C. Reinsch, S. M. Marinakos, G. V. Lowry and H. Hsu-Kim, Cysteine-induced modifications of zero-valent silver nanomaterials: implications for particle surface chemistry, aggregation, dissolution, and silver speciation, Environ. Sci. Technol., 2012, 46(13), 7037–7045 CrossRef CAS PubMed.
- T. Xia, M. Kovochich, M. Liong, L. Madler, B. Gilbert, H. Shi, J. I. Yeh, J. I. Zink and A. E. Nel, Comparison of the mechanism of toxicity of zinc oxide and cerium oxide nanoparticles based on dissolution and oxidative stress properties, ACS Nano, 2008, 2(10), 2121–2134 CrossRef CAS PubMed.
- S. Oikawa and S. Kawanishi, Distinct mechanisms of site-specific DNA damage induced by endogenous reductants in the presence of iron(III) and copper(II), Biochim. Biophys. Acta, 1998, 1399(1), 19–30 CrossRef CAS.
- D. Bagchi, P. J. Vuchetich, M. Bagchi, E. A. Hassoun, M. X. Tran, L. Tang and S. J. Stohs, Induction of oxidative stress by chronic administration of sodium dichromate [chromium VI] and cadmium chloride [cadmium II] to rats, Free Radical Biol. Med., 1997, 22(3), 471–478 CrossRef CAS PubMed.
- E. Cholewinska, K. Ognik, B. Fotschki, Z. Zdunczyk and J. Juskiewicz, Comparison of the effect of dietary copper nanoparticles and one copper (II) salt on the copper biodistribution and gastrointestinal and hepatic morphology and function in a rat model, PLoS One, 2018, 13(5), e0197083 CrossRef PubMed.
- C. Ning, X. Wang, L. Li, Y. Zhu, M. Li, P. Yu, L. Zhou, Z. Zhou, J. Chen, G. Tan, Y. Zhang, Y. Wang and C. Mao, Concentration ranges of antibacterial cations for showing the highest antibacterial efficacy but the least cytotoxicity against mammalian cells: implications for a new antibacterial mechanism, Chem. Res. Toxicol., 2015, 28(9), 1815–1822 Search PubMed.
- J. R. Morones, J. L. Elechiguerra, A. Camacho, K. Holt, J. B. Kouri, J. T. Ramirez and M. J. Yacaman, The bactericidal effect of silver nanoparticles, Nanotechnology, 2005, 16(10), 2346–2353 CrossRef CAS PubMed.
- A. Sukhanova, S. Bozrova, P. Sokolov, M. Berestovoy, A. Karaulov and I. Nabiev, Dependence of Nanoparticle Toxicity on Their Physical and Chemical Properties, Nanoscale Res. Lett., 2018, 13(1), 44 CrossRef PubMed.
- W. Norde, Colloids and interfaces in life sciences, Marcel Dekker, New York, 2003 Search PubMed.
- A. E. Wright, S. R. Douglas and J. B. Sanderson, An experimental investigation of the role of the blood fluids in connection with phagocytosis, Rev. Infect. Dis., 1989, 11(5), 827–834 CrossRef CAS PubMed.
- L. Vroman, Effect of absorbed proteins on the wettability of hydrophilic and hydrophobic solids, Nature, 1962, 196, 476–477 CrossRef CAS PubMed.
- M. Holmberg and X. Hou, Competitive protein adsorption–multilayer adsorption and surface induced protein aggregation, Langmuir, 2009, 25(4), 2081–2089 CrossRef CAS PubMed.
- D. Dell'Orco, M. Lundqvist, C. Oslakovic, T. Cedervall and S. Linse, Modeling the time evolution of the nanoparticle-protein corona in a body fluid, PLoS One, 2010, 5(6), e10949 CrossRef PubMed.
- D. Docter, D. Westmeier, M. Markiewicz, S. Stolte, S. K. Knauer and R. H. Stauber, The nanoparticle biomolecule corona: lessons learned – challenge accepted?, Chem. Soc. Rev., 2015, 44(17), 6094–6121 RSC.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(38), 14265–14270 CrossRef CAS PubMed.
- J. H. Shannahan, X. Lai, P. C. Ke, R. Podila, J. M. Brown and F. A. Witzmann, Silver nanoparticle protein corona composition in cell culture media, PLoS One, 2013, 8(9), e74001 CrossRef CAS.
- J. Klein, Probing the interactions of proteins and nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(7), 2029–2030 CrossRef CAS.
- E. Casals, T. Pfaller, A. Duschl, G. J. Oostingh and V. Puntes, Time evolution of the nanoparticle protein corona, ACS Nano, 2010, 4(7), 3623–3632 CrossRef CAS.
- M. A. Dobrovolskaia, A. K. Patri, J. Zheng, J. D. Clogston, N. Ayub, P. Aggarwal, B. W. Neun, J. B. Hall and S. E. McNeil, Interaction of colloidal gold nanoparticles with human blood: effects on particle size and analysis of plasma protein binding profiles, Nanomedicine, 2009, 5(2), 106–117 CrossRef CAS PubMed.
- M. Lundqvist, I. Sethson and B. H. Jonsson, Protein adsorption onto silica nanoparticles: conformational changes depend on the particles' curvature and the protein stability, Langmuir, 2004, 20(24), 10639–10647 CrossRef CAS PubMed.
- W. Shang, J. H. Nuffer, J. S. Dordick and R. W. Siegel, Unfolding of ribonuclease A on silica nanoparticle surfaces, Nano Lett., 2007, 7(7), 1991–1995 CrossRef CAS PubMed.
- S. R. Saptarshi, A. Duschl and A. L. Lopata, Interaction of nanoparticles with proteins: relation to bio-reactivity of the nanoparticle, J. Nanobiotechnol., 2013, 11, 26 CrossRef CAS PubMed.
- J. E. Gagner, M. D. Lopez, J. S. Dordick and R. W. Siegel, Effect of gold nanoparticle morphology on adsorbed protein structure and function, Biomaterials, 2011, 32(29), 7241–7252 CrossRef CAS PubMed.
- Z. J. Deng, M. Liang, I. Toth, M. Monteiro and R. F. Minchin, Plasma protein binding of positively and negatively charged polymer-coated gold nanoparticles elicits different biological responses, Nanotoxicology, 2013, 7(3), 314–322 CrossRef CAS PubMed.
- S. Nandhakumar, M. D. Dhanaraju, V. D. Sundar and B. Heera, Influence of surface charge on the in vitro protein adsorption and cell cytotoxicity of paclitaxel loaded poly(ε-caprolactone) nanoparticles, Bull. Fac. Pharm., 2017, 55(2), 249–258 Search PubMed.
- K. L. Prime and G. M. Whitesides, Self-assembled organic monolayers: model systems for studying adsorption of proteins at surfaces, Science, 1991, 252(5009), 1164–1167 CrossRef CAS PubMed.
- G. Maiorano, S. Sabella, B. Sorce, V. Brunetti, M. A. Malvindi, R. Cingolani and P. P. Pompa, Effects of cell culture media on the dynamic formation of protein-nanoparticle complexes and influence on the cellular response, ACS Nano, 2010, 4(12), 7481–7491 CrossRef CAS PubMed.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. B. Bombelli and K. A. Dawson, Physical-chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles, J. Am. Chem. Soc., 2011, 133(8), 2525–2534 CrossRef CAS PubMed.
- J. O'Brien and K. J. Shea, Tuning the Protein Corona of Hydrogel Nanoparticles: The Synthesis of Abiotic Protein and Peptide Affinity Reagents, Acc. Chem. Res., 2016, 49(6), 1200–1210 CrossRef PubMed.
- V. H. Nguyen and B. J. Lee, Protein corona: a new approach for nanomedicine design, Int. J. Nanomed., 2017, 12, 3137–3151 CrossRef CAS.
- F. Wang, L. Yu, M. P. Monopoli, P. Sandin, E. Mahon, A. Salvati and K. A. Dawson, The biomolecular corona is retained during nanoparticle uptake and protects the cells from the damage induced by cationic nanoparticles until degraded in the lysosomes, Nanomedicine, 2013, 9(8), 1159–1168 CrossRef CAS.
- E. Casals, T. Pfaller, A. Duschl, G. J. Oostingh and V. F. Puntes, Hardening of the nanoparticle-protein corona in metal (Au, Ag) and oxide (Fe3O4, CoO, and CeO2) nanoparticles, Small, 2011, 7(24), 3479–3486 CrossRef CAS.
- D. Riethorst, P. Baatsen, C. Remijn, A. Mitra, J. Tack, J. Brouwers and P. Augustijns, An In-Depth View into Human Intestinal Fluid Colloids: Intersubject Variability in Relation to Composition, Mol. Pharm., 2016, 13(10), 3484–3493 CrossRef CAS PubMed.
- M. Vertzoni, P. Augustijns, M. Grimm, M. Koziolek, G. Lemmens, N. Parrott, C. Pentafragka, C. Reppas, J. Rubbens, J. Van Den Alphabeele, T. Vanuytsel, W. Weitschies and C. G. Wilson, Impact of regional differences along the gastrointestinal tract of healthy adults on oral drug absorption: an UNGAP review, Eur. J. Pharm. Sci., 2019, 134, 153–175 CrossRef CAS PubMed.
- D. M. Mudie, G. L. Amidon and G. E. Amidon, Physiological parameters for oral delivery and in vitro testing, Mol. Pharm., 2010, 7(5), 1388–1405 CrossRef CAS PubMed.
- D. Lichtenstein, J. Ebmeyer, P. Knappe, S. Juling, L. Bohmert, S. Selve, B. Niemann, A. Braeuning, A. F. Thunemann and A. Lampen, Impact of food components during in vitro digestion of silver nanoparticles on cellular uptake and cytotoxicity in intestinal cells, Biol. Chem., 2015, 396(11), 1255–1264 CAS.
- H. Sinnecker, K. Ramaker and A. Frey, Coating with luminal gut-constituents alters adherence of nanoparticles to intestinal epithelial cells, Beilstein J. Nanotechnol., 2014, 5, 2308–2315 CrossRef.
- K. Gerloff, D. I. Pereira, N. Faria, A. W. Boots, J. Kolling, I. Forster, C. Albrecht, J. J. Powell and R. P. Schins, Influence of simulated gastrointestinal conditions on particle-induced cytotoxicity and interleukin-8 regulation in differentiated and undifferentiated Caco-2 cells, Nanotoxicology, 2013, 7(4), 353–366 CrossRef CAS PubMed.
- S. A. Adibi and D. W. Mercer, Protein digestion in human intestine as reflected in luminal, mucosal, and plasma amino acid concentrations after meals, J. Clin. Invest., 1973, 52(7), 1586–1594 CrossRef CAS PubMed.
- K. A. Lentz, Current methods for predicting human food effect, AAPS J., 2008, 10(2), 282–288 CrossRef CAS PubMed.
- C. P. Gayer and M. D. Basson, The effects of mechanical forces on intestinal physiology and pathology, Cell. Signalling, 2009, 21(8), 1237–1244 CrossRef CAS PubMed.
- S. Bellmann, D. Carlander, A. Fasano, D. Momcilovic, J. A. Scimeca, W. J. Waldman, L. Gombau, L. Tsytsikova, R. Canady, D. I. Pereira and D. E. Lefebvre, Mammalian gastrointestinal tract parameters modulating the integrity, surface properties, and absorption of food-relevant nanomaterials, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2015, 7(5), 609–622 CAS.
- D. F. Evans, G. Pye, R. Bramley, A. G. Clark, T. J. Dyson and J. D. Hardcastle, Measurement of gastrointestinal pH profiles in normal ambulant human subjects, Gut, 1988, 29(8), 1035–1041 CrossRef CAS PubMed.
- M. Koziolek, M. Grimm, D. Becker, V. Iordanov, H. Zou, J. Shimizu, C. Wanke, G. Garbacz and W. Weitschies, Investigation of pH and Temperature Profiles in the GI Tract of Fasted Human Subjects Using the Intellicap® System, J. Pharm. Sci., 2015, 104(9), 2855–2863 CrossRef CAS PubMed.
- S. Linden, J. Mahdavi, J. Hedenbro, T. Boren and I. Carlstedt, Effects of pH on Helicobacter pylori binding to human gastric mucins: identification of binding to non-MUC5AC mucins, Biochem. J., 2004, 384(Pt 2), 263–270 CrossRef CAS PubMed.
- I. G. Godinez and C. J. Darnault, Aggregation and transport of nano-TiO2 in saturated porous media: effects of pH, surfactants and flow velocity, Water Res., 2011, 45(2), 839–851 CrossRef CAS PubMed.
- M. B. Romanello and M. M. Fidalgo de Cortalezzi, An experimental study on the aggregation of TiO2 nanoparticles under environmentally relevant conditions, Water Res., 2013, 47(12), 3887–3898 CrossRef CAS PubMed.
- K. Brake, A. Gumireddy, A. Tiwari, H. Chauhan and D. Kumari, In vivo Studies for Drug Development via Oral Delivery: Challenges, Animal Models and Techniques, Pharm. Anal. Acta, 2017, 8(9), 560 Search PubMed.
