Open Access Article
Open Access ArticleA label-free aptamer-based biosensor for microRNA detection by the RNA-regulated fluorescence of malachite green†
Honghong Wang ,
Hui Wang
,
Hui Wang ,
Mai Zhang,
Yuting Jia and
Zhengping Li
,
Mai Zhang,
Yuting Jia and
Zhengping Li *
*
School of Chemistry and Biology Engineering, University of Science and Technology Beijing, Beijing, 100083, P. R. China. E-mail: lzpbd@ustb.edu.cn
First published on 15th October 2019
Abstract
MicroRNAs (miRNAs) have been considered as promising molecular biomarkers for disease diagnosis, prognosis, as well as drug development. Herein, we wish to report a low background and label-free aptamer-based biosensor for miRNA assay by RNA-regulated fluorescence of malachite green (MG). In this biosensor-based strategy, target miRNA can specifically hybridize with the DNA extension template to form the T7 in vitro transcription system. Then the following transcription amplification produces a large number of MG RNA aptamers (MGAs) which light up the fluorescence of the MG, achieving significant fluorescence enhancement for miRNA quantitative analysis. The aptamer-based biosensor exhibits high sensitivity with a quite low detection limit of 10 amol target miRNA and high specificity to clearly discriminate very similar miRNA family members, even only one base difference. Furthermore, we have demonstrated that the biosensor is practical and reliable for the quantitative detection of miRNA in complex real samples.
1 Introduction
MicroRNAs (miRNAs) are a group of small, noncoding RNAs with the length of 18–25 nucleotides (nt) that play a critical role in the process of gene expression.1,2 Accumulating evidence has shown that abnormal expression levels of miRNAs are closely associated with the development of a variety of diseases including cancers.3,4 So miRNAs have been considered as promising biomarkers for disease diagnosis, prognosis, as well as drug development.5 Therefore, sensitive and selective detection of miRNAs is of great significance for clinical diagnosis and therapy. Up to now, various strategies have been developed for miRNA detection. Typically, the northern blotting and DNA microarray are widely used as gold-standard methods for miRNA expression profiling. But they have several instinctive drawbacks including limited sensitivity and specificity, time- and sample-consuming.2 In order to achieve high sensitivities, a variety of nucleic acid amplification strategies are developed for miRNA detection, such as reverse transcription-based or ligase-depended PCR,6,7 ligase-mediated rolling circle amplification (RCA),8,9 loop-mediated isothermal amplification (LAMP),10,11 and isothermal exponential amplification (EXPAR)12–14 which efficiently improve the sensitivity and selectivity of miRNAs assay. However, some of these methods require the complex design of multiple probes/primers or multiple operating steps such as reverse transcription/ligation reaction and subsequent amplification and detection processes. Among these methods, EXPAR can realize miRNA detection by one step-reaction with only one amplification template, but the sequence dependence of the EXPAR has limited its widespread use.15 The duplex specific nuclease (DSN)-assisted signal amplification16,17 and circular template-depended RCA9,18 can also realize miRNA detection with high sensitivity and relatively simplified steps, in which labeled probes are indispensable for signal readout. Therefore, a sensitive and label-free miRNA detection method with simple design and operation steps is still in great demand.Aptamers are single-stranded DNA/RNA molecules that can specifically bind a defined ligand with high affinity, which can be selected by vitro selection technique known as the Systematic Evolution of Ligands by Exponential enrichment (SELEX).19,20 SELEX is a powerful technique that mimics natural evolution in the laboratory and has selected various aptamers specifically binding to a wide-ranging of targets from small organic molecules to proteins, even cells and viruses.21 The affinity between the aptamer and the target can be comparable to that of the antibody (typically with pM affinities).22 Furthermore, aptamers can be screened under non-physiological conditions, and also synthesized easily.19–21
Due to the excellent characteristics of aptamers, many research fields about aptamer-based sensors have developed rapidly.19,20 The most design concepts of the aptamer-based sensors are to convert binding events between the aptamers and ligands into detectable signal changes, such as colorimetric,23 electrochemical,24 chemiluminescent,25 fluorescent,26,27 and phase changes.28 Among these aptamer-based sensors, fluorescent sensors have attracted more attention in biosensing because of their high sensitivity and the feasibility of quantification. However, most fluorescent aptamer sensors need to label fluorescent groups and quencher groups at the ends or active sites of aptamer molecules.29 These labeled groups will reduce the affinity of aptamers and increases the measurement cost. In addition, the quencher group cannot completely quench the fluorescence from the fluorescent group, resulting in a high background signal.30 Malachite green aptamer (MGA) is a kind of RNA aptamer that can bind with malachite green (MG) and light up the fluorescence more than 2000-fold compared to negligible fluorescence of free MG.31 The MGA-MG system represents a promising platform for the construction of low background and label-free fluorescence biosensors.
In this study, combining the MGA-MG system and T7 in vitro transcriptional amplification, we attempt to develop a low background and label-free aptamer-based biosensor for the quantitative detection of microRNA (miRNA). In this assay, target miRNA can specifically generate the T7 transcription system, which ensures the specificity of the biosensor-based miRNA detection. The miRNA-generated T7 transcription system could produce a large number of MGA resulting in fluorescence signal amplification, which guarantees the sensitivity for miRNA assay. Compared to the previously reported the Spinach aptamer-based biosensor for miRNA assay,14,32,33 the novel system requires only one DNA template, which will greatly simplify the design principle and reduce the measurement cost.
2 Results and discussion
2.1 Principle of the aptamer-based biosensor for miRNA assay
The principle of the aptamer-based biosensor for miRNA detection is schematically illustrated in Fig. 1. Let-7 miRNA acts as a tumor suppressor in human lung cancer34 and its reduced expression is closely associated with shortened postoperative survival.35 In this work, let-7a was chosen as a proof-of-concept target. First, an extension DNA template is reasonably designed containing a complementary sequence of miRNA (Anti-miRNA, red), a single-stranded T7 RNA polymerase promoter sequence (T7 RNA Pro, pink) and a complementary sequence of MG RNA aptamer (Anti-MGA, orange). The sequences of miRNA and extension DNA template are listed in Table S1 (ESI).† In the presence of the miRNA target (green), miRNAs hybridize to the anti-miRNA sequence of the extension DNA template and extension reaction is initiated by using miRNA as a primer under the action of the Bst DNA polymerase. The extension can generate a double-stranded template for T7 RNA polymerase transcription system, which can produce a large number of MG RNA aptamers (MGA) by the transcription amplification of T7 RNA polymerase. MGA can specifically bind to malachite green (MG) and restrict the vibrational de-excitation of the MG to produce fluorescence. In this way, one miRNA target can produce amounts of MGA molecules, thereby generating a signal amplification mechanism for miRNA detection. In the absence of the miRNA target, no extension reaction and transcription amplification will occur, with only free MG, thus effectively eliminating non-specific fluorescence background. In addition, the extension reaction of Bst DNA polymerase and transcription amplification of T7 RNA polymerase can be efficiently carried out under the same reaction conditions, which greatly simplifies the operation steps. Hence the measurements can be accomplished in one reaction tube.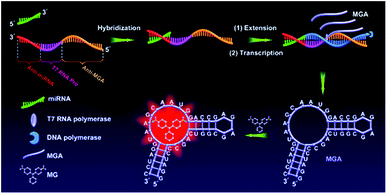 | ||
| Fig. 1 Schematic illustration of the label-free aptamer-based biosensor for miRNA detection by the RNA-regulated fluorescence of malachite green. | ||
2.2 Evaluation of the feasibility of the proposed biosensor
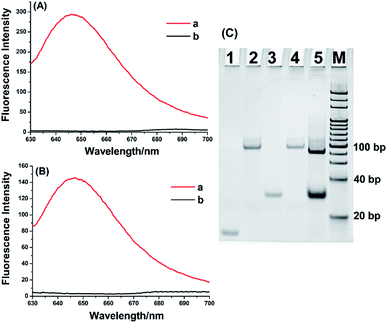
Several verification experiments were constructed to confirm the feasibility of the aptamer-based biosensor for miRNA detection. As exhibited in Fig. 2A, when synthetic MGA binding to MG, a significantly enhanced fluorescence signal can be observed (curve a). In contrast, only MG is present, no fluorescence signal can be generated (curve b). These results clearly prove that MGA can effectively limit the self-vibration de-excitation of MG and greatly enhance the fluorescence of MG. Furthermore, the proposed biosensor-based system was further verified by the fluorescence measurement after the extension reaction and transcription amplification. As demonstrated in Fig. 2B, in the presence of let-7a, the obviously enhanced fluorescence signal was observed (curve a). In contrast, no significant fluorescence signal was observed in the blank control. The system is also validated by polyacrylamide gel electrophoresis (PAGE). As shown in Fig. 2C, the bands in lane 1, lane 2, and lane 3 represent let-7a (22 nucleotides), extension DNA template (83 nucleotides), and MGA (38 nucleotides), respectively. After let-7a-primed extension and T7 RNA polymerase-catalyzed transcription, an obvious band appeared in lane 5 at the same position of lane 3, indicating that the new band was the transcribed MGA. At the same time, because the extension DNA template was extended to form an RNA–DNA duplex, a brighter and slightly faster band appeared in lane 5 near the position of the extension DNA template. All of these results indicated that the enhanced fluorescence signal is specifically derived from target miRNA and the proposed biosensor is unquestionably feasible for label-free miRNA assay.2.3 Analytical performance of the aptamer-based biosensor for miRNA detection
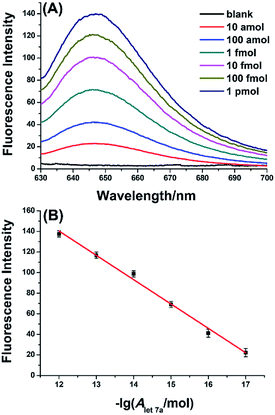
The analytical performance of the aptamer-based biosensor for miRNA detection was investigated by detecting 10-fold serial dilutions of let-7a. As depicted in Fig. 3A, the fluorescence signal significantly increases with increasing the amount of let-7a in the range from 10 amol to 1 pmol, indicating that the fluorescence signals come from the presence of the miRNA target. More importantly, Fig. 3B displays that the fluorescence intensities at 650 nm (IF) are linearly dependent on the logarithm (lg) of let-7a miRNA amount with the correlation equation of IF = 424.0 + 23.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AmiRNA(mol), where the correlation coefficient is R2 = 0.9909. The results demonstrate that the aptamer-based biosensor can quantitatively detect target miRNA with high sensitivity and a wide dynamic range spanning over 5 orders of magnitude. It is noteworthy that the sensitivity of the proposed biosensor-based miRNA assay is superior to the ligation-mediated Spinach-based fluorescent light-up biosensor,32 and is comparable to that of ligation-mediated RCA assay8 and amplified tandem Spinach-based aptamer transcription assay (AmptSpi assay).33 Unfortunately, the ligation-mediated RCA assay and the AmptSpi assay require multiple operation steps and longer analysis time. Certainly, the sensitivity is inferior to that of the duplex specific nuclease (DSN)-assisted signal amplification,16,17 circular template-depended RCA,9,18 and electrocatalysis-assisted biosensor.36 Fortunately, the sensitivity of the proposed biosensor can satisfy the needs of most miRNA-related biological studies without labeled probes.
AmiRNA(mol), where the correlation coefficient is R2 = 0.9909. The results demonstrate that the aptamer-based biosensor can quantitatively detect target miRNA with high sensitivity and a wide dynamic range spanning over 5 orders of magnitude. It is noteworthy that the sensitivity of the proposed biosensor-based miRNA assay is superior to the ligation-mediated Spinach-based fluorescent light-up biosensor,32 and is comparable to that of ligation-mediated RCA assay8 and amplified tandem Spinach-based aptamer transcription assay (AmptSpi assay).33 Unfortunately, the ligation-mediated RCA assay and the AmptSpi assay require multiple operation steps and longer analysis time. Certainly, the sensitivity is inferior to that of the duplex specific nuclease (DSN)-assisted signal amplification,16,17 circular template-depended RCA,9,18 and electrocatalysis-assisted biosensor.36 Fortunately, the sensitivity of the proposed biosensor can satisfy the needs of most miRNA-related biological studies without labeled probes.
2.4 Specificity evaluation
Owing to the similarity of miRNA sequences, especially for miRNA family members, the specificity is another essential aspect to assess a practical miRNA assay. Human let-7 family consisting of eight highly homologous miRNAs (let-7a to 7g, and let-7i), they differ from each other with only 1–4 nucleotide, which provides an ideal model system to evaluate the specificity of the proposed miRNA assay. In this study, the same amount (10 fmol) of let-7 family members were simultaneously detected by using the let-7a-specific extension DNA template. As can be seen from Fig. 4A, the fluorescence signal produced by let-7a could be well distinguished completely from the other let-7 family members with a one-base difference (let-7c, let-7e, and let-7f), two-base difference (let-7b, let-7c, let-7d, and let-7g), or four-base difference (let-7i). For this assay, the specificity depends on the target miRNA-primed specific extension reaction along with the extension DNA template. Compared with let-7a, only one base is different with let-7e and the mismatched base is located near the 5′-terminus of let-7e sequence, so let-7e produced the maximum nonspecific signal (19.5%). Meanwhile, let-7c and let-7f also have a base difference with let-7a and produce relatively small nonspecific signals (1.0% and 7.2%, respectively). In addition, except that let-7d produces 2.1% nonspecific signal, the rest of let-7 family members (let-7b, let-7g, and let-7i) all produce negligible nonspecific signals (less than 0.1%). Therefore, the proposed aptamer-based biosensor assay has high specificity for miRNA detection and can clearly discriminate very similar miRNA family members, even only one base difference.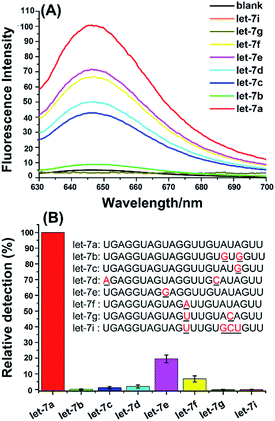 | ||
| Fig. 4 Evaluation of the specificity for miRNA detection by using the aptamer-based biosensor. (A) Fluorescence spectra of let-7 family members. The blank was treated as the same as let-7a detection except without adding any miRNAs. (B) The relative detection of human let-7 miRNA family members. The relative detection of let-7a is normalized to 100%, and the relative detection of other miRNAs is calculated by the correlation equation from Fig. 3B. Each of the miRNAs was 10 fmol. Error bars represent the standard deviation of five repetitive tests of each data. | ||
2.5 Detection of miRNA in the total RNA sample
As a promising biomarker, miRNAs expression levels have been applied for cancer diagnosis.3–5 To test the practicality of the aptamer-based biosensor for miRNA detection in complex biological samples, the aptamer-based biosensor was applied to detect the amount of let-7a in the total RNA samples extracted from A549 lung adenocarcinoma cells. As shown in Fig. 5, a well-defined fluorescence signal can be produced by the total RNA sample. With the calibration curve (Fig. 3B), the average amount of let-7a in 1 μg of the total RNA sample was estimated to be 0.29 fmol. To further confirm the reliability of the aptamer-based biosensor for miRNA detection, a spiked sample (1.0 fmol synthetic let-7a spiked into 1 μg total RNA sample) was detected by using the proposed biosensor and the determined let-7a in the spiked sample was 1.23 fmol with a satisfactory recovery of 94.2%. These results suggested that the aptamer-based biosensor is practical and reliable for quantitative detection of miRNA in complex real samples.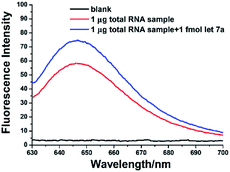 | ||
| Fig. 5 Fluorescence spectra for let-7a miRNA detection in 1 μg total RNA sample and 1.0 fmol synthetic let-7a miRNA + 1 μg total RNA sample. | ||
3 Conclusions
In conclusion, we have developed a low background label-free aptamer-based biosensor by the RNA-regulated fluorescence of MG and the novel sensor has been successfully applied to the quantitative detection of miRNA. With the amplification mechanism of the T7 in vitro transcription reaction, the proposed biosensor-based miRNA assay has high sensitivity and allows to detect as low as 10 amol miRNA target, which can generally satisfy the needs for miRNA-related biological studies. In addition, the proposed biosensor also exhibits remarkably specificity to discriminate highly similar family miRNAs, which is enough reliable to determinate the miRNA target in the complex samples. This new strategy provides an ultra-low background and fluorescent light-up platform for miRNA assay without complex nucleic acids amplification, multiple operating steps, and labeled DNA/RNA probes. By virtue of its various advantages, we believe this aptamer-based biosensor assay holds great potential in miRNA-related fundamental research. More importantly, the MGA-MG can be used as a universal signal reporting system, and thus the aptamer-based biosensor will open new opportunities for the design of more novel sensing protocols and expansion its application in different fields.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China [21705008 and 21775012], the China Postdoctoral Science Foundation [2017M620605], and the Fundamental Research Funds for the Central Universities [FRF-TP-17-046A1].Notes and references
- L. F. R. Gebert and I. J. MacRae, Nat. Rev. Mol. Cell Biol., 2019, 20, 21–37 CrossRef CAS.
- H. Dong, J. Lei, L. Ding, Y. Wen, H. Ju and X. Zhang, Chem. Rev., 2013, 113, 6207–6233 CrossRef CAS.
- M. Esteller, Nat. Rev. Genet., 2011, 12, 861–874 CrossRef CAS.
- C. P. Bracken, H. S. Scott and G. J. Goodall, Nat. Rev. Genet., 2016, 17, 719–732 CrossRef CAS.
- M. Chen, G. A. Calin and Q. H. Meng, Adv. Clin. Chem., 2014, 67, 189–214 CAS.
- C. Chen, D. A. Ridzon, A. J. Broomer, Z. Zhou, D. H. Lee, J. T. Nguyen, M. Barbisin, N. L. Xu, V. R. Mahuvakar and M. R. Andersen, Nucleic Acids Res., 2005, 33, e179 CrossRef PubMed.
- J. Zhang, Z. Li, H. Wang, Y. Wang, H. Jia and J. Yan, Chem. Commun., 2011, 47, 9465–9467 RSC.
- Y. Cheng, X. Zhang, Z. Li, X. Jiao, Y. Wang and Y. Zhang, Angew. Chem., Int. Ed., 2009, 48, 3268–3272 CrossRef CAS PubMed.
- R. Deng, L. Tang, Q. Tian, Y. Wang, L. Lin and J. Li, Angew. Chem., Int. Ed., 2014, 53, 2389–2393 CrossRef CAS.
- C. Li, Z. Li, H. Jia and J. Yan, Chem. Commun., 2011, 47, 2595–2597 RSC.
- W. Du, M. Lv, J. Li, R. Yu and J. Jiang, Chem. Commun., 2016, 52, 12721–12724 RSC.
- H. Jia, Z. Li, C. Liu and Y. Cheng, Angew. Chem., Int. Ed., 2010, 49, 5498–5501 CrossRef CAS PubMed.
- G. L. Wang and C. Y. Zhang, Anal. Chem., 2012, 84, 7037–7042 CrossRef CAS.
- M. Zhou, X. Teng, Y. Li, R. Deng and J. Li, Anal. Chem., 2019, 91, 5295–5302 CrossRef CAS.
- J. Qian, T. M. Ferguson, D. N. Shinde, A. J. Ramirez-Borrero, A. Hintze, C. Adami and A. Niemz, Nucleic Acids Res., 2012, 40, e87 CrossRef CAS.
- B. C. Yin, Y. Q. Liu and B. C. Ye, J. Am. Chem. Soc., 2012, 134, 5064–5067 CrossRef CAS.
- F. Ma, W. J. Liu, Q. Zhang and C. Y. Zhang, Chem. Commun., 2017, 53, 10596–10599 RSC.
- J. Hu, M. H. Liu and C. Y. Zhang, Chem. Sci., 2018, 9, 4258–4267 RSC.
- F. Michael, J. R. S. Hartig and M. Günter, Chem. Rev., 2007, 107, 3715–3743 CrossRef PubMed.
- J. D. Munzar, A. Ng and D. Juncker, Chem. Soc. Rev., 2019, 48, 1390–1419 RSC.
- S. M. Shamah, J. M. Healy and S. T. Cload, Acc. Chem. Res., 2008, 41, 130–138 CrossRef CAS PubMed.
- K. N. Morris, K. B. Jensen, C. M. Julin, M. Weil and L. Gold, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 2902–2907 CrossRef CAS PubMed.
- L. Juewen and L. Yi, Angew. Chem., Int. Ed., 2010, 45, 90–94 Search PubMed.
- Z. Xiaolei, X. Yi and K. W. Plaxco, J. Am. Chem. Soc., 2009, 131, 6944–6945 CrossRef PubMed.
- L. Di, S. Bella, E. Johann and W. Itamar, J. Am. Chem. Soc., 2007, 129, 5804–5805 CrossRef.
- R. Pei, J. Rothman, Y. Xie and M. N. Stojanovic, Nucleic Acids Res., 2009, 37, e59 CrossRef.
- C. C. Huang and H. T. Chang, Chem. Commun., 2008, 1461–1463 RSC.
- H. Yang, H. Liu, H. Kang and W. Tan, J. Am. Chem. Soc., 2008, 130, 6320–6321 CrossRef CAS.
- S. D. Jhaveri, R. Kirby, R. Conrad, E. J. Maglott, M. Bowser, R. T. Kennedy, G. Glick and A. D. Ellington, J. Am. Chem. Soc., 2000, 122, 2469–2473 CrossRef CAS.
- R. Nutiu and Y. Li, Methods, 2005, 37, 16–25 CrossRef CAS PubMed.
- J. R. Babendure, S. R. Adams and R. Y. Tsien, J. Am. Chem. Soc., 2003, 125, 14716–14717 CrossRef CAS.
- Z. M. Ying, B. Tu, L. Liu, H. Tang, L. J. Tang and J. H. Jiang, Chem. Commun., 2018, 54, 3010–3013 RSC.
- X. Tang, R. Deng, Y. Sun, X. Ren, M. Zhou and J. Li, Anal. Chem., 2018, 90, 10001–10008 CrossRef CAS.
- C. D. Johnson, A. Esquela-Kerscher, G. Stefani, M. Byrom, K. Kelnar, D. Ovcharenko, M. Wilson, X. Wang, J. Shelton, J. Shingara, L. Chin, D. Brown and F. J. Slack, Cancer Res., 2007, 67, 7713–7722 CrossRef CAS.
- T. Junichi, K. Hiroyuki, Y. Kiyoshi, T. Shuta, O. Hirotaka, E. Hideki, H. Tomoko, Y. Yasushi, N. Masato and N. Yuji, Cancer Res., 2004, 64, 3753–3756 CrossRef.
- L. Cui, M. Wang, B. Sun, S. Ai, S. Wang and C. Y. Zhang, Chem. Commun., 2019, 55, 1172–1175 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details. See DOI: 10.1039/c9ra07552f |
| This journal is © The Royal Society of Chemistry 2019 |