Open Access Article
Open Access ArticleGreen synthesis of palladium nanoparticles using lentinan for catalytic activity and biological applications†
Zengsheng Hana,
Le Donga,
Jin Zhangb,
Tianming Cuia,
Shengfu Chen c,
Guanglong Mac,
Xiaolei Guoa and
Longgang Wang
c,
Guanglong Mac,
Xiaolei Guoa and
Longgang Wang *a
*a
aKey Laboratory of Applied Chemistry, College of Environmental and Chemical Engineering, Yanshan University, Qinhuangdao, 066004, China. E-mail: lgwang@ysu.edu.cn
bCollege of Chemistry and Environmental Engineering, Shanxi Datong University, Datong, 037009, China
cKey Laboratory of Biomass Chemical Engineering of Ministry of Education, College of Chemical and Biological Engineering, Zhejiang University, Hangzhou, 310027, China
First published on 22nd November 2019
Abstract
The green synthesis of palladium nanoparticles (Pd NPs) for catalysis and biological applications has been gaining great interest. To replace complex plant extracts, lentinan (LNT) may be a good reducing and stabilizing agent. In this work, a simple and green method using LNT to reduce and stabilize palladium Pd NPs was verified. The resulting LNT stabilized palladium nanoparticles (Pdn-LNT NPs) were characterized by UV-Vis spectroscopy, DLS, TEM, and XPS. The results indicated that Pd NPs inside of Pdn-LNT NPs had a small size (2.35–3.32 nm). Pdn-LNT NPs were stable in solution for 7 days. In addition, Pdn-LNT NPs had higher catalytic activity towards the reduction of 4-nitrophenol than other catalysts. More importantly, Pdn-LNT NPs had negligible cytotoxicity towards cells and showed good antioxidant activity. Taken together, the prepared Pdn-LNT NPs have great potential bio-related applications.
1 Introduction
Metal nanoparticles play an increasingly important role in many areas such as catalysis, sensors, and controlled drug delivery.1–3 The various applications of metal nanoparticles are often related to their size and stability in solutions. Metal nanoparticles have been extensively prepared by using many kinds of materials. To tune their size, many agents such as sodium borohydride (NaBH4), and dimethylamine borane (DMAB) are used to reduce metal ions to produce zerovalent metal nanoparticles. In addition, to increase their stability, many polymers such as polyvinyl pyrrolidone (PVP), and polyethyleneimine (PEI) are employed to stabilize metal nanoparticles. However, most of these chemicals are toxic and harmful to biological applications of prepared metal nanoparticles.In order to minimize the usage of hazardous chemicals, the green methods for the preparation of biocompatible metal nanoparticles are drawing great attention. The green methods of using various plant extracts have many advantages such as eco-friendly, low cost and low energy consumption. These plant extracts include turmeric extracts,4 pomegranate extract,5 geranium leaf aqueous extract,6 Sapium sebiferum leaf extract,7 and Thymus kotschyanus extract.8 However, these plant extracts contain many different components. These components of each plant extract are different because of different growth stages and conditions for plants, which leads to different reducing power and stabilizing ability for plant extracts. Thus, it may be difficult to precisely control the size and morphology of metal nanoparticles. Specific component of plant extracts has fixed composition. Polyphenols in the plant extracts have highest levels of antioxidant properties.9 Polyphenols should be the main component to produce metal nanoparticle.9,10 Fu and coworkers found that the size distribution of proanthocyanidin-functionalized gold nanoparticles was smaller than that of gold nanoparticles prepared using water extract.11 Compared with polyphenols, polysaccharides have mild antioxidant properties and high stability in water. Lentinan (LNT) is soluble and biologically active biomacromolecule. LNT has potential immune regulation, anti-viral and anti-cancer bioactivities.12 LNT is composed of one β-(1→6)-D-glucopyranoside branches for every three β-(1→3)-glucopyranoside linear linkages.12 LNT was used to prepare silver nanoparticles and investigated their antibacterial activity.13
Palladium nanoparticles (Pd NPs) have been widely used in various catalytic processes. Herein, it is the first time that a green method to prepare LNT stabilized palladium nanoparticles (Pdn-LNT NPs) was demonstrated. Pdn-LNT NPs had small size for Pd NPs and high stability in different solutions. The catalytic activity of Pdn-LNT NPs was evaluated by the catalytic reduction of 4-nitrophenol (4-NP). Moreover, the biocompatibility of Pdn-LNT NPs was measured by MTT method. The antioxidant property of Pdn-LNT NPs was evaluated by the DPPH free radical scavenging activity assay.
2 Results and discussion
2.1 UV-Vis spectra analysis
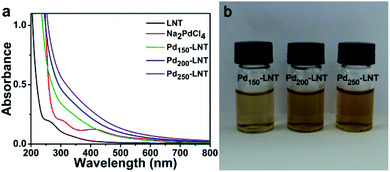
LNT had good antioxidant activity reported by Tu and coworkers,14 leading to their potential ability to reduce the Na2PdCl4. To evaluate the formation of Pd NPs, the UV-Vis spectra and pictures of forming solution were studied. Fig. 1a showed that Na2PdCl4 had an obvious peak at 415 nm. After the incubation of Na2PdCl4 with LNT at 50 °C for 6 h, the peak of Na2PdCl4 at 415 nm disappeared, and a very broad peak was observed. In addition, as the molar ratio of Na2PdCl4 to LNT increased, the absorbance was bigger and the color of the solution (Fig. 1b) gradually deepened. This was due to the increased content of Pd elements in solution when the concentration of LNT was 1.82 μM. The results were consistent with zwitterionic dendrimer-encapsulated Pd NPs15 and Pd NPs prepared using Dioscorea bulbifera tuber extract16 in previous reports. Both results demonstrated the successful formation of Pd NPs.2.2 TEM observation
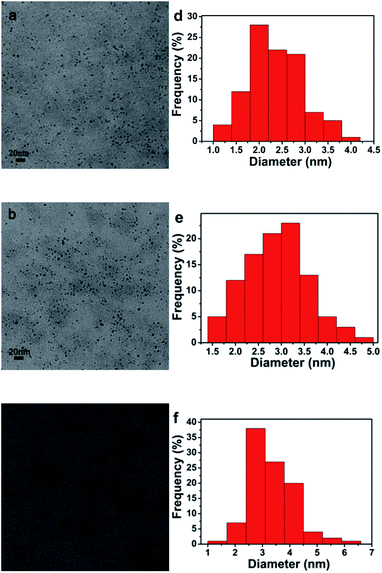
The size and dispersion of Pd NPs inside of Pdn-LNT NPs were detected by TEM. As shown in Fig. 2, TEM image indicated that Pd NPs had a nearly spherical shape with highly dispersed states. The sizes of Pd NPs inside of Pd150-LNT NPs, Pd200-LNT NPs, Pd250-LNT NPs were 2.35 ± 0.57, 2.90 ± 0.69, 3.32 ± 0.83 nm, respectively. Thus, Pd NPs inside of Pdn-LNT NPs had small size with narrow size distribution. The sizes of Pd NPs inside of Pd150-LNT NPs was smaller than Pd NPs (12.85 nm) prepared using fritillaria imperialis flower extract by Veisi and coworkers,17 indicating that Pdn-LNT had larger specific surface area, which often led to higher catalytic activity.2.3 FTIR spectra analysis
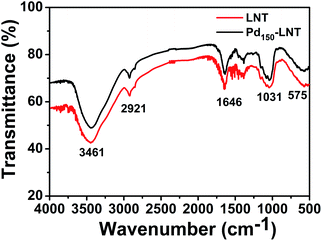
LNT and Pd150-LNT NPs were further characterized by FTIR spectra.18 Fig. 3 showed there were five obvious peaks (575, 1031, 1646, 2921 and 3461 cm−1) in the FTIR spectra of LNT and Pd150-LNT NPs. The peak at 3461 cm−1 was due to the stretching vibration absorption peak of O–H. The peak at 2921 cm−1 was attributed to the stretching vibration absorption peak of C–H bond. The characteristic of carbonyl stretching was at 1646 cm−1. 1031 cm−1 was assigned to the stretching vibration absorption peak of C–O–C on the sugar ring. The absorption peak at 575 cm−1 was attributed to β-pyranose in the sugar.19 There was slight difference between LNT and Pd150-LNT NPs, indicating that the structure of LNT was stable.2.4 XPS analysis
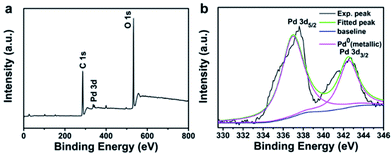
Elemental composition of Pd150-LNT NPs measured by XPS was used to further study the formation of Pdn-LNT NPs. Fig. 4a showed that the peaks at 284.6 and 531.36 eV were assigned to C 1 s and O 1 s, respectively. Both elements were derived from LNT. Fig. 4b showed the Pd binding energy of Pdn-LNT NPs had two obvious peaks at 342.5 and 337.2 eV, which corresponded to Pd 3d3/2 and Pd 3d5/2, respectively. These values were consistent with Pd(0) binding energy, indicating that Pd(II) has been successfully reduced to Pd(0) by LNT.20 Kardanpour and coworkers prepared highly dispersed Pd NPs which had similar binding energy compared with Pd150-LNT NPs.21 Therefore, XPS results confirmed the successful preparation of Pdn-LNT NPs.2.5 Stability
Besides the size of Pd NPs, the states of Pdn-LNT NPs in solution were related to their catalytic performance. The hydrodynamic size and zeta potential of Pdn-LNT NPs were determined by DLS. Fig. 5a showed that the hydrodynamic size of LNT was about 20 nm, and the hydrodynamic size of Pdn-LNT NPs was 25, 27, 29 nm (n = 150, 200, 250), respectively. The hydrodynamic size of Pdn-LNT NPs was about 10 times larger than that measured by TEM. The difference was due to the different methods of sample preparation. The sample tested by TEM was in a dry state, while the sample measured by DLS was in the hydrated state. Thus, Pd NPs are surrounded by LNT in the hydrated state. Fig. 5b showed the zeta potential of LNT was −4 mV, and the zeta potential of Pdn-LNT NPs was −6, −7, −8 mV (n = 150, 200, 250), respectively. Fig. 5c showed that the hydrodynamic size of Pd150-LNT NPs was between 25–30 nm from pH 4 to 10 for 7 days, indicating they maintained good stability in a wide pH range for 7 days. Fig. 5d showed the zeta potential of Pd150-LNT NPs in different pH buffers was between −4 and −8 mV. The zeta potential is an important parameter which can be used to assess the stability of nanoparticles.22 The zeta potential of nanoparticles is above +30 mV which results in good stability of nanoparticles. Moreover, the metal nanoparticles can be stabilized by macromolecules such as dendrimer to inhibit agglomeration of nanoparticles due to steric effects.23,24 In our case, LNT was used both as a reducing and stabilizing agent to prepare Pdn-LNT NPs. Therefore, electrostatic repulsion and steric hindrance between Pd150-LNT NPs should be main reasons for their high stability. The high stability of Pdn-LNT NPs was good for efficient degradation of organic pollutants in complex environments.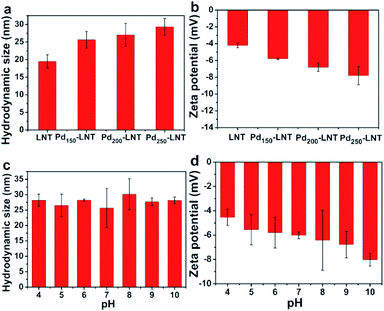 | ||
| Fig. 5 (a) Hydrodynamic size and (b) zeta potential of Pdn-LNT NPs. (c) The stability and (d) zeta potential of Pd150-LNT NPs with pH from 4 to 10 within 7 days. | ||
2.6 Catalytic performance
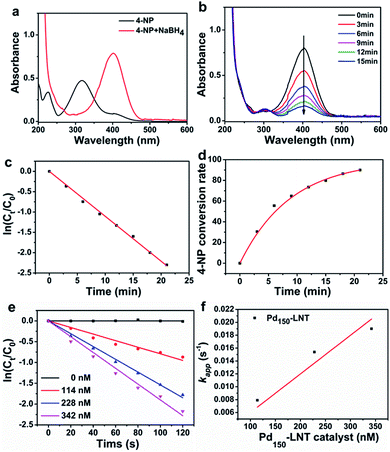
Metal nanoparticles have been wildly used in treatment of various organic pollutants.25–27 As discussed above, the prepared Pdn-LNT NPs had good stability and large specific surface area. These advantages are beneficial to high catalytic efficiency. Pd150-LNT NPs were used to study the catalytic performance on 4-NP. 4-NP is common organic pollutants and is usually found in insecticides and synthetic dyes.28 4-Aminophenol (4-AP) is widely used as a developer, lubricant and dye.29 Therefore, the conversion of 4-NP to 4-AP has good value. Fig. 6a showed 4-NP solution had an absorption peak at 317 nm, and this peak shifted to 400 nm after addition of NaBH4, which corresponded to newly generated 4-hydroxyaminophenol (4-Hx).30 After the addition of the Pd150-LNT NPs, Fig. 6b showed the peak at 400 nm quickly decreased and the peak of 4-AP at 300 nm increased, which indicated the formation of 4-AP. Fig. 6c showed that the relationship between ln(Ct/C0) of Pd150-LNT NPs and reaction time was linear. Thus, when NaBH4 was excessive, this catalytic reaction followed pseudo first-order kinetic equation (eqn (1)).31 After 21 min, the conversion of 4-NP was 90% (Fig. 6d), and the color of the reaction system changed from yellow to colorless. These indicated that Pd150-LNT NPs had good catalytic properties towards the reduction of 4-NP. Fig. 6f showed that the kapp of the catalytic reaction gradually increased as the increased concentration of Pd NPs which offered more active sites. The catalytic reduction of 4-NP by Pd150-LNT NPs should follow the Langmuir–Hinshelwood kinetic model.32–34 4-Hx and BH4− were first adsorbed onto the surface of the Pd NPs. The 4-Hx was converted to 4-AP and the product 4-AP left the catalyst surface. Therefore, the active sites provided on the surface of the catalyst were the key factors affecting the catalytic reaction.
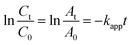 | (1) |
In order to compare the catalytic performance of Pd150-LNT NPs with other catalysts, normalized rate constant (knor = kapp/CPd)35 and turn over frequency (TOF)36 were calculated. The knor and TOF of Pd150-LNT NPs were 69.39 s−1 mM−1 and 678.18 h−1, respectively. Table 1 showed the comparison of Pd150-LNT NPs with other catalysts. Pd150-LNT NPs had the highest knor and TOF, indicating they had the highest catalytic activity. Morère and coworkers deposited Pd on mesoporous silica which had the knor and TOF for 0.188 s−1 mM−1 and 19.08 h−1, respectively. Zhao and coworkers prepared Pd NPs loaded on carbon nanospheres which had the knor and TOF for 1.4 s−1 mM−1 and 504 h−1, respectively. Their catalytic parameters were lower than those of Pd150-LNT NPs. The high catalytic activity of Pd150-LNT NPs should be due to their small particle size of 2.35 ± 0.57 nm and the corresponding big specific surface area. More importantly, Pd150-LNT NPs remained stable in aqueous solutions in a pH range of 4–10. This high stability and high specific surface area provided more exposed Pd atoms which acted as active sites during catalytic reduction of 4-NP.
2.7 DPPH free radical scavenging assay
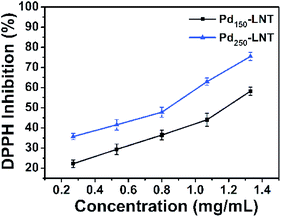
Besides the catalytic activity of Pd150-LNT NPs, the antioxidant activity of Pd150-LNT NPs and Pd250-LNT NPs were also measured. DPPH was a stable free radical compound and was employed to evaluate the antioxidant activity. Antioxidant activity plays an important role in the protection of our body from harm of free radical.43,44 Fig. 7 showed that when the concentration of Pd150-LNT NPs increased from 0.27 to 1.33 mg mL−1, the antioxidant activity increased from 20% to 60%, indicating effective free radical inhibition by Pd150-LNT NPs. Thus, Pd150-LNT NPs and Pd250-LNT NPs had obvious antioxidant activity. Dimocarpus longan seed extract was also used as a reducing and stabilizing agent to prepare silver nanoparticles which had good antioxidant activity.452.8 Biocompatibility
The presence of microorganisms in sewage has a significant reverse effect on the catalytic degradation of organic pollutants, which should be due to strong interaction between catalysts and microorganisms. This interaction always results in high cytotoxicity of nanoparticles.46 Here, the cytotoxicity of Pdn-LNT NPs (n = 150, 200 and 250) was determined by MTT assay on A549 cells and HCT116 cells. MTT is commonly used method to evaluate the cytotoxicity of nanoparticles.47–49 As illustrated in Fig. 8, the cell viability was higher than 90% after incubation with Pdn-LNT NPs and LNT at a concentration within 1000 μg mL−1. Both Pdn-LNT NPs and LNT had good biocompatibility. In contrast, polyethyleneimine (PEI) had high cytotoxicity because of highly positive charges on their surface. Taken together, Pdn-LNT NPs had good biocompatibility in vitro. This should be due to biocompatible LNT and the negative surface charge of Pdn-LNT NPs. Both factors were good for reduced the interaction between Pdn-LNT NPs and cells. The high biocompatibility of Pdn-LNT NPs was good for bio-related catalytic applications in nature.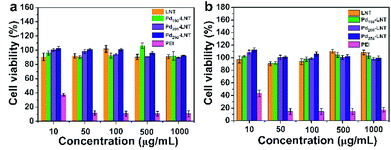 | ||
| Fig. 8 Cell viabilities of (a) A549 cells and (b) HCT116 cells for Pdn-LNT NPs (n = 150, 200, 250) and LNT. | ||
3 Conclusion
In summary, we have prepared Pdn-LNT NPs, where LNT was used both as a reducing and stabilizing agent. This method was simple, eco-friendly and easy to scaled up. Compared with plants extracts, LNT had fixed components and weaker reducing ability. Pdn-LNT NPs had nearly spherical shape for Pd NPs and showed high stability in a wide pH range for 7 days. Pdn-LNT NPs had the hydrodynamic size about 25–29 nm and slightly negative zeta potential. In addition, Pdn-LNT NPs had high catalytic activity for 4-NP to generate 4-AP due to the high specific surface area and stability. Furthermore, Pdn-LNT NPs showed no cytotoxicity towards cells and good antioxidant activity. Pdn-LNT NPs have great potential application in bio-related catalysis and nanomedicine.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors appreciate financial support from National Nature Science Foundation of China (21474085, 21674092 and 21975216), and National Development Project on Key Basic Research (973 Project, 2015CB655303), Natural Science Foundation of Hebei Province (B2017203229), China Postdoctoral Science Foundation (2016M601284).Notes and references
- N. Tsumori, L. Chen, Q. Wang, Q.-L. Zhu, M. Kitta and Q. Xu, Chem, 2018, 4, 845–856 CAS.
- C. Wang, Y. Shi, Y.-Y. Dan, X.-G. Nie, J. Li and X.-H. Xia, Chem.–Eur. J., 2017, 23, 6717–6723 CrossRef CAS PubMed.
- J. Prasad Rao, P. Gruenberg and K. E. Geckeler, Prog. Polym. Sci., 2015, 40, 138–147 CrossRef CAS.
- F. K. Alsammarraie, W. Wang, P. Zhou, A. Mustapha and M. Lin, Colloids Surf., B, 2018, 171, 398–405 CrossRef CAS PubMed.
- B. Şahin, A. Aygün, H. Gündüz, K. Şahin, E. Demir, S. Akocak and F. Şen, Colloids Surf., B, 2018, 163, 119–124 CrossRef PubMed.
- R. D. Rivera-Rangel, M. P. González-Muñoz, M. Avila-Rodriguez, T. A. Razo-Lazcano and C. Solans, Colloids Surf., A, 2018, 536, 60–67 CrossRef CAS.
- K. Tahir, S. Nazir, B. Li, A. Ahmad, T. Nasir, A. U. Khan, S. A. A. Shah, Z. U. H. Khan, G. Yasin and M. U. Hameed, J. Photochem. Photobiol., B, 2016, 164, 164–173 CrossRef CAS PubMed.
- M. Hamelian, M. M. Zangeneh, A. Amisama, K. Varmira and H. Veisi, Appl. Organomet. Chem., 2018, 32, 4458 CrossRef.
- F. U. Khan, Y. Chen, N. U. Khan, Z. U. H. Khan, A. U. Khan, A. Ahmad, K. Tahir, L. Wang, M. R. Khan and P. Wan, J. Photochem. Photobiol., B, 2016, 164, 344–351 CrossRef PubMed.
- S. Lebaschi, M. Hekmati and H. Veisi, J. Colloid Interface Sci., 2017, 485, 223–231 CrossRef CAS PubMed.
- L. Biao, S. Tan, Q. Meng, J. Gao, X. Zhang, Z. Liu and Y. Fu, Nanomaterials, 2018, 8, 53 CrossRef PubMed.
- Y. Zhang, S. Li, X. Wang, L. Zhang and P. C. K. Cheung, Food Hydrocolloids, 2011, 25, 196–206 CrossRef CAS.
- D. Guo, G. Liu, X. Li, X. Tang, J. Zhang, X. Zhu and S. Jiang, J. Bionanosci., 2015, 9, 325–329 CrossRef CAS.
- X. Huang, Z. Tu, Y. Jiang, H. Xiao, Q. Zhang and H. Wang, Int. J. Biol. Macromol., 2012, 51, 926–932 CrossRef CAS.
- L. Wang, J. Zhang, X. Guo, S. Chen, Y. Cui, Q. Yu, L. Yang, H. Sun, D. Gao and D. Xie, New J. Chem., 2018, 42, 19740–19748 RSC.
- S. Ghosh, R. Nitnavare, A. Dewle, G. B. Tomar, R. Chippalkatti, P. More, R. Kitture, S. Kale, J. Bellare and B. A. Chopade, Int. J. Nanomed., 2015, 10, 7477–7490 CAS.
- M. Yazdankhah, H. Veisi and S. Hemmati, J. Taiwan Inst. Chem. Eng., 2018, 91, 38–46 CrossRef CAS.
- K. Ma, R. Wang, T. Jiao, J. Zhou, L. Zhang, J. Li, Z. Bai and Q. Peng, Colloids Surf., A, 2020, 584, 124023 CrossRef CAS.
- Z. Ruan, J. Su, H. Dai and M. Wu, Int. Immunopharmacol., 2005, 5, 811–820 CrossRef CAS.
- Y. Zhao, J. Feng, L. Hong, Y. Li, C. Wang and S. Ye, Inorg. Chem. Front., 2018, 5, 1133–1138 RSC.
- R. Kardanpour, S. Tangestaninejad, V. Mirkhani, M. Moghadam, I. Mohammadpoor-Baltork, A. R. Khosropour and F. Zadehahmadi, J. Organomet. Chem., 2014, 761, 127–133 CrossRef CAS.
- D. Silvestri, S. Wacławek, B. Sobel, R. Torres-Mendieta, V. Novotný, N. H. A. Nguyen, A. Ševců, V. V. T. Padil, J. Müllerová, M. Stuchlík, M. P. Papini, M. Černík and R. S. Varma, Green Chem., 2018, 20, 4975–4982 RSC.
- R. M. Crooks, M. Zhao, L. Sun, V. Chechik and L. K. Yeung, Acc. Chem. Res., 2001, 34, 181–190 CrossRef CAS PubMed.
- H. Sun, F. M. Haque, Y. Zhang, A. Commisso, M. A. Mohamed, M. Tsianou, H. Cui, S. M. Grayson and C. Cheng, Angew. Chem., Int. Ed., 2019, 58, 10572–10576 CrossRef CAS PubMed.
- R. El-Shabasy, N. Yosri, H. El-Seedi, K. Shoueir and M. El-Kemary, Optik, 2019, 192, 162943 CrossRef CAS.
- D. Yang, R. Zhang, T. Zhao, T. Sun, X. Chu, S. Liu, E. Tang and X. Xu, Catal. Sci. Technol., 2019, 9, 6145–6151 RSC.
- Y. Cui, B. Liang, J. Zhang, R. Wang, H. Sun, L. Wang and D. Gao, Transition Met. Chem., 2019, 44, 655–662 CrossRef CAS.
- W. Shen, Y. Qu, X. Pei, S. Li, S. You, J. Wang, Z. Zhang and J. Zhou, J. Hazard. Mater., 2017, 321, 299–306 CrossRef CAS PubMed.
- M. Guo, J. He, Y. Li, S. Ma and X. Sun, J. Hazard. Mater., 2016, 310, 89–97 CrossRef CAS PubMed.
- S. Gu, J. Kaiser, G. Marzun, A. Ott, Y. Lu, M. Ballauff, A. Zaccone, S. Barcikowski and P. Wagener, Catal. Lett., 2015, 145, 1105–1112 CrossRef CAS.
- X. Guo, J. Zhang, Y. Cui, S. Chen, H. Sun, Q. Yang, G. Ma, L. Wang and J. Kang, New J. Chem., 2019, 43, 7646–7652 RSC.
- H. Cheng, X. Qian, Y. Kuwahara, K. Mori and H. Yamashita, Adv. Mater., 2015, 27, 4616–4621 CrossRef CAS PubMed.
- R. Begum, Z. H. Farooqi, E. Ahmed, K. Naseem, S. Ashraf, A. Sharif and R. Rehan, Appl. Organomet. Chem., 2017, 31, 3563 CrossRef.
- L. Wang, X. Zhang, Y. Cui, X. Guo, S. Chen, H. Sun, D. Gao, Q. Yang and J. Kang, Transition Met. Chem., 2019 DOI:10.1007/s11243-019-00353-z.
- X. Le, Z. Dong, Y. Liu, Z. Jin, T.-D. Huy, M. Le and J. Ma, J. Mater. Chem. A, 2014, 2, 19696–19706 RSC.
- L. Wang, Q. Yang, Y. Cui, D. Gao, J. Kang, H. Sun, L. Zhu and S. Chen, New J. Chem., 2017, 41, 8399–8406 RSC.
- J. Morère, M. J. Tenorio, M. J. Torralvo, C. Pando, J. A. R. Renuncio and A. Cabañas, J. Supercrit. Fluids, 2011, 56, 213–222 CrossRef.
- M. Yu, L. Yan, P. Frank and B. Matthias, Chem. Mater., 2007, 19, 1062–1069 CrossRef.
- Z. Zhao, X. Ma, X. Wang, Y. Ma, C. Liu, H. Hang, Y. Zhang, Y. Du and W. Ye, Appl. Surf. Sci., 2018, 457, 1009–1017 CrossRef CAS.
- S. Arora, P. Kapoor and M. L. Singla, React. Kinet., Mech. Catal., 2010, 99, 157–165 CAS.
- H. Li, L. Han, J. Cooper-White and I. Kim, Green Chem., 2012, 14, 586–591 RSC.
- H. He and C. Gao, Molecules, 2010, 15, 4679–4694 CrossRef CAS PubMed.
- M. Hamelian, K. Varmira and H. Veisi, J. Photochem. Photobiol., B, 2018, 184, 71–79 CrossRef CAS PubMed.
- D. Dong, Q. Li, W. Hou and H. Zhang, J. Mol. Struct., 2020, 1199, 127002 CrossRef CAS.
- F. U. Khan, Y. Chen, N. U. Khan, Z. U. H. Khan, A. U. Khan, A. Ahmad, K. Tahir, L. Wang, M. R. Khan and P. Wan, J. Photochem. Photobiol., B, 2016, 164, 344–351 CrossRef PubMed.
- Y. Cui, J. Zhang, Q. Yu, X. Guo, S. Chen, H. Sun, S. Liu, L. Wang, X. Lai and D. Gao, New J. Chem., 2019, 43, 9076–9083 RSC.
- X. W. Zhang, L. Y. Luo, L. Li, Y. C. He, W. W. Cao, H. Liu, K. Niu and D. W. Gao, Nanomedicine, 2019, 15, 142–152 CrossRef CAS PubMed.
- L. Luo, Y. Bian, Y. Liu, X. Zhang, M. Wang, S. Xing, L. Li and D. Gao, Small, 2016, 12, 4103–4112 CrossRef CAS PubMed.
- Y. He, C. Cong, Y. He, Z. Hao, C. Li, S. Wang, Q. Zhao, H. He, R. Zhu, X. Li and D. Gao, Chem. Eng. J., 2019, 375, 122079 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra08051a |
| This journal is © The Royal Society of Chemistry 2019 |