- I. Khadra, Z. Zhou, C. Dunn, C. G. Wilson and G. Halbert, Statistical investigation of simulated intestinal fluid composition on the equilibrium solubility of biopharmaceutics classification system class II drugs, Eur. J. Pharm. Sci., 2015, 67, 65–75 CrossRef CAS PubMed.
- E. L. McConnell, H. M. Fadda and A. W. Basit, Gut instincts: explorations in intestinal physiology and drug delivery, Int. J. Pharm., 2008, 364(2), 213–226 CrossRef CAS PubMed.
- B. Wuyts, D. Riethorst, J. Brouwers, J. Tack, P. Annaert and P. Augustijns, Evaluation of fasted and fed state simulated and human intestinal fluids as solvent system in the Using chambers model to explore food effects on intestinal permeability, Int. J. Pharm., 2015, 478(2), 736–744 CrossRef CAS PubMed.
- D. Riethorst, J. Brouwers, J. Motmans and P. Augustijns, Human intestinal fluid factors affecting intestinal drug permeation in vitro, Eur. J. Pharm. Sci., 2018, 121, 338–346 CrossRef CAS PubMed.
- E. Jantratid, N. Janssen, C. Reppas and J. B. Dressman, Dissolution media simulating conditions in the proximal human gastrointestinal tract: an update, Pharm. Res., 2008, 25(7), 1663–1676 CrossRef CAS PubMed.
- M. Vertzoni, N. Fotaki, E. Kostewicz, E. Stippler, C. Leuner, E. Nicolaides, J. Dressman and C. Reppas, Dissolution media simulating the intralumenal composition of the small intestine: physiological issues and practical aspects, J. Pharm. Pharmacol., 2004, 56(4), 453–462 CrossRef CAS PubMed.
- D. P. Chopra, A. A. Dombkowski, P. M. Stemmer and G. C. Parker, Intestinal epithelial cells in vitro, Stem Cells Dev., 2010, 19(1), 131–142 CrossRef PubMed.
- J. Christensen, S. El-Gebali, M. Natoli, T. Sengstag, M. Delorenzi, S. Bentz, H. Bouzourene, M. Rumbo, A. Felsani, S. Siissalo, J. Hirvonen, M. R. Vila, P. Saletti, M. Aguet and P. Anderle, Defining new criteria for selection of cell-based intestinal models using publicly available databases, BMC Genomics, 2012, 13, 274 CrossRef.
- H. Y. Tan, S. Trier, U. L. Rahbek, M. Dufva, J. P. Kutter and T. L. Andresen, A multi-chamber microfluidic intestinal barrier model using Caco-2 cells for drug transport studies, PLoS One, 2018, 13(5), e0197101 CrossRef.
- J. M. Yong, J. Mantaj, Y. Cheng and D. Vllasaliu, Delivery of Nanoparticles across the Intestinal Epithelium via the Transferrin Transport Pathway, Pharmaceutics, 2019, 11(7), 298 CrossRef.
- L. Hashem, M. Swedrowska and D. Vllasaliu, Intestinal uptake and transport of albumin nanoparticles: potential for oral delivery, Nanomedicine, 2018, 13(11), 1255–1265 CrossRef CAS PubMed.
- P. Guo, A. M. Weinstein and S. Weinbaum, A hydrodynamic mechanosensory hypothesis for brush border microvilli, Am. J. Physiol. Renal. Physiol., 2000, 279(4), F698–F712 CrossRef CAS.
- D. E. Lefebvre, K. Venema, L. Gombau, L. G. Valerio Jr, J. Raju, G. S. Bondy, H. Bouwmeester, R. P. Singh, A. J. Clippinger, E. M. Collnot, R. Mehta and V. Stone, Utility of models of the gastrointestinal tract for assessment of the digestion and absorption of engineered nanomaterials released from food matrices, Nanotoxicology, 2015, 9(4), 523–542 CrossRef CAS PubMed.
- N. Patel, B. Forbes, S. Eskola and J. Murray, Use of simulated intestinal fluids with Caco-2 cells and rat ileum, Drug Dev. Ind. Pharm., 2006, 32(2), 151–161 CrossRef CAS PubMed.
- E. Frohlich, Comparison of conventional and advanced in vitro models in the toxicity testing of nanoparticles, Artif. Cells, Nanomed., Biotechnol., 2018, 46(sup2), 1091–1107 CrossRef PubMed.
- F. Kong and R. P. Singh, A human gastric simulator (HGS) to study food digestion in human stomach, J. Food Sci., 2010, 75(9), E627–E635 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |
