Open Access Article
Open Access ArticleGenome-based analysis of the type II PKS biosynthesis pathway of xanthones in Streptomyces caelestis and their antifungal activity†
Ling-Li Liu a,
Hong-Fei Liua,
Hua-Hua Gaoa,
Zheng-Zhong Yanga,
Xiao-Lan Fenga,
Jin-Ming Gao
a,
Hong-Fei Liua,
Hua-Hua Gaoa,
Zheng-Zhong Yanga,
Xiao-Lan Fenga,
Jin-Ming Gao *a and
Jian-Bang Zhao
*a and
Jian-Bang Zhao *b
*b
aShaanxi Key Laboratory of Natural Products & Chemical Biology, College of Chemistry & Pharmacy, Northwest A&F University, Yangling 712100, Shaanxi, People's Republic of China. E-mail: jinminggao@nwafu.edu.cn
bCollege of Information Engineering, Northwest A&F University, Yangling 712100, Shaanxi, People's Republic of China. E-mail: zhaojianbang@nwafu.edu.cn
First published on 15th November 2019
Abstract
The ethyl acetate extract from the liquid fermentation of S. caelestis Aw99c exhibited high and broad antifungal activities against plant pathogenic fungi. Bioassay guide fractionation led to the discovery of two xanthones, citreamicin ε and θ. The draft genome sequence of S. caelestis Aw99c was analyzed by a similarity-based approach to elucidate the pathway for the citreamicins. A 48 kb citreamicin (cit) gene cluster with 51 open reading frames encoding type II polyketide synthases and unique polyketide tailoring enzymes was proposed based on the genome analysis and the chemical structure derivation. In vitro antifungal assay showed that citreamicin ε exhibited significant growth inhibition against the plant pathogenic fungi with MIC values ranging from 1.56 to 12.5 μM. The cellular structural change of M. grisea treated with citreamicin ε was detected by SEM and the result showed that citreamicin ε caused disruptive surface of the mycelia.
1. Introduction
Actinomycetes are a well known microbial source for antibiotics discovery. Compared with the other source of antibiotics, the microbial source possesses much flexibility for further biotechnological developments.1,2 Two-thirds of the known antibiotics produced by microorganisms are indeed from actinomycetes, especially from the Streptomyces genus.3,4 However, more and more research indicates that such organisms have the potential to produce a far greater number of bioactive compounds than have been identified.5–7 The analysis of the gene clusters encoding various secondary metabolites provides us with a new approach to explore the potential production of bioactive compounds in actinomycetes. Recent genome projects of actinomycetes revealed that most biosynthetic pathways of bioactive compounds were associated with non-ribosomal peptide synthase (NRPS) and polyketide synthase (PKS).8–10Polyketides are a large family of secondary metabolites with a range of biological and pharmacological activities. Most polyketides are produced from short chain carboxylic molecules, such as acyl coenzyme A by actinomycetes and fungi.11,12 A typical polyketide biosynthesis gene cluster would include the core PKS synthase gene for producing the backbone of compounds and various additional genes for modification.13 Citreamicins, belonging to the family of polycyclic xanthone antibiotics, were isolated from Streptomyces sp. and active against a variety of Gram-positive bacteria, including MRSA and vancomycin-resistant Enterococcus faecalis (VRE).14–18
Plant pathogenic fungi making specific organs infection are responsible for the heavy economic loss in agriculture. Thus, different strategies have been used for the control of phytopathogenic fungi, mainly by using synthetic fungicides. Our group has been committed to find environmental friendly fungicides from natural products for many years19,20 and in our attempts to search suitable antifungal agents for control of plant pathogenic fungi, we found that the Streptomyces caelestis Aw99c isolated from the Red-Sea showed significant antifungal activity. In this study, 20 L scale fermentation of S. caelestis Aw99c in SPY media was carried out and the EtOAc extracts of the S. caelestis Aw99c was used to detect bioactivities against six plant pathogenic fungi. The followed by antifungal bioassay guide separation led to the discovery of two antifungal agents citreamicin ε and θ. We sequenced the draft genomes of the strain S. caelestis Aw99c. Herein, we reported the genome-based analysis, together with the metabolite analysis of the strain S. caelestis Aw99c and proposed a possible biosynthetic pathway for citreamicins. The antifungal activity of citreamicins was also discussed.
2. Results and discussion
2.1. Characterization of the isolated compounds
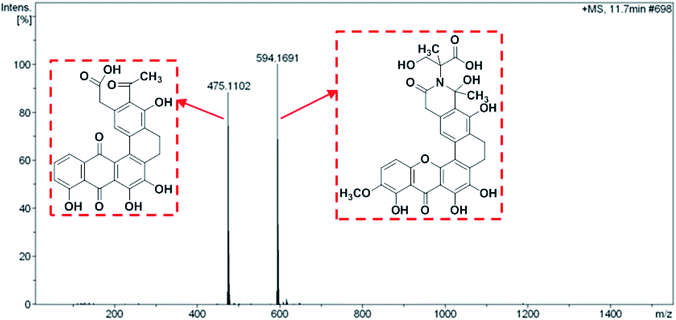
Both compound 1 and 2 were purified as a yellow powder from the extract of broth culture of S. caelestis Aw99c. The molecular formulas of both compounds were determined to be the same (C30H25NO11) based on HRESIMS, which indicated that compound 1 and 2 were isomers. In comparison with the 1H and 13C NMR spectral data of both compounds with the reported one, the structures were determined to be the polycyclic xanthones, citreamicin ε and θ, respectively.The UPLCMS analysis of the strain extract revealed mass spectra with m/z 475.1102 and 594.1691 adduct ions (Fig. 1) which provided intermediates information for proposing the biosynthetic pathway of citreamicins. Most polycyclic xanthones feature an amide ring, which is otherwise rare among aromatic polyketides. However, different polycyclic xanthones possess various substituents on the amide nitrogen, such as an amino group in simaomicin,21 or in lysolipin.22 Citreamicin ε and its derivatives feature more complicated heterocyclic structures at amide ring. Among the remaining citreamicin family compounds, citreamicin ε is unique for several reasons: the phenolic oxygen atom on the A-ring is at regioisomeric position compared to the other family members, and the D-ring contains a unit of saturation. Citreamicin θ possesses the same molecular weight with citreamicin ε and the only different is that citreamicin θ contains a unique ring system that bending with an acute angle. We suspected that they shared same biosynthetic pathway.
2.2. Antifungal activity of citreamicins against various fungal stains
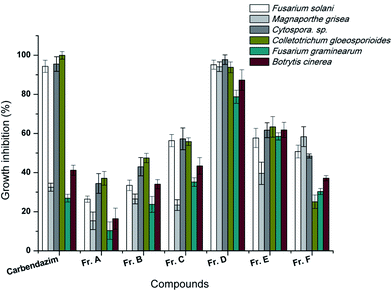
The ethyl acetate extract of the broth culture of Aw99c was fractionated to six parts by reverse phase chromatography, and then all of the six fractions were submitted to test the antifungal activity. The result showed Fr. D possessed significant activity against the six plant pathogenic fungi (Fig. 2). Over 90% of the growth of F. solani, C. sp., C. gloeosporioides, M. grisea and B. cinerea was inhibited by the treatment of Fr. D at the concentration of 50 μg mL−1, which was similar with the positive control carbendazim. The antifungal activity of Fr. D leaded to the further purification of two compounds, citreamicin ε and θ.The inhibition effects of the purified citreamicin ε and θ against growth of six plant pathogenic fungi were further detected. The result showed that fungistatic activity of citreamicin ε with the minimal inhibitory concentration (MIC) ranged from 1.56 to 12.5 μM was greater than citreamicin θ (Table 1). Citreamicin ε completely prevented the growth of Cytospora sp. and C. gloeosporioides at the concentration of 1.56 and 3.12 μM, respectively and the corresponding concentration of the positive control carbendazim was 12.5 μM. Citreamicin θ showed growth inhibition only against to F. solani and Cytospora sp. With the MIC of 50 and 25 μM, respectively. Compared with positive control carbendazim, citreamicin ε was more potent against M. grisea. Therefore, we selected M. grisea as a model organism to investigate the morphological change of the mycelia.
| Phytopathogenic fungi | Citreamicin ε (MIC, μM) | Citreamicin θ (MIC, μM) | Carbendazim (MIC, μM) |
|---|---|---|---|
| F. solani | 6.25 | 50 | 25 |
| M. grisea | 6.25 | NA | >50 |
| Cytospora sp. | 1.56 | 25 | 12.5 |
| C. gloeosporioides | 3.12 | NA | 12.5 |
| F. graminearum | 12.5 | NA | >50 |
| B. cinerea | 12.5 | NA | >50 |
2.3. Morphological change of M. grisea after treatment of citreamicin ε
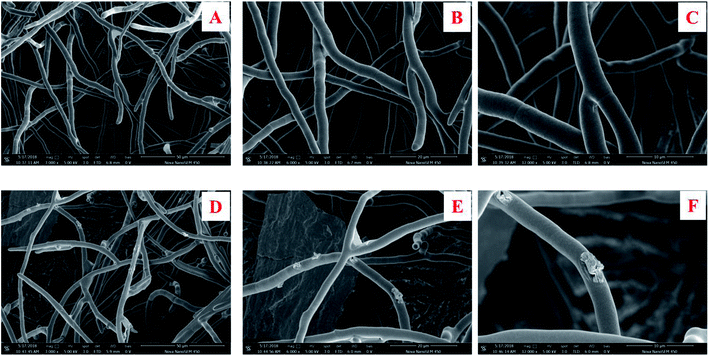
The morphology change in the surface of the M. grisea treated with citreamicin ε was further studied. M. grisea belongs to the filamentous fungi family which is characterized by the growth of hyphae. The hyphae are comprised of hypha which consisted of a chain of elongated cells encapsulated for cell wall protection. The formation of hyphae in filamentous fungi contributes to adhesion and invasion of host cells.23 The broken down of the hyphae may lead to the death of the pathogenic fungi cells.24 Since hyphae are the most common morphology observed during infections, we applied the scanning electron microscopic (SEM) to observe the surface change of the hyphae that cause by the treatment of citreamicin ε. The result of SEM on the control showed a normal smooth surface (Fig. 3A–C). Treatment of citreamicin ε on the M. grisea caused irregular branching of hyphae and disruptive surface (Fig. 3D–F). Unusual pattern of hyphal growth, as well as lyses of hyphal wall by the treatment of citreamicin ε was confirmed.2.4. The proposal of biosynthetic pathway and the identification of citreamicin related gene clusters
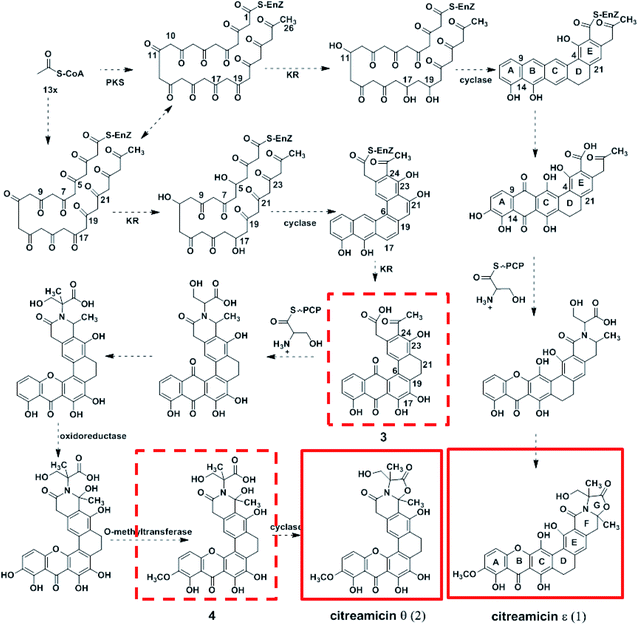
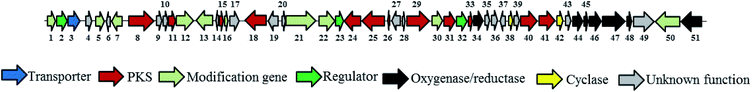
Citreamicin ε possessed basic chemical structure of seven condensed rings, including six polycyclic aromatic rings and an oxazolidone ring. Previous study showed that the nitrogen and oxygen-containing heterocycle oxazolidone ring in citreamicins may play an important role in bioactivity.18 The introduction and formation of the oxazolidone ring in polycyclic aromatic compounds attracted us to study the biosynthetic pathway of citreamicins. Based on the chemical structures of the identified compounds, a proposal regarding to the biosynthetic pathway is raised as shown in Fig. 4. The presence of aromatic rings is supposed to be catalyzed by polyketide synthase (PKS), which start with 13 acetyl-CoA. The combination of the product catalyzed by PKS and a serine give the core structure of citreamicins. The post-modification of methylation, oxidoreductase and cyclization provide the final structures of citreamicin ε and θ.In order to get more evident to support the proposal, the draft genome of the S. caelestis Aw99c was sequenced. The 7 Mb draft genome was assembled into 297 contigs with the coverage approximately >200. The G + C content was up to 74%. The antiSMASH analysis of the draft genome sequence of S. caelestis Aw99c revealed 27 gene clusters encoding secondary metabolite biosynthesis (Table S1, ESI file†). A putative type II PKS gene clusters with 34% similarity to xantholipin cluster were identified.25 Both citreamicin and xantholipin possess an amide ring and the formation of the amide ring might be catalyzed by an asparagine synthetase homolog. Thus, we searched the asparagine synthetase against to the whole draft genome of S. caelestis Aw99c. Only one hit located at contig 12 was obtained. Further analysis of the surroundings of the asparagine synthetase homolog was lead to the discovery of a 48 kb gene cluster which might associate with the production of citreamicins.
Putative functions are assigned to the 48 kb gene cluster that contains 51 ORFs as shown in Fig. 5 and Table S2.† Three cit genes encode a type II minimal PKS consisting of the polyketide bata-ketoacyl synthase alpha (KSα, cit-40), polyketide bata-ketoacyl synthase bata (KSβ, cit-41), and acyl carrier protein (ACP, cit-33). Besides, nine cit genes are supposed to relate to the PKS synthesis and two cyclases (cit-38 and 42) are surrounding the type II minimal PKS. Ten dispersed genes are suspected for post-PKS tailoring, including an asparagine synthase homolog (cit-50) for the formation of the amide ring. Four monooxygenase (cit-45, 47, 48, and 51) and three oxidoreductase genes (cit-34, 44, and 46) are identified which is consistent with the highly oxidized structure of citreamicins. In addition to the biosynthetic machinery, three regulators (cit-2, 23 and 32) and a transporter are also identified.
In type II PKS pathway, the biosynthesis of the polyketide backbone is catalyzed by the minimal polyketide synthase, which includes three components, the KSα, KSβ, and ACP. These enzymes probably duplicate enzymes from the primary metabolism, which supply malonyl-CoA for fatty acid biosynthesis.26 The minimal PKS (cit 33, 40, and 41) accepts an acetyl-CoA starter unit and condenses 13 equivalents of acetyl to afford a backbone (Fig. 4). Studies on the biosynthesis of lysolipin indicated that a Baeyer–Villiger oxidation was involved in the formation of xanthone.22 The biosynthesis study of antibiotic FD-594 provided the further evidence for the involvement of Baeyer–Villiger oxidation during the formation of xanthone.27 Therefore the dehydrogenase/reductase might catalyze these reactions. Cit-47 and Cit-51 are homologs of XanO4 which is identified to be Baeyer–Villiger monooxygenases (BVMOs).28,29 The identical between XanO4 and Cit-47 and Cit-51 is 39.7% and 39.4% respectively.
Cyclases are required to suppress spontaneous cyclization and to promote regioselectivity of intramolecular aldolcondensations.30 Due to the KSα–KSβ promotion of the regioselectivity of the first cyclization, most minimal PKSs produce C-7–C-12 cyclized polyketides as major products in the absence of additional tailoring enzymes.31 The difference between citreamicin ε and citreamicin θ is the cyclization position of the polyketide backbone. Based on isotope-feeding experiments of citreamicin, three carbonyl reductions at C-11, C-17, and C-19 were postulated to produce the hypothetical intermediate compound.32 We proposed that the carbonyl reductions of citreamicin θ were at C-5, C-11, and C-17. The cyclases then catalyze the cyclization of the polyketide backbone. As shown in Fig. 4, C-9–C-14, C-7–C-16, C-5–C-18 and C-4–C-21 cyclization make the backbone of citreamicin ε, while C-9–C-14, C-7–C-16, C-6–C-19 and C-5–C-22 cyclization make the backbone of citreamicin θ. The cyclization of citreamicin θ is different from most angucycline antibiotic gene clusters.33
Based on the structure of citreamicins, the pentacyclic intermediate undergo three hydