Open Access Article
Open Access ArticleThe cotreatment of landfill leachate and high-nitrate wastewater using SBRs: evaluation of denitrification performance and microbial analysis†
Huaguang Liu a,
Xingyu Zhou
a,
Xingyu Zhou *bc,
Chaoshen Zhanga and
Jinsong Zhangabc
*bc,
Chaoshen Zhanga and
Jinsong Zhangabc
aSchool of Civil Engineering, Guangzhou University, 510006, Guangzhou, China
bShenzhen Municipal Wastewater Treatment and Recycling Engineering Laboratory, Shenzhen Water (Group) Co. Ltd., 518031, Guangdong, China. E-mail: zhouxingyu2017@163.com
cSchool of Environment, Harbin Institute of Technology, 150090, Harbin, China
First published on 2nd December 2019
Abstract
Resourceful disposal of landfill leachate has always been an intractable worldwide problem. This study was conducted to investigate the feasibility of biologically treating a combined waste stream of landfill leachate and high-concentration nitrate nitrogen (high-nitrate) wastewater. Raw landfill leachate was pretreated using anaerobic fermentation and ammonia stripping to improve biodegradability. The control sequencing batch reactor (SBR, named R0) was fed only with synthetic high-nitrate wastewater with sodium acetate as the carbon source, whereas the other experimental SBR (named R1) was loaded with mixtures containing leachates. Excessive increase in leachate adversely affected the cotreatment, and it was concluded that the landfill leachate volume ratio should never exceed 7.5% of the total wastewater (14% of the initial COD) based on further batch experiments. The maximum specific denitrification rate of 58.05 mg NO3−-N (gVSS h)−1 was attained in R1, while that of 32.32 mg NO3−-N (gVSS h)−1 was obtained in R0. Illumina MiSeq sequencing revealed that adding landfill leachate did not change the fact that Pseudomonas, Thauera, and Pannonibacter dominant in the sodium acetate supported the denitrification systems, but led to the adjustment of their relative abundance. Moreover, the narG, nirK, nirS, and norB denitrifying genes exhibited increased abundance by 138–980% in the cotreated system, which was confirmed by q-PCR analyses. These findings reveal that the denitrification efficiency of activated sludge in SBR cotreated with landfill leachate and high-nitrate wastewater significantly improved, and this may contribute toward the understanding of the molecular mechanisms of biological denitrification under the blending treatment of leachate and high-nitrate wastewater.
1. Introduction
In China, the increasing amounts of municipal solid waste (MSW) are putting tremendous pressure on the treatment of landfill leachates. According to the survey data from the Professional Committee of Urban Domestic Refuse Treatment of the China Association of Environmental Protection Industry, the amount of MSW disposed by the Chinese government exceeded 191.1 million tons in 2015 (ref. 1) and is expected to reach 266.8 million tons by 2025.2 In China, more than 60% MSW is disposed of in sanitary landfills, which produce large amounts of landfill leachates (Fig. S1†). However, due to the complex structure and high cost of landfill leachates, the treatment capacity of mainstream advanced technologies such as nanofiltration, reverse osmosis, advanced oxidation, and activated carbon adsorption are unable to keep pace with the generation rate of landfill leachates, which places substantial pressure on sanitary landfills for follow-up treatments. Therefore, it is imperative to find a reasonable strategy to treat landfill leachate.As a cost-effective potential carbon source, blending landfill leachate with domestic sewage for treatment has become a popular research topic. Organic carbon concentration in raw wastewater of most municipal wastewater treatment plants (WWTPs) is very low,3,4 but the denitrification process needs a large number of carbon sources for consumption as electron donors. Therefore, to prevent incomplete denitrification and nitrite nitrogen accumulation caused by insufficient carbon sources, supplementary carbon sources such as methanol, acetic acid, or glucose need to be added to the denitrification tank to ensure that the total nitrogen content in the effluent meets the emission standards.5,6 Moreover, rapidly biodegradable forms of organic matter, such as short-chain fatty acids (SCFAs) and volatile fatty acids (VFAs), contained in landfill leachates can provide a carbon source that can be easily assimilated by microorganisms during the denitrification process. Consequently, the combined treatment of landfill leachates and domestic sewage in WWTPs has been extensively studied around the world.7–10 Yu et al. designed an anaerobic–anoxic–oxic (A2/O) bioreactor system to conduct a field test on the combined treatment of domestic wastewater with landfill leachates in the Datangsha WWTP (Guangzhou, China); they found that the removal rates of chemical oxygen demand (COD), ammonia nitrogen (NH4+-N), and total nitrogen (TN) were 82.65%, 92.69%, and 57.10%, respectively, at the optimal volume ratio of 0.2%.11 Ferraz et al. studied the optimal mixing ratio for the cotreatment of landfill leachate and domestic sewage in a Brazilian WWTP using a pilot-scale submerged aerobic biofilter (SAB). The results showed that the removal rates of biological oxygen demand (BOD), COD, and dissolved organic carbon (DOC) were 98%, 80%, and 80%, respectively, when the leachate volume ratio was 2%.12 Landfill leachate cotreatment with sanitary wastewater was investigated at the bench scale in an upflow anaerobic sludge blanket (UASB), and the COD removal rate was greater than 70% for leachate volume ratios of up to 5%.13 A combined system of a long-term lab-scale reactor was used for a more comprehensive cotreatment of landfill leachate and municipal sewage at volumetric ratios of 1%, 3%, and 5%, and the excess sludge could be reduced by 20–48%, resulting in no inhibitory effect on the denitrification process.14 Three Irish WWTPs were selected for field testing by Brennan et al., who deemed that the coprocessing of landfill leachate with municipal sewage may be the most sustainable leachate treatment solution that is currently available.15 However, research concerning cotreatment with landfill leachate has been limited to municipal domestic sewage, and reports regarding the cotreatment of landfill leachate and high-nitrate wastewater are rare.
Wastewater containing high nitrate concentrations is widely derived from various types of industrial applications, such as fertilizer production,16 stainless steel manufacturing and fish canning,17 cellophane finishing,18 wet lime–gypsum desulfurization production,19 nuclear applications,20 and ion exchange processes.21 When compared with domestic sewage treatment, the denitrification process consumes more carbon sources when a biological process that can completely remove nitrogen is used to treat such high-nitrate wastewater. Fernández-Nava et al.22 studied denitrification in a sequencing batch reactor (SBR) fed with nitrate (2500 mg L−1) using wastewater from a candy factory, residue from a soft drink factory, and residue from a dairy plant as the organic carbon sources. Nevertheless, it is still unclear whether landfill leachate can be used as an alternative carbon source for the treatment of high-nitrate wastewater or not.
The current study aimed to evaluate the behavior of landfill leachate when cotreated with high-nitrate wastewater in a lab-scale SBR. To better assess the organic matter and nitrogen removal characteristics, the denitrification rates of activated sludge in the treatment of high-nitrate nitrogen at different landfill leachate volume ratios were compared. The effects of landfill leachate on the denitrification efficiency of activated sludge systems were revealed from the microbial structure and denitrification functional genes by combining 16S rDNA sequencing and quantitative real-time PCR (q-PCR) analyses. A new resource utilization model of landfill leachate was comprehensively explored, which could provide a technical reference for the selection of economical and efficient additional carbon sources for the treatment of high-nitrate wastewater.
2. Materials and methods
2.1 Experimental materials
2.2 Experimental procedures
The experiment was divided into domestication phases, denitrification rate contrast phases, and optimal volume ratio research phases. Prior to introducing the landfill leachate into the SBR, the activated sludge underwent a 6-week acclimation period (85 operating days with 8 h anoxic reaction cycles). During the startup, the reactor was fed with synthetic high-nitrate wastewater at increasing concentrations step by step in the range of 50–500 mg L−1 with NaAc as the carbon source and COD/NO3−-N ratio (C/N ratio) of 5.0. After this domestication stage, R0 was used as the control system and operated in the original acclimation mode, while R1 was used as the experimental system and operated to cotreat the landfill leachate and high-nitrate wastewater (NaAc was partially replaced by the landfill leachate); the denitrification rates in the two bioreactors were then compared. To explore the optimum mixing ratio of the landfill leachate, batch experiments with C/N ratios of 2.5, 3.2, 4.5, 4.9, 5.6, and 6.1 were performed. The volume ratios of the landfill leachate in each batch of the 6 reactors were 0%, 2.5%, 5%, 7.5%, 10%, and 20%, and the amount of NaAc added (Table 1) was correspondingly decreased.| Volume ratio of landfill leachate (%) | Sodium acetate dosage (g L−1) | |||||
|---|---|---|---|---|---|---|
| C/N = 2.5 | C/N = 3.2 | C/N = 4.5 | C/N = 5.0 | C/N = 5.6 | C/N = 6.1 | |
| 0 | 1.79 | 2.29 | 3.21 | 3.50 | 4.00 | 4.36 |
| 2.5 | 1.65 | 2.15 | 3.07 | 3.36 | 3.86 | 4.22 |
| 5.0 | 1.51 | 2.01 | 2.93 | 3.22 | 3.72 | 4.08 |
| 7.5 | 1.37 | 1.87 | 2.79 | 3.08 | 3.58 | 3.94 |
| 10.0 | 1.23 | 1.73 | 2.65 | 2.94 | 3.44 | 3.80 |
| 20.0 | 1.09 | 1.59 | 2.51 | 2.80 | 3.30 | 3.66 |
2.3 Chemical analysis of wastewater samples and landfill leachate
Samples (10 mL) were collected from a fixed sampling port and diluted a certain number of times, after which the supernatant was filtered through a 0.22 μm filter membrane. SS, COD, BOD, NO2−-N, and NO3−-N in the landfill leachate and bioreactor samples were measured according to the Standard Methods for the Examination of Water and Wastewater.24 The concentrations of heavy metals in the landfill leachate (Cd, Cr, As, Pb, Hg, and Mo) were analyzed. Details of the above methods and instruments are listed in Table S3.† The concentration of dissolved oxygen (DO) and pH were measured using signal feedback DO and pH equipment (Hach, USA). The mixed liquid volatile suspended solids (VSS) content of the activated sludge samples was determined by the muffle furnace combustion gravimetric method, and the maximum specific denitrification rate was defined as the amount of nitrate removed per unit weight of VSS per unit time.2.4 DNA extraction and PCR
To qualitatively analyze the denitrification function, genes in the two bioreactors and DNA in the sludge samples were extracted for performing the polymerase chain reaction (PCR) analysis. Sludge samples were centrifuged at a speed of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 5 min before supernatant removal. All the DNA was extracted from the concentrated sludge using a FastDNA soil kit (MP Biomedicals, USA), and the DNA concentration and purity were measured by a Nanodrop nd-1000 instrument (Nanodrop Technologies, USA). The nitrate reductase gene narG, nitrite reductase genes nirK and nirS, nitric oxide reductase gene norB, and nitrous oxide reductase gene nosZ were amplified using the PCR primers (as listed in Table S4†). The PCR system contained 10 μL hot-start Premix Ex Taq™ version (Tiangen Biotech, Beijing), 1 μL upstream primer, 1 μL downstream primer, 0.5 μL DNA template, and 7.5 μL ddH2O. The PCR products were detected by agarose gel electrophoresis at a mass concentration of 1.5% (w/v) in 10× TAE buffer. The PCR was repeated three times for each sample, and sterile water was used as a negative control to check repeatability.
000 × g for 5 min before supernatant removal. All the DNA was extracted from the concentrated sludge using a FastDNA soil kit (MP Biomedicals, USA), and the DNA concentration and purity were measured by a Nanodrop nd-1000 instrument (Nanodrop Technologies, USA). The nitrate reductase gene narG, nitrite reductase genes nirK and nirS, nitric oxide reductase gene norB, and nitrous oxide reductase gene nosZ were amplified using the PCR primers (as listed in Table S4†). The PCR system contained 10 μL hot-start Premix Ex Taq™ version (Tiangen Biotech, Beijing), 1 μL upstream primer, 1 μL downstream primer, 0.5 μL DNA template, and 7.5 μL ddH2O. The PCR products were detected by agarose gel electrophoresis at a mass concentration of 1.5% (w/v) in 10× TAE buffer. The PCR was repeated three times for each sample, and sterile water was used as a negative control to check repeatability.
2.5 MetaVx™ library construction and Illumina MiSeq sequencing
In order to analyze the microbial communities in the two bioreactors, 15 mL of the mixture was taken from the bottom of the R0 and R1 reactors on the 0th (inoculated sludge) and 65th (acclimated activated sludge) days of system operation, respectively, and the sludge samples were cryopreserved and sent to GENEWIZ (Suzhou, China) for 16S rRNA gene sequencing. The construction of a high-throughput sequencing library and sequencing based on the Illumina MiSeq platform were completed by GENEWIZ. A Qubit 2.0 fluorometer (Invitrogen, USA) was used to determine the concentration of DNA samples, and a sequencing library was constructed using the MetaVx™ Library Construction Kit (GENEWIZ, USA). Using a 30–50 ng DNA sample as the template, two highly variable regions of prokaryotic 16S rDNA, namely, V3 and V4, were amplified by a series of PCR primers. The V3 and V4 regions were amplified using an upstream primer containing the “CCTACGGRRBGCASCAGKVRVGAAT” sequence and downstream primer containing the “GGACTACNVGGGTWTCTAATCC” sequence. In addition, a primer with an index connector was added to the end of the PCR product of 16S rDNA by PCR for NGS sequencing. The quality of the library was determined using an Agilent 2100 bioanalyzer (Agilent Technologies, USA), and the library concentrations were determined by a Qubit 2.0 fluorometer. After the DNA library was mixed, the Illumina MiSeq platform (San Diego, USA) was used for dual-end sequencing, and the sequence information was read by the MiSeq control software (MCS).2.6 RNA extraction and q-PCR
In order to further quantify the differences in the denitrifying functional genes for both the bioreactors, five target denitrifying functional genes were analyzed using q-PCR. The total RNA was extracted from activated sludge by using the TRIzol reagent (Invitrogen, Shanghai); further, cDNA was synthesized using a reverse transcription kit (Promega, Beijing). Here, q-PCR was performed using a SYBR green qPCR kit (Genestar, Beijing) and detected in a LightCycler 480 II system (Roche, Switzerland).The q-PCR primers and amplification procedures for each gene are listed in Table S4.†
2.7 Analytical methods
The forward and reverse reads obtained by double-end sequencing were connected via two assemblies: filtering sequences containing N in the splicing result and retaining sequences with a sequence length greater than 200 bp. After mass filtration, the chimeric sequences were removed, and the resulting sequences were used for operational taxonomic unit (OTU) analysis. Sequence clustering was performed using VSEARCH (1.9.6) (sequence similarity was set to 97%), and the aligned 16S rRNA reference database was Silva 132. Then, the Ribosomal Database Project (RDP) classifier based on the Bayesian algorithm was used to perform species taxonomic analysis on the OTU representative sequences. Further, the community composition of each sample was calculated at different species classification levels. The water quality indicator data were calculated using Excel 2007 and OriginPro 8.0 (OriginLab, Northampton) software.3. Results and discussion
3.1 Reactor performance
The denitrification performance of the bioreactor is shown in Fig. 2. During the startup period (operating days: 0–45), the acclimation of the activated sludge with anhydrous NaAc as the carbon source improved its denitrification efficiency. The initial influent NO3−-N concentration was set at 50 mg L−1 and then successively increased in 5 stages (50 → 100 → 200 → 300 → 400 → 500 mg L−1). The concentration level was raised when the nitrate removal rate reached 80%. When the final concentration was increased to approximately 500 mg L−1, the removal rate remained above 90%, and the effect remained stable for one week; at this point, the reactors were considered to be successfully started. After this acclimation stage, different volume ratios of landfill leachates were coprocessed with high-nitrate wastewater. Landfill leachates were added to the R1 system at volume ratios of 20%, 10%, and 5% from the 46th, 61st, and 71st operating days, respectively. The removal rate of nitrate nitrogen and COD sharply decreased (average rates: 78.8% and 55%, respectively) when the volume ratio was more than 10%, while the average removal rate of COD increased to 68.2% and the denitrification efficiency of the reactor could be restored to its original level after 3 operating cycles when the volume ratio was less than 10%. After acclimatization for 39 operating days (∼46–85), the nitrate removal rate gradually increased to 98 ± 1%. The denitrifying bacteria were effectively adapted to the high concentration of NO3−-N, which met the requirements for subsequent batch experiments to study the denitrification efficiency when landfill leachate was added as the carbon source under high load conditions.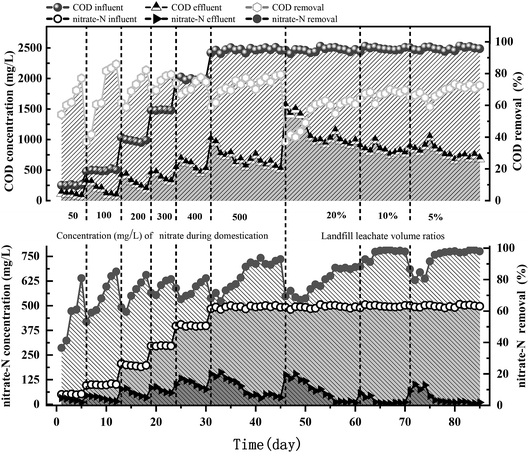 | ||
| Fig. 2 Concentration and removal evolution of nitrate-N and COD in the SBR fed with different volume ratios of landfill leachates. | ||
3.2 Effect of landfill leachate addition on denitrification
NaAc was used as the carbon source in R0, and 10% landfill leachate in R1 was mixed with high-concentration nitrate nitrogen wastewater. The C/N ratio of each reactor was controlled at 5.0 in order to compare the denitrification rates in a typical cycle (Fig. 3) with sufficient electron donors. It took approximately 6 h for the R0 system to remove nitrate nitrogen almost completely (the effluent concentration was lower than the detection limit); the denitrification rate was 32.32 mg NO3−-N (gVSS h)−1, while the denitrification rate of the R1 system was 58.05 mg NO3−-N (gVSS h)−1, which was 1.79 times higher than that of R0. This difference was mainly due to the diversity of COD sources in the landfill leachate. When compared with the single carbon source for R0, the mixed carbon sources for R1 facilitate accelerating the denitrification process of SBR, thereby improving the denitrification rate.25,26 Elefsiniotis27 and Wareham28 suggested that when there are many carbon sources in a system, microorganisms can use many kinds of electron donors at the same time. It can be inferred that there was no zero-order reaction in the R1 system, but there was a coupling reaction involving multiple carbon sources; therefore, the denitrification rate was faster than that in R0, which had only one carbon source. In addition, leachates rich in Ca and Mg facilitate the rapid growth of biomass. Fernández-Nava et al.29 reported that increasing the calcium concentration from 50 to 150 mg Ca2+ L−1 yielded a 1.4-fold increase in the denitrifying microorganism growth rate.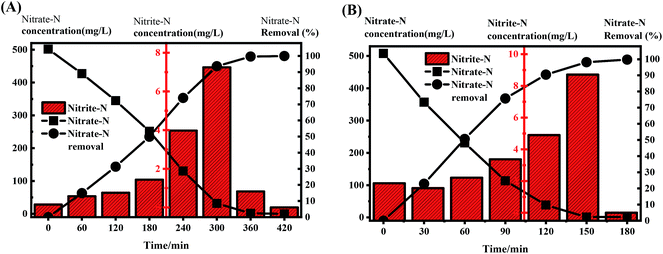 | ||
| Fig. 3 Concentration evolution of nitrate-N and nitrite-N in a typical cycle at a C/N ratio of 5.0. (A) R0 (no landfill leachate); (B) R1 (landfill leachate cotreatment). | ||
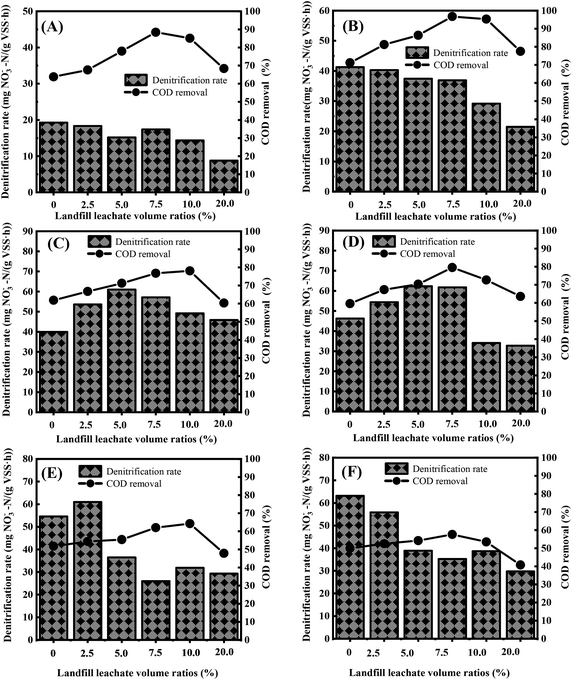
3.3 Determination of optimum volume ratio of landfill leachate
To investigate the removal of carbon and nitrogen at different volume ratios of landfill leachates, batch experiments were performed. Fig. 4A and B show that the denitrification rate was inversely proportional to the volume ratio of landfill leachate when the C/N ratios were equal to 2.5 and 3.2. This relationship was due to the high proportion of macromolecule organic matter at a low carbon ratio. With increased leachate addition, the relative content of macromolecular organic matter exceeded the degradation capability for specific organic matter macromolecules. As a result, the electron donors could not completely reduce the high nitrate concentration in a given cycle, which decreased the reduction amount of nitrate nitrogen per unit time. When the landfill leachate volume ratio was less than 7.5%, the denitrification rate accelerated with the increased addition amount at C/N ratios of 4.5 and 5.0 (Fig. 4C and D), and the maximum values were 61.03 and 62.28 mg NO3−-N (gVSS h)−1, respectively. When the addition ratio was more than 7.5%, the denitrification rate decreased with increasing addition amount and reached the lowest value when the addition ratio was 20%, resulting in values of 45.82 and 32.71 mg NO3−-N (gVSS h)−1, respectively. This result could be attributed to the balance between the relative content of small-molecule carbon sources and complex carbon sources in the system, as well as the demand of different bacteria for various carbon sources under these conditions. Synergy between the microorganisms accelerates the reduction of nitrate nitrogen. In contrast, from Fig. 4E and F, it is evident that the denitrification rate at C/N ratios of 5.6 and 6.1 decreased with increasing landfill leachate volume fraction, which is related to the toxicity of higher concentrations of free ammonia toward microorganisms30–32 and is probably due to the cometabolism in the R1 system. Under the cometabolism mechanism, a high mass concentration ratio of the growth matrix to nongrowth matrix can exert competitive inhibition on the production of key enzymes.33 In the R1 system, NaAc and nitrate acted as a part of the growth matrix, and complex organic matter (humic acid, etc.) in the landfill leachate acted as a nongrowth matrix; when the C/N ratio was 5.6 and 6.1, the dosage of NaAc was very high; therefore, it can be assumed that the mass concentration ratio of the growth matrix to the nongrowth matrix exceeded the demand range for cometabolism.34In addition, converting the landfill leachate content into COD concentration reveals that the corresponding COD proportion was 4.10–13.5% when the landfill leachate volume fraction was 2.5–7.5% and the C/N ratio was 3.2–5.0. Landfill leachate COD ratios in the range of 4.10–13.5% promoted the reduction rate of nitrate nitrogen. As shown in Fig. 4C–F, when C/N was 4.5–6.1, the COD removal rate improved with increased landfill leachate addition, which indicates that the cotreatment of landfill leachate can promote the utilization of NaAc and reduce the dosage of NaAc. However, for C/N = 5.6 and 6.1, the COD removal rates were less than 64.96%, while for C/N = 3.2, 4.5, and 5.0, the COD removal rates reached 96.77%, 84.38%, and 82.67%, respectively. Therefore, considering both COD utilization and denitrification rate, it was suggested that the volume fraction of landfill leachate should be controlled at 2.5–7.5% (COD concentration: 4.10–13.5%) under the conditions of C/N of 3.2–5.0, which accounts for both COD removal rate and denitrification rate.
3.4 Molecular biological analysis
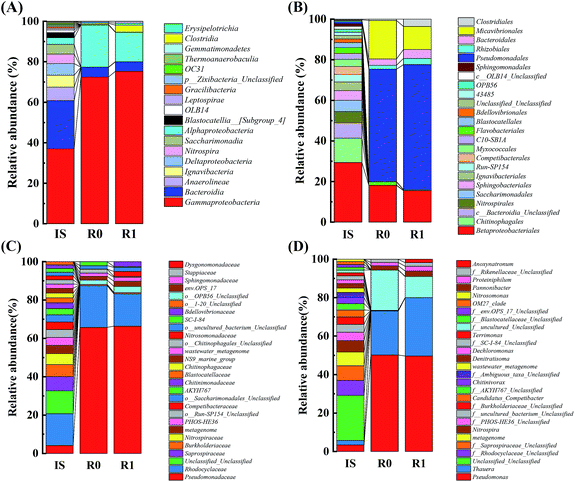
At the order level (Fig. 5B), the five dominant bacteria in R0 were Pseudomonadales (54.75%), Betaproteobacteriales (17.97%), Micavibrionales (18.72%), Bacteroidales (3.09%), and Rhizobiales (1.91%), accounting for 96.44% of the total content. The six dominant bacteria in R1 were Pseudomonadales (60.08%), Betaproteobacteriales (15.2%), Micavibrionales (11.05%), Bacteroidales (4.33%), Rhizobiales (2.93%), and Clostridiales (3.51%), accounting for 97.1% of the total content. These results indicated that adding landfill leachate not only increased the number of dominant functional bacteria at the order level but also stimulated the growth of Pseudomonadales and Clostridiales to change the community structure. The main families in R0 and R1 included Pseudomonadaceae, Rhodocyclaceae, Stappiaceae, Dysgonomonadaceae, and Rikenellaceae, with relative abundances of 54.63%, 18.13%, 1.91%, 1.59%, and 1.49% in R0, respectively, and 60.08%, 15.24%, 2.93%, 2.44% and 1.89% in R1, respectively (Fig. 5C). It is noteworthy that Clostridiaceae_2 increased from 0.05% to 2.33% (R1). At the genus level (Fig. 5D), 28 genera were detected in IS, while 17 genera were detected in R0 and R1, out of which Pseudomonas, Thauera, Pannonibacter, and Proteiniphilum were the dominant genera in the two reactors, with relative abundances of 44.75%, 20.49%, 1.91%, and 1.59% in R0, respectively, and 50.08%, 30.59%, 2.93%, and 2.44%, in R1, respectively. These four denitrifying bacteria were enhanced and enriched in R1, while Pseudomonas, Thauera, and Pannonibacter had good denitrification characteristics.38–40 The capability of the Pseudomonas aeruginosa PCN-2 strain for nitrate reduction was confirmed by the PCR analyses of the ChrR, napA, nirS, cnorB and nosZ genes.41 In addition, Gram-negative bacillus Thauera sp. K11 was previously isolated from petrochemical wastewater by Qiao et al.,42 who found that the bacteria could denitrify by using more than 10 phenolic derivatives as electron donors. Furthermore, the abundance of Anoxynatronum in R1 was 1.85%, which was 46.25-fold higher than that in R0 (only 0.04%), indicating that landfill leachate was beneficial to the growth of Anoxynatronum. Garnova et al.43 isolated an alkalophilic anaerobic bacterium, namely, Anoxynatronum, from Lake Baikal and found that it can use cellulose and other macromolecular organic matter as a carbon source to achieve denitrification and can decompose more complex compounds, which explains the increase in the COD removal rate in batch experiments as the amount of leachate addition increased.
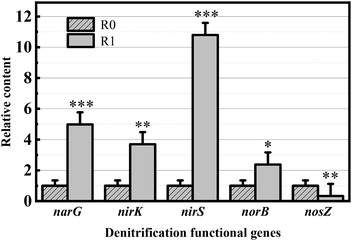
The q-PCR results are shown in Fig. 6. Except for the NOS-encoding gene nosZ, the relative abundance of the narG, nirS, nirK, and norB genes increased by 4.98-, 3.69-, 10.80-, and 2.38-folds, respectively, in R1, indicating that the addition of landfill leachate can increase the relative abundance of the denitrification functional genes. The nir gene consisted of the copper-containing nitrite reductase (Cu-NIR) coding gene nirK and heme-containing nitrite reductase (cd1-NIR) coding gene nirS. The abundant copper ions in the landfill leachate provide abundant substrate binding sites for type II copper of the copper-NIR monomer, thereby enhancing the relative abundance of nirK. Here, cd1-NIR is a homodimeric bifunctional enzyme that acts as a catalyst with azurin, pseudoazurin, or cytochrome c551 as electron donors; therefore, it can be speculated that landfill leachates provide the required electron donors to increase the relative expression of nirK. In addition, NAR activity can be enhanced by adding appropriate concentrations of Fe(III) and Mo(VI),44 and the presence of Fe(II) can increase the copy numbers of nirS and nosZ to facilitate NO2− reduction and N2O formation.45 The results of the relative quantitative analysis of functional genes obtained by q-PCR help elucidate the difference in the denitrification efficiencies between the two reactors and enable the analysis of the feasibility of the cotreatment of landfill leachate with high-nitrate wastewater from the perspective of functional genes.
4. Conclusions
Landfill leachate and high-nitrate wastewater were cotreated to obtain efficient activated sludge for denitrification with a maximum denitrification rate of 62.28 mg NO3−-N (gVSS h)−1. The cotreatment of landfill leachate and high-nitrate wastewater improved the utilization rate of NaAc (the COD removal rate reached 96%). The addition of landfill leachate changed the relative abundances of dominant bacteria (Pseudomonas, Thauera, Pannonibacter, and Proteiniphilum increased, while Alphaproteobacteria and Bacteroidia decreased). After the cotreatment, the relative contents of the denitrification functional genes narG, nirK, nirS, and norB increased by 398%, 269%, 980%, and 138%, respectively.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the China Post-doctoral Science Foundation (2019M653086), the Scientific Research Foundation of Shenzhen Water (Group) Co. Ltd. (20180004) and the Key Technology Assessment and Standardization of Urban Water Supply System Operation Management (2017ZX07501002). The authors would like to acknowledge the use of the the Light Cycler 480 II system at the School of Life Sciences, Sun Yat-Sen University.References
- CAEPI, Development Report on Treatment Industry of Urban Domestic Refuse in 2017, China Environmental Protection Industry, 2017, vol. 4, pp. 9–15 Search PubMed.
- H. Guo, J. M. Li, M. J. Qiu, J. L. Liu and W. C. Zhang, Multivariate linear regression prediction of domestic waste yield in China, Environ. & Dev., 2018, 30(1), 61–63 Search PubMed.
- Y. Pan, B. Ni, P. L. Bond, L. Ye and Z. Yuan, Electron competition among nitrogen oxides reduction during methanol-utilizing denitrification in wastewater treatment, Water Res., 2013, 47, 3273–3281 CrossRef CAS PubMed.
- Y. Peng, Y. Ma and S. Wang, Denitrification potential enhancement by addition of external carbon sources in a pre-denitrification process, J. Environ. Sci., 2007, 19, 284–289 CrossRef CAS.
- H. J. Hamlin, J. T. Michaels, C. M. Beaulaton, W. F. Graham, W. Dutt, P. Steinbach, T. M. Losordo, K. K. Schrader and K. L. Main, Comparing denitrification rates and carbon sources in commercial scale upflow denitrification biological filters in aquaculture, Aquacult. Eng., 2008, 38, 79–92 CrossRef.
- P. J. Robinson and C. A. Woolhead, Post-translational membrane insertion of an endogenous YidC substrate, Biochim. Biophys. Acta, 2013, 1833(12), 2781–2788 CrossRef CAS PubMed.
- E. M. de Albuquerque, E. Pozzi, I. K. Sakamoto and P. Jurandyr, Treatability of landfill leachate combined with sanitary sewage in an activated sludge system, Journal of Water Process Engineering, 2018, 23, 119–128 CrossRef.
- F. M. Ferraz, A. T. Bruni, J. Povinelli and E. M. Vieira, Leachate/domestic wastewater aerobic co-treatment: a pilot-scale study using multivariate analysis, J. Environ. Manage., 2016, 166, 414–419 CrossRef CAS PubMed.
- J. Kalka, Landfill Leachate Toxicity Removal in Combined Treatment with Municipal Wastewater, Sci. World J., 2012, 2012, 202897 CAS.
- Z. L. Ye, X. Xie, L. Dai, Z. Wang, W. Wu, F. Zhao, X. Xie, S. Huang, M. Liu and S. Chen, Full-scale blending treatment of fresh MSWI leachate with municipal wastewater in a wastewater treatment plant, Waste Manag., 2014, 34(11), 2305–2311 CrossRef CAS PubMed.
- J. Yu, S. Zhou and W. Wang, Combined treatment of domestic wastewater with landfill leachate by using A2/O process, J. Hazard. Mater., 2010, 178(1–3), 81–88 CrossRef CAS PubMed.
- F. M. Ferraz, J. Povinelli, E. Pozzi, E. M. Vieira and J. C. Trofino, Co-treatment of landfill leachate and domestic wastewater using a submerged aerobic biofilter, J. Environ. Manage., 2014, 141, 9–15 CrossRef CAS PubMed.
- P. Torres, J. A. Rodríguez, L. E. Barba, L. F. Marmolejo and C. A. Pizarro, Combined treatment of leachate from sanitary landfill and municipal wastewater by UASB reactors, Water Sci. Technol., 2009, 60(2), 491–495 CrossRef CAS PubMed.
- M. Zheng, S. Li, Q. Dong, X. Huang and Y. Liu, Effect of blending landfill leachate with activated sludge on the domestic wastewater treatment process, Environ. Sci.: Water Res. Technol., 2019, 5, 268–276 RSC.
- R. B. Brennan, E. Clifford, C. Devroedt, L. Morrison and M. G. Healy, Treatment of landfill leachate in municipal wastewater treatment plants and impacts on effluent ammonium concentrations, J. Environ. Manage., 2017, 188, 64–72 CrossRef CAS PubMed.
- T. Osaka, K. Shirotani, S. Yoshie and S. Tsuneda, Effects of carbon source on denitrification efficiency and microbial community structure in a saline wastewater treatment process, Water Res., 2008, 42(14), 3709–3718 CrossRef CAS PubMed.
- J. L. Campos, A. Mosquera-Corral, M. Sánchez, R. Méndez and J. M. Lema, Nitrification in saline wastewater with high ammonia concentration in an activated sludge unit, Water Res., 2002, 36(10), 2555–2560 CrossRef CAS PubMed.
- C. Glass and J. Silverstein, Denitrification of high-nitrate, high-salinity wastewater, Water Res., 1999, 33(1), 223–229 CrossRef CAS.
- K. Windey, B. I. De and W. Verstraete, Oxygen – limited autotrophic nitrification – denitrification (OLAND) in a rotating biological contactor treating high – salinity wastewater, Water Res., 2005, 39(18), 4512–4520 CrossRef CAS PubMed.
- C. Glass and J. Silverstein, Denitrification kinetics of high nitrate concentration water: pH effect on inhibition and nitrite accumulation, Water Res., 1998, 32(3), 831–839 CrossRef CAS.
- B. M. Peyton, M. R. Mormile and J. N. Petersen, Nitrate reduction with Halomonas campisalis: kinetics of denitrification at pH 9 and 12.5% NaCl, Water Res., 2001, 35(17), 4237–4242 CrossRef CAS PubMed.
- Y. Fernández-Nava, E. Marañón, J. Soons and L. Castrillón, Denitrification of high nitrate concentration wastewater using alternative carbon sources, J. Hazard. Mater., 2010, 173(1–3), 682–688 CrossRef PubMed.
- F. A. El-Gohary and G. Kamel, Characterization and biological treatment of pre-treated landfill leachate, Ecol. Eng., 2016, 94, 268–274 CrossRef.
- APHA, Standard Methods for the Examination of Water and Wastewater, American Public Health Association/American Water Works Association/Water Environment Federation, Washington, DC, 22nd edn, 2012 Search PubMed.
- J. Sun, M. Guo, L. Zhao, Y. G. Gao, M. C. She and Z. lian, The effects of denitrification with sludge alkaline fermentation liquid and thermal hydrolysis liquid as carbon sources, RSC Adv., 2016, 6(76), 72333–72341 RSC.
- F. Liu, Y. Tian, Y. Ding and Z. Li, The use of fermentation liquid of wastewater primary sedimentation sludge as supplemental carbon source for denitrification based on enhanced anaerobic fermentation, Bioresour. Technol., 2016, 219, 6–13 CrossRef CAS PubMed.
- P. Elefsiniotis and D. G. Wareham, Utilization patterns of volatile fatty acids in the denitrification reaction, Enzym. Microb. Technol., 2007, 41(1–2), 92–97 CrossRef CAS.
- P. Elefsiniotis, D. G. Wareham and M. O. Smith, Use of volatile fatty acids from an acid-phase digester for denitrification, J. Biotechnol., 2004, 114(3), 289–297 CrossRef CAS PubMed.
- Y. Fernández-Nava, E. Marañón, J. Soons and L. Castrillón, Denitrification of wastewater containing high nitrate and calcium concentrations, Bioresour. Technol., 2008, 99(17), 7976–7981 CrossRef PubMed.
- F. Zhang, H. Yang, J. W. Wang, Z. Q. Liu and Q. K. Guan, Effect of free ammonia inhibition on NOB activity in high nitrifying performance of sludge, RSC Adv., 2018, 8(56), 31987–31995 RSC.
- W. Qian, Y. Peng, X. Li, Q. Zhang and B. Ma, The inhibitory effects of free ammonia on ammonia oxidizing bacteria and nitrite oxidizing bacteria under anaerobic condition, Bioresour. Technol., 2017, 243, 1247–1250 CrossRef CAS PubMed.
- J. A. Torà, J. Lafuente, J. A. Baeza and J. Carrera, Combined effect of inorganic carbon limitation and inhibition by free ammonia and free nitrous acid on ammonia oxidizing bacteria, Bioresour. Technol., 2010, 101(15), 6051–6058 CrossRef PubMed.
- M. Nsenga Kumwimba and F. Meng, Roles of ammonia-oxidizing bacteria in improving metabolism and cometabolism of trace organic chemicals in biological wastewater treatment processes: a review, Sci. Total Environ., 2019, 659, 419–441 CrossRef CAS PubMed.
- M. Xiao, X. Yin, H. Gai, H. Ma, Y. Qi, K. Li, X. Hua, M. Sun and H. Song, Effect of hydroxypropyl-β-cyclodextrin on the cometabolism of phenol and phenanthrene by a novel Chryseobacterium sp, Bioresour. Technol., 2019, 273, 56–62 CrossRef CAS PubMed.
- S. L. S. Rollemberg, L. Q. de Oliveira, A. R. M. Barros, V. M. M. Melo, P. I. M. Firmino and A. B. Dos Santos, Effects of carbon source on the formation, stability, bioactivity and biodiversity of the aerobic granule sludge, Bioresour. Technol., 2019, 278, 195–204 CrossRef CAS PubMed.
- Z. Xu, X. Dai and X. Chai, Effect of different carbon sources on denitrification performance, microbial community structure and denitrification genes, Sci. Total Environ., 2018, 634, 195–204 CrossRef CAS PubMed.
- L. Yan, M. Zhang, Y. Liu, C. Liu, Y. Zhang, S. Liu, L. Yu, G. Hao, Z. Chen and Y. Zhang, Enhanced nitrogen removal in an aerobic granular sequencing batch reactor under low DO concentration: role of extracellular polymeric substances and microbial community structure, Bioresour. Technol., 2019, 289, 121651 CrossRef CAS PubMed.
- H. Bai, S. Liao, A. Wang, J. Huang, W. Shu and J. Ye, High-efficiency inorganic nitrogen removal by newly isolated Pannonibacter phragmitetus B1, Bioresour. Technol., 2019, 271, 91–99 CrossRef CAS PubMed.
- Y. Mao, Y. Xia and T. Zhang, Characterization of Thauera-dominated hydrogen-oxidizing autotrophic denitrifying microbial communities by using high-throughput sequencing, Bioresour. Technol., 2013, 128, 703–710 CrossRef CAS PubMed.
- L. Yang, X. H. Wang, S. Cui, Y. X. Ren, J. Yu, N. Chen, Q. Xiao, L. K. Guo and R. H. Wang, Simultaneous removal of nitrogen and phosphorous by heterotrophic nitrification-aerobic denitrification of a metal resistant bacterium Pseudomonas putida strain NP5, Bioresour. Technol., 2019, 285, 121360 CrossRef CAS PubMed.
- D. He, M. Zheng, T. Ma, C. Li and J. Ni, Interaction of Cr(VI) reduction and denitrification by strain Pseudomonas aeruginosa PCN-2 under aerobic conditions, Bioresour. Technol., 2015, 185, 346–352 CrossRef CAS PubMed.
- N. Qiao, L. Xi, J. Zhang, D. Liu, B. Ge and J. Liu, Thauera sinica sp. nov., a phenol derivative-degrading bacterium isolated from activated sludge, Antonie Leeuwenhoek, 2018, 111(6), 945–954 CrossRef CAS PubMed.
- E. S. Garnova, T. N. Zhilina, T. P. Tourova and A. M. Lysenko, Anoxynatronum sibiricum gen.nov., sp.nov. alkaliphilic saccharolytic anaerobe from cellulolytic community of Nizhnee Beloe (Transbaikal region), Extremophiles, 2003, 7(3), 213–220 CrossRef CAS PubMed.
- P. Pintathong, D. J. Richardson, S. Spiro and W. Choorit, Influence of metal ions and organic carbons on denitrification activity of the halotolerant bacterium, Paracoccus pantotrophus P16 a strain from shrimp pond, Electron. J. Biotechnol., 2009, 12(2), 1–9 Search PubMed.
- S. Li, X. M. Li and F. B. Li, Effect of Fe(II) on denitrification and associated functional microbial communities, China Environ. Sci., 2018, 38(1), 263–274 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra07966a |
| This journal is © The Royal Society of Chemistry 2019 |