Open Access Article
Open Access ArticleSensitive colorimetric detection of ochratoxin A by a dual-functional Au/Fe3O4 nanohybrid-based aptasensor
Rong Huang,
Lu Lu Xiong,
Hui Hui Chai,
Jing Jing Fu,
Zhisong Lu * and
Ling Yu
* and
Ling Yu *
*
Key Laboratory of Luminescent and Real-Time Analytical Chemistry, Ministry of Education, Institute for Clean Energy and Advanced Materials, School of Materials and Energy, Southwest University, Chongqing 400715, PR China. E-mail: lingyu12@swu.edu.cn; zslu@swu.edu.cn
First published on 25th November 2019
Abstract
A novel colorimetric aptasensor based on a Au/Fe3O4 nanohybrid was developed to detect ochratoxin A (OTA). The aptasensor is composed of a free OTA aptamer, a Au/Fe3O4 nanohybrid coated with biotinylated complementary DNA of the OTA aptamer (biotin-cDNA-Au/Fe3O4), and free alkaline-phosphatase-labeled streptavidin (SA-ALP). The Au/Fe3O4 nanohybrid not only immobilizes biotin-cDNA but also magnetically separates SA-ALP from the sample solution. One part of the OTA aptamer sequence hybridizes with biotin-cDNA immobilized on Au/Fe3O4, and the left part of the OTA aptamer sequence covers the biotin and blocks the specific interaction between biotin and SA-ALP. OTA can interrupt the interaction of OTA aptamer binding to biotin-cDNA-Au/Fe3O4 and can inhibit the shielding effect of the OTA aptamer on biotin. The amount of SA-ALP that can be captured by biotin-cDNA-Au/Fe3O4 thus increases with increasing OTA concentration. Through a simple magnetic separation, the collected SA-ALP-linked Au/Fe3O4 can produce a yellow-colored solution in the presence of p-nitrophenyl phosphate (p-NPP). This colorimetric aptasensor can detect OTA as low as 1.15 ng mL−1 with high specificity.
Introduction
Ochratoxin A (OTA) is a ubiquitous mycotoxin and is produced by various species of Aspergillus and Penicillium. It can potentially contaminate cereals (wheat, corn, and barley) and wines.1,2 Several studies have suggested that OTA has nephrotoxicity, hepatotoxicity, neurotoxicity, and immunotoxicity.2,3 Gastrointestinal damage, lymphatic lesions, bone marrow toxicity, and digestive disorders have also been reported in animal studies.4,5 OTA has been classified as a group 2B “possibly carcinogenic to humans” by the International Agency for Research on Cancer (IARC).2 The European Commission has established regulatory limits for raw cereal grains (5 μg kg−1), roasted coffee (5 μg kg−1), grape juice and all types of wine (2 μg kg−1), and dried fruits (10 μg kg−1).6,7 Therefore, it is crucial to have a suitable analysis technique for high sensitivity detection of OTA.Conventional methods for OTA quantification are based on instruments such as high-performance liquid chromatography8,9 (HPLC) connected to fluorescence10 or tandem mass spectrometry.11–13 However, the high cost and sophisticated equipment as well as the time and labor costs and need for trained operators have hindered their applications.14,15 Thus, low-cost, convenient, and user-friendly detection of OTA is highly desired.
Colorimetric analysis can be easily monitored with the naked eye. It is affordable and simple, especially for field analysis.16,17 Aptamers are single-stranded oligonucleotides that can specifically bind to their target molecules with high binding affinity.18–22 Nanoparticle (NP)-based bioanalysis has attracted tremendous attention due to its high specific surface area and size-dependent optical properties.3,23–26 Previously, OTA has been detected with gold nanoparticles (AuNPs) and nano-graphite homogenous reaction systems.27 In those studies, the samples were directly mixed with the sensing solution,28,29 and the change in fluorescent signal or color can be read without further separation. In one example of colorimetric detection, Xiao reported a OTA detection aptasensor based on analyte-induced assembling of oriented AuNP dimers.28 Asymmetric functionalization of Au by DNA prevented the formation of large Au aggregates, and a Y-shaped DNA duplex was utilized to minimize the inter-particle distance. However, the sample solution properties such as color (wine, peanut oil, etc.) would interfere with the signal readout—especially for a colorimetric detection scheme. Thus, the isolation or separation of trace amounts of target from complex samples as well as their specific detection would potentially minimize the interference induced by the sample color.
Centrifugation is common in NP separation, but Fe3O4 NPs can be separated under a magnetic field. However, immobilization of probes on Fe3O4 NPs was compromised because they often lack functional groups for covalent conjugation; thus, functionalization of Fe3O4 NPs was often required. For instance, a silicon oxide layer was coated on Fe3O4 NPs. Then, a SiO2 layer was built with 3-aminopropyltrimethoxysilane (APTES) to introduce NH2 groups for covalent probe immobilization.30 The interaction between amino and aldehyde groups allows researchers to link DNA to Fe3O4. However, this process requires multi-step reactions and cross-linking reagents. For instance, Wang et al. reported a colorimetric aptasensor of OTA using Au@Fe3O4 and glass beads with a sophisticated reaction. The amino group was first coupled with the exposed hydroxyl groups on the Au@Fe3O4 surface to yield an amino-terminated self-assembled monolayer. The distal amino groups on Au@Fe3O4 surfaces were then reacted with aldehyde groups to immobilize amino-modified cDNA. While, additional glass beads were used to immobilize the aptamers.31
The intrinsic interaction between thiol group and gold has motivated researchers to immobilize DNA on Au. Here, we propose an Au/Fe3O4 aptasensor to achieve fast separation and colorimetric detection of OTA. The Au/Fe3O4 nanohybrids offer magnetic separation and immobilize probes for OTA detection. The 3′-end and 5′-end of the OTA aptamer recombinant cDNA fragment are tagged with a biotin and a thiol group, respectively (biotin-cDNAOTA-SH). The mechanism is that binding of OTA aptamer with biotin-cDNAOTA to shield the biotin and thus inhibit the interaction of biotin with streptavidin-alkaline phosphatase (SA-ALP). The binding of OTA with OTA aptamer will set free the cDNA-biotin, leading to an increase in SA-ALP capture. The ALP can specifically hydrolyze its substrate p-nitrophenyl phosphate (p-NPP) to p-nitrophenol, which is a yellow complex with an intense absorption band at ∼405 nm. This colorimetric change is proportional to the OTA concentration. We first optimized the amount of biotin-cDNAOTA-SH and OTA aptamer in the sensing system and then evaluated the working range and detection limit. Next, the potential of the proposed Au/Fe3O4-based aptasensors was demonstrated by assaying OTA in peanut, corn, and wine samples. The results were compared with commercial OTA ELISA kit to prove its potential in real sample testing.
Materials and methods
Chemical and reagents
The sequence of OTA aptamer was 5′-GAT CGG GTG TGG GTGGCG TAA AGG GAG CAT CGG ACA-3′32 and the sequence of its partially complementary oligonucleotide was 5′-SH-TGTCCG ATG CTC CCT TTA-C6-biotin-3′ (biotin-cDNA-SH). Both of them were chemically synthesized and purified by Sangon Biotech Co. Ltd. (Shanghai, China). FeCl3·6H2O, FeCl2·4H2O were ordered from Aladdin Chemistry Co. Ltd. (Shanghai, China). Ammonio solution was purchased from Sinopharm Chemical Reagent Co., Ltd. HAuCl4 was purchased from Rhawn reagent (Shanghai, China), and trisodium citrate dihydrate was purchased from Kelong Chemical Reagent Factory (Chengdu, China). OTA was obtained from Cayman chemical (USA). Aflatoxin (AFB1), fumonisin B1 (FB1), and deoxynivalenol (DON) from Fermentek Ltd. (Israel). Phosphate buffered solution (PBS) was purchased from Beijing Dingguo Changsheng Biotechnology Co., Ltd. (Beijing, China). Tris was purchased from Genview (USA), and p-NPP, SA-ALP were purchased from Sigma-Aldrich (USA). A commercial OTA sandwich ELISA kit was purchased from Shanghai Youlong Biotech Co., Ltd. (Shanghai, China).Synthesis of Au/Fe3O4 nanohybrid
First, Fe3O4 NPs were synthesized with a coprecipitation method.33 In brief, FeCl3·6H2O (3.244 g) and FeCl2·4H2O (1.192 g) were dissolved in water (80 mL), and then 15 mL ammonia solution (28%, w/v%) was added. The solution was left undisturbed for 10 min at room temperature. The mixture was incubated at 90 °C with constant mechanical stirring under nitrogen gas protection. After 30 min, the Fe3O4 NPs were isolated with a permanent magnetic and washed several times with DI water. Finally, the Fe3O4 NPs were dried at 45 °C under vacuum for 24 h. To synthesize Au/Fe3O4 nanohybrid, the HAuCl4 solution (0.04%, 50 mL) was boiled with constant stirring. Then, 500 μL Fe3O4 NPs and 1 mL 5% sodium citrate solution were added. The color of the solution changed from brown to purple and finally turned red. Stirring was continued for 10 min after the color change stopped. The heating source was removed, and stirring was continued for 15 min at room temperature. After the reaction, the Au/Fe3O4 hybrids were harvested by magnetic separation. The pellet was then washed several times and re-dispersed in DI water and stored at 4 °C.Characterization of Au/Fe3O4 nanohybrid
The X-ray diffraction (XRD) patterns of the synthesized Au/Fe3O4 nanohybrid were obtained using an XRD-7000 (Shimadzu, Japan) with Cu-Kα source radiation. The morphology of the Au/Fe3O4 was investigated by a transmission electron microscopy (TEM, Tecnai G2 F20, FEI, USA). The chemical status of the Au/Fe3O4 was characterized by X-ray photoelectron spectroscopy (XPS, Escalab 250xi, Thermo Fisher, USA). The size of Fe3O4 NP and Au/Fe3O4 nanohybrids was measured using a Malvern Zetasizer Nano ZS 90 (Malvern, UK). The magnetic properties of Fe3O4 NP and Au/Fe3O4 nanohybrids were characterized by a vibrating sample magnetometer (VSM, MPMS Squid-VSM, USA).Construction of biotin-cDNAOTA-Au/Fe3O4 NP-based OTA aptasensor for colorimetric detection of OTA
A vial of received biotin-cDNAOTA-SH was dissolved with 0.01 M PBS to obtain the stock solution with a concentration of 100 μM. Next, 5 μL 100 μM biotin-cDNAOTA-SH was mixed with Au/Fe3O4, and then incubated at 37 °C for 40 h. To remove the unconjugated oligonucleotides, biotin-cDNAOTA-Au/Fe3O4 NPs conjugates were collected and washed three times with magnetic separation. The collected biotin-cDNAOTA-Au/Fe3O4 NPs were used for the OTA aptasensors construction.OTA was dissolved in methanol for a stock solution with a concentration of 2 mg mL−1. PBS was then used as a solvent to prepare working solutions with various concentrations of OTA as indicated. Different concentrations of OTA solution were mixed with excess aptamer (20 μM) and allowed to react at room temperature for 30 min. The biotin-cDNAOTA-Au/Fe3O4 was heated to 45 °C for 30 min and then incubated with the OTA-aptamer mixture overnight at room temperature. Then 50 μL of 1.5 mg L−1 SA-ALP (dissolved in pH = 8.0, 0.01 M Tris–HCl buffer) was added to 50 μL of the reaction solution and incubated at 37 °C for 1 h. The nano-hybrids pellet was washed three times with Tris–HCl buffer via magnetic separation. Next, 50 μL p-NPP was added to the collected Au/Fe3O4 pellet and incubated for 1 h at 37 °C. Finally, the absorption spectrum was recorded after adding 100 μL of 3 M NaOH stop solution. OTA at 100 pg mL−1, 1 ng mL−1, 10 ng mL−1, 100 ng mL−1, 1 μg mL−1, 10 μg mL−1, and 100 μg mL−1 were tested. The OTA induced colorimetric change was characterized by calculating the change of absorbance at 405 nm as follows:
| ΔA = |(AOTA − A0)/A0|, |
Specificity of biotin-cDNAOTA-Au/Fe3O4 NP-based OTA aptasensor
To evaluate the specificity of the as proposed OTA aptasensor, other mycotoxicins [AFB1 (100 ng mL−1), FB1 (100 ng mL−1), and DON (100 ng mL−1)] were measured following the procedure as detailed above.In addition, peanut, corn extracts and wine were used to test the feasibility of the biotin-cDNAOTA-Au/Fe3O4 NP-based on aptasensor as described previously. The extracts were obtained following a protocol from literature.34 Then the extracts were passed through a 0.22 μm syringe filter, and the OTA was added into the peanut, corn and wine samples at a final concentration of 5, 10, and 20 ng mL−1 and measured by the biotin-cDNAOTA-Au/Fe3O4 NP-based OTA aptasensor. At the same time, the samples were measured by the commercial ELISA kit according to product instructions to validate the results obtained from as prepared OTA aptasensor.
Statistics analysis
All experiments were repeated three times and in triplicate. Data are expressed as the mean ± standard deviation. Experiment results were analyzed with the Student's t-test using Origin Statistic software (OriginLab, USA). P values less than 0.05 were considered statistically significant.Results and discussion
Working principle of biotin-cDNAOTA-Au/Fe3O4 NP-based OTA aptasensor
The colorimetric detection of OTA based on Au/Fe3O4 NPs is illustrated in Scheme 1. Here, the OTA aptamer works as a shielding cap to modulate the interaction between SA and biotin. Specifically, in the absence of OTA, the aptamer binds to its partially cDNA probes that immobilized on Au/Fe3O4, while the unpaired bases at the 5′-end of OTA aptamer cover the biotin molecule at the end of cDNA. This inhibits the biotin–streptavidin interaction and further blocks the conjunction of ALP. After magnetic separation, there are fewer ALP captured by biotin-cDNA probes resulting in a lighter yellow color when the p-NPP substrate is added. However, in the presence of OTA, the hybridized aptamer is detached from its complementary DNA because the aptamer prefers to bind with OTA. The biotin molecule at the end of the complementary DNA is uncovered leading to high accessibility for the SA-ALP in solution. The increased ALP leads to a stronger p-NPP color change. The NPs could be removed magnetically, and the absorbance of the supernatant reflects the OTA concentration.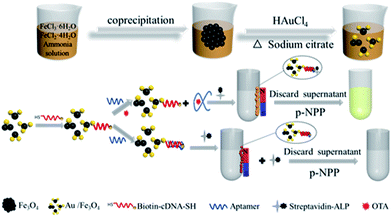 | ||
| Scheme 1 Schematic illustration of the biotin-cDNAOTA-Au/Fe3O4 NP-based OTA aptasensor for colorimetric detection of OTA. | ||
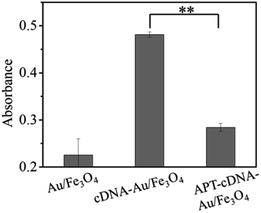
First, the shielding effect of OTA aptamer on biotin–streptavidin interaction was studied. Fig. 1 shows that less p-NPP is catalyzed with lower 405 nm absorbance when directly adding SA-ALP to the Au/Fe3O4 hybrids. However, adding SA-ALP into biotin-cDNAOTA-Au/Fe3O4 hybrids can increase the amount of SA-ALP immobilization and thus significantly increase the absorbance to 0.48 ± 0.006. Adding OTA aptamer to the biotin-cDNAOTA-Au/Fe3O4 hybrides can reduce the absorbance to 0.28 ± 0.008 because the unpaired base at the 5′ end blocks the biotin molecule at the end of the cDNA and reduces the binding of SA-ALP.
Characterization of the Au/Fe3O4 hybrids
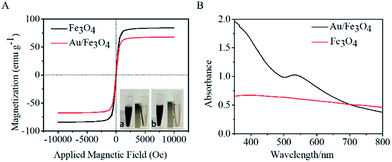
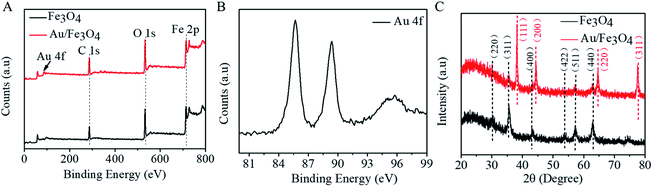
Magnetic properties of Fe3O4 and nanohybrid were demonstrated by VSM. The magnetization loops are shown in Fig. 2A. Low coercivity of remanence existing at 300 K indicated the superparamagnetic behaviors of the two nanoparticles. The saturation magnetization value of Fe3O4 was calculated to be 84.4 emu g−1. The value decreased to 67.9 emu g−1 after reacting with gold. The color of the Fe3O4 was black, and that of Au/Fe3O4 nanohybrid was dark red (inset of Fig. 2A). Though the magnetic strength of Au/Fe3O4 decreased, it can be easily attracted by an external magnetic field, which shows its significant magnetic separation capacity. The UV-visible absorption spectra of Fe3O4 and nanohybrid were measured because the Au was incorporated to provide a base for probe immobilization via the thiol group and Au interactions. A new peak at 532 nm was observed for Au/Fe3O4 (Fig. 2B) versus pure Fe3O4 NPs indicating the existence of Au in the hybrids.The chemical composition of Au/Fe3O4 nanohybrid was further ascertained by XPS measurements. The peaks of C, O, Au, and Fe elements were successfully observed in Au/Fe3O4 nanohybrid. In contrast, the peak of Au was not found in the XPS image of Fe3O4 (Fig. 3A). In the Au 4f core-level photoelectron spectrum of the Au/Fe3O4 nanohybrid (Fig. 3B), the peaks at 85.1 and 89.2 eV can be attributed to the Au 4d7/2 and Au 4f5/2 of metallic Au, respectively.35 XRD characterization showed typical XRD diffraction peaks at 2θ values of 30.1°, 35.5°, 43.1°, 53.5°, 57.0°, and 62.6° indexed to (220), (311), (400), (422), (511), and (440) planes of a cubic structure of Fe3O4 (JCPDS no. 19-0629), respectively. These were observed from both the pure Fe3O4 NP and Au/Fe3O4 nanohybrids (Fig. 3C). Four additional peaks centered at 38.1°, 44.3°, 64.5°, and 77.6° appeared for the Au/Fe3O4 nanohybrid, which can be attributed to the (111), (200), (220), and (311) planes for the face-centered-cubic gold (JCPDS no. 04-0784), respectively.31
The size of Fe3O4 NP and Au/Fe3O4 nanohybrids was characterized by dynamic light scattering. The hydrated particle size of Fe3O4 is more than 100 nm, and even several hundred nm, suggesting aggregation of Fe3O4 NP (Fig. 4A). However, the Au/Fe3O4 nanohybrid scatters with hydrated particle size of tens of nanometers (Fig. 4B). The TEM images shown in Fig. 4C and D further prove that incorporation of Au onto Fe3O4 NP can reduce the aggregation of the magnetic nanoparticle. The uniform dispersion of the Au/Fe3O4 nanohybrid will benefit immobilization of probes and subsequent sensing.
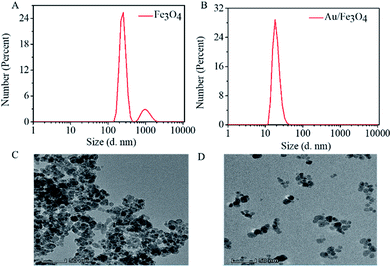 | ||
| Fig. 4 Dispersion of AuNP/Fe3O4 nanohybrid. Hydration particle size of (A) Fe3O4 and (B) Au/Fe3O4; TEM image of (C) Fe3O4 and (D) Au/Fe3O4. | ||
Optimization of biotin-cDNAOTA-Au/Fe3O4 NP-based OTA aptasensor
Scheme 1 shows that binding of the OTA aptamer with its corresponding cDNA inhibits biotin–SA-ALP bonding thus leading to less p-NPP hydrolysis. To achieve sound sensing performance, the amount of cDNA probes on Au/Fe3O4 nanohybrid, OTA aptamers, and SA-ALP were optimized. Different concentrations of biotin-cDNA-SH were applied to prepare biotin-cDNA-Au/Fe3O4 nanohybrid. Then, SA-ALP was added. The pellet obtained via magnetic separation was re-dispersed in p-NPP solution. The stronger yellow color indicates that a larger amount of biotin-cDNA is successfully immobilized on Au/Fe3O4 nanohybrid for OTA sensing. First, the amount of biotin-cDNA used in the reaction system was optimized. Fig. 5A shows that the relative UV-visible absorbance intensity (ΔA) increases with the concentration of biotin-cDNA and then reaches its maximum at a biotin-cDNA concentration of 0.1 μM. Additional biotin-cDNA does not enhance the signal further. In contrast, when the DNA concentration is higher than 0.1 μM, the absorbance at 405 nm decreases again probably due to the steric hindrance. Hence, the optimal concentration of biotin-cDNA-SH was selected to be 0.1 μM.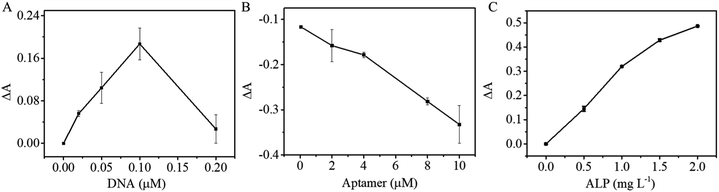 | ||
| Fig. 5 Optimization experimental conditions of OTA detection: (A) concentration of DNA; (B) concentration of APT; (C) concentration of SA-ALP. | ||
The OTA competes for binding to the OTA aptamer and its cDNA; thus, the amount of OTA aptamer applied to the sensing system was optimized next. When the concentration of biotin-cDNA used in the reaction is 0.1 μM, the relative absorbance ΔA decreases with OTA aptamer concentration increases, indicating that the aptamer pairs with the complementary DNA. Thus, this complex covers the biotin molecule at the end of the DNA and inhibits binding between biotin and SA-ALP (Fig. 5B). A concentration of 10 μM aptamer was selected from a reaction efficiency and reagent saving perspective.
The colorimetric signal is largely dependent on the enzymatic reaction of ALP and its substrate; thus, the alkaline phosphatase concentration is also an important parameter for colorimetric biosensors. Then with the optimized cDNA and OTA aptamer concentration, the SA-ALP concentration was titrated. Fig. 5C shows a significant increase in UV-visible absorbance with increasing ALP concentration. A high alkaline phosphatase concentration leads to a higher background; thus, 1.5 mg L−1 was used for subsequent reactions.
Analytical performance of biotin-cDNAOTA-Au/Fe3O4 NP-based OTA aptasensor
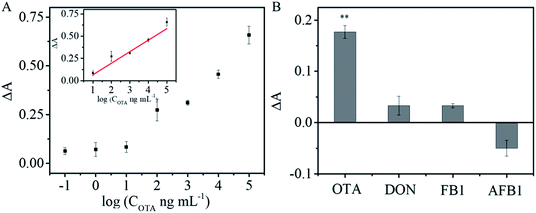
Different concentrations of OTA were introduced into a mixture of OTA aptamer, SA-ALP, and biotin-cDNA-AuNPs/Fe3O4. Fig. 6A shows that the intensity of the absorption at 405 nm increases successively upon incubation with an increasing amount of OTA. A linear relationship was obtained between ΔA and the logarithm of the OTA concentrations (inset in Fig. 6A): ΔA = 0.064 + 0.13 log(COTA ng mL−1) (R2 = 0.95147) from 10 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ng mL−1. The detection limit of 1.15 ng mL−1 was calculated as 3 times the standard deviation (s.d.) of the blank sample signal (s.d. = 3.13 × 10−3) divided by slope of the regression equation (slope = 0.13). The analytical performance of the proposed OTA aptasensor was compared with the previously reported colorimetric aptasensors (Table 1). This LOD is comparable with or better than those obtained using other nanomaterial-based sensors for OTA.
000 ng mL−1. The detection limit of 1.15 ng mL−1 was calculated as 3 times the standard deviation (s.d.) of the blank sample signal (s.d. = 3.13 × 10−3) divided by slope of the regression equation (slope = 0.13). The analytical performance of the proposed OTA aptasensor was compared with the previously reported colorimetric aptasensors (Table 1). This LOD is comparable with or better than those obtained using other nanomaterial-based sensors for OTA.
| Nanomaterials | Separation method | Linear range | Limit of detection | Reference |
|---|---|---|---|---|
| G-rich DNA | — | 1.1–4.03 ng mL−1 | 1.1 ng mL−1 | 36 |
| Au NPs | — | 8.06–251.88 ng mL−1 | 8.06 ng mL−1 | 37 |
| Au NPs | Centrifugal separation | — | 0.02 ng mL−1 | 28 |
| Hairpin DNA | — | 0.004–0.128 ng mL−1 | 0.004 ng mL−1 | 1 |
| G-rich DNA | — | 1.61–12.11 ng mL−1 | 1.61 ng mL−1 | 38 |
| Au/Fe3O4 | Magnetic separation | 10–100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ng mL−1 000 ng mL−1 |
1.15 ng mL−1 | This work |
Specificity is of great importance for practical applications of sensors. To evaluate the specificity of the developed aptasensor, interferences from three other common mycotoxins (AFB1, FB1, and DON) were tested under identical conditions. Negligible responses were observed from FB1, AFB1, and DON, but strong OTA signal was obvious (Fig. 6B; p < 0.01). The results demonstrate that the colorimetric aptasensor has good specificity for OTA detection due to the high binding affinity and specificity between OTA and its aptamer.
To verify applicability and accuracy, trace amounts of OTA were spiked into peanut, corn extracts and wine, and measured via the proposed colorimetric aptasensor. Table 2 shows that the recovery rates of the peanut, corn and wine samples range from 104.1% to 107.9% with coefficient of variation (CV) of 3.25–6.44%, 98.6–111.2% with the CV of 7.48–7.50%, 95.5–110.14% with the CV of 6.24–10.51%, respectively. At the same time, the samples were further tested by using commercial ELISA kit. The corresponding recovery rates are 94.2–101.1% with CV of 16.71–24.51% for peanut extracts, 95.9–103.2% with CV of 4.56–9.47% for corn extracts, respectively. However, the analytical performance with wine sample is not as good as those in testing peanut and corn extracts. The recovery rates of wine sample range from 78.5%–143.2% with the CV of 7.97–17.66%. The comparatively poor performance in testing wine sample may be caused by the polyphenolic compounds in wine, which potentially disturb signal readings. Successful detecting trace amount of OTA in peanut, corn and wine samples further demonstrated the potential of proposed aptasensor in real sample analysing.
| Sample | Spiked OTA (ng mL−1) | This method | Commercial ELISA | ||||
|---|---|---|---|---|---|---|---|
| Detected (ng mL−1) | Recovery (%) | CV (%) | Detected (ng mL−1) | Recovery (%) | CV (%) | ||
| Peanuts | 5 | 5.205 ± 0.331 | 104.1% | 6.36% | 5.054 ± 1.239 | 101.1% | 24.51% |
| 10 | 10.791 ± 0.351 | 107.9% | 3.25% | 10.269 ± 1.715 | 102.7% | 16.71% | |
| 20 | 21.081 ± 1.357 | 105.4% | 6.44% | 18.837 ± 4.017 | 94.2% | 21.33% | |
| Corns | 5 | 5.019 ± 0.376 | 100.4% | 7.50% | 4.796 ± 0.454 | 95.9% | 9.47% |
| 10 | 11.208 ± 0.839 | 112.1% | 7.48% | 10.075 ± 0.459 | 100.8% | 4.56% | |
| 20 | 19.755 ± 1.479 | 98.6% | 7.49% | 20.633 ± 1.477 | 103.2% | 7.16% | |
| Wine | 5 | 5.520 ± 0.361 | 110.4% | 6.53% | 7.159 ± 1.912 | 143.2% | 26.71% |
| 10 | 10.252 ± 0.640 | 102.5% | 6.24% | 9.230 ± 0.735 | 92.3% | 7.97% | |
| 20 | 19.100 ± 2.007 | 95.5% | 10.51% | 15.700 ± 2.722 | 78.25% | 17.66% | |
Conclusions
A biotin-cDNAOTA-Au/Fe3O4 nanohybrid-based OTA aptasensor was prepared for the colorimetric detection of OTA. The Au/Fe3O4 nanohybrid offered magnetic separation and immobilized biotin-cDNA as a probe for OTA detection. Under optimized conditions, the linear working range was 10 ng mL−1 to 100 μg mL−1, and the LOD was 1.15 ng mL−1. Trace amounts of OTA were easily detected in peanut and corn extracts confirming the potential of this technique for real sample analysis.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by National Natural Science Foundation of China (No. 31872753), the China Scholarship Council (No. 201906995004), Natural Science Foundation of Chongqing (cstc2019jcyj-msxmX0211), Applied Basic Research Program of Sichuan Province (19YJC0975, 2018GZYZF0008).Notes and references
- C. Wang, X. Dong, Q. Liu and K. Wang, Anal. Chim. Acta, 2015, 860, 83–88 CrossRef CAS PubMed.
- A. el Khoury and A. Atoui, Toxins, 2010, 2, 461–493 CrossRef CAS PubMed.
- A. Hayat, C. Yang, A. Rhouati and J. L. Marty, Sensors, 2013, 13, 15187–15208 CrossRef CAS PubMed.
- A. Pfohl-Leszkowicz and R. A. Manderville, Mol. Nutr. Food Res., 2007, 51, 61–99 CrossRef CAS PubMed.
- H. Badie Bostan, N. M. Danesh, G. Karimi, M. Ramezani, S. A. Mousavi Shaegh, K. Youssefi, F. Charbgoo, K. Abnous and S. M. Taghdisi, Biosens. Bioelectron., 2017, 98, 168–179 CrossRef CAS PubMed.
- R. K. Mishra, A. Hayat, G. Catanante, G. Istamboulie and J. L. Marty, Food Chem., 2016, 192, 799–804 CrossRef CAS PubMed.
- S. M. Sanzani, M. Reverberi, C. Fanelli and A. Ippolito, Toxins, 2015, 7, 812–820 CrossRef CAS PubMed.
- M. Blank and M. Blind, Curr. Opin. Chem. Biol., 2005, 9, 336–342 CrossRef CAS PubMed.
- L. Pussemier, J. Y. Piérard, M. Anselme, E. K. Tangni, J. C. Motte and Y. Larondelle, Food Addit. Contam., 2006, 23, 1208–1218 CrossRef CAS PubMed.
- X. Zhao, Y. Yuan, X. Zhang and T. Yue, Food Control, 2014, 46, 332–337 CrossRef CAS.
- L. C. Huang, N. Zheng, B. Q. Zheng, F. Wen, J. B. Cheng, R. W. Han, X. M. Xu, S. L. Li and J. Q. Wang, Food Chem., 2014, 146, 242–249 CrossRef CAS PubMed.
- I. Bazin, E. Nabais and M. Lopez-Ferber, Toxins, 2010, 2, 2230–2241 CrossRef CAS PubMed.
- A. Roland, P. Bros, A. Bouisseau, F. Cavelier and R. Schneider, Anal. Chim. Acta, 2014, 818, 39–45 CrossRef CAS PubMed.
- L. Yang, Y. Zhang, R. Li, C. Lin, L. Guo, B. Qiu, Z. Lin and G. Chen, Biosens. Bioelectron., 2015, 70, 268–274 CrossRef CAS PubMed.
- S. Wu, N. Duan, X. Ma, Y. Xia, H. Wang, Z. Wang and Q. Zhang, Anal. Chem., 2012, 84, 6263–6270 CrossRef CAS PubMed.
- J. Qian, X. Yang, Z. Yang, G. Zhu, H. Mao and K. Wang, J. Mater. Chem. B, 2015, 3, 1624–1632 RSC.
- Y. Song, W. Wei and X. Qu, Adv. Mater., 2011, 23, 4215–4236 CrossRef CAS PubMed.
- M. Liu, Z. Wang, T. Tan, Z. Chen, X. Mou, X. Yu, Y. Deng, G. Lu and N. He, Theranostics, 2018, 8, 5772–5783 CrossRef CAS PubMed.
- R. Huang, L. He, Y. Xia, H. Xu, C. Liu, H. Xie, S. Wang, L. Peng, Y. Liu, Y. Liu, N. He and Z. Li, Small, 2019, 15, e1900735 CrossRef PubMed.
- M. Wang, D. Yue, Q. Qiao, L. Miao, H. Zhao and Z. Xu, Chin. Chem. Lett., 2018, 29, 703–706 CrossRef CAS.
- M. Liu, A. Khan, Z. Wang, Y. Liu, G. Yang, Y. Deng and N. He, Biosens. Bioelectron., 2019, 130, 174–184 CrossRef CAS PubMed.
- Y. Liu, Y. Deng, T. Li, Z. Chen, H. Chen, S. Li and H. Liu, J. Biomed. Nanotechnol., 2018, 14, 2156–2161 CrossRef CAS PubMed.
- Q. Yuan, D. Lu, X. Zhang, Z. Chen and W. Tan, TrAC, Trends Anal. Chem., 2012, 39, 72–86 CrossRef CAS.
- S. Jiang, K. Y. Win, S. Liu, C. P. Teng, Y. Zheng and M.-Y. Han, Nanoscale, 2013, 5, 3127 RSC.
- J. Yan, L. Wang, L. Tang, L. Lin, Y. Liu and J. Li, Biosens. Bioelectron., 2015, 70, 404–410 CrossRef CAS PubMed.
- M. Gu, J. Liu, D. Li, M. Wang, K. Chi, X. Zhang, Y. Deng, Y. Ma, R. Hu and Y. Yang, Nanosci. Nanotechnol. Lett., 2019, 11, 1139–1144 CrossRef.
- Z. Wang and L. Ma, Coord. Chem. Rev., 2009, 253, 1607–1618 CrossRef CAS.
- R. Xiao, D. Wang, Z. Lin, B. Qiu, M. Liu, L. Guo and G. Chen, Anal. Methods, 2015, 7, 842–845 RSC.
- Y. Wei, J. Zhang, X. Wang and Y. Duan, Biosens. Bioelectron., 2015, 65, 16–22 CrossRef CAS PubMed.
- Z. Xi, B. Zheng and C. Wang, Nanosci. Nanotechnol. Lett., 2016, 8, 1061–1066 CrossRef.
- C. Wang, J. Qian, K. Wang, X. Yang, Q. Liu, N. Hao, C. Wang, X. Dong and X. Huang, Biosens. Bioelectron., 2016, 77, 1183–1191 CrossRef CAS PubMed.
- Y. Liu, J. Yu, Y. Wang, Z. Liu and Z. Lu, Sens. Actuators, B, 2016, 222, 797–803 CrossRef CAS.
- Y. Xing, Y. Jin, J. Si, M. Peng, X. Wang, C. Chen and Y. Cui, J. Magn. Magn. Mater., 2015, 380, 150–156 CrossRef CAS.
- X. Ren, P. Lu, R. Feng, T. Zhang, Y. Zhang, D. Wu and Q. Wei, Talanta, 2018, 188, 593–599 CrossRef CAS PubMed.
- T. Zeng, X. Zhang, Y. Ma, H. Niu and Y. Cai, J. Mater. Chem., 2012, 22, 18658 RSC.
- C. Yang, V. Lates, B. Prieto-Simon, J. L. Marty and X. Yang, Biosens. Bioelectron., 2012, 32, 208–212 CrossRef CAS PubMed.
- C. Yang, Y. Wang, J. L. Marty and X. Yang, Biosens. Bioelectron., 2011, 26, 2724–2727 CrossRef CAS PubMed.
- C. Yang, V. Lates, B. Prieto-Simon, J. L. Marty and X. Yang, Talanta, 2013, 116, 520–526 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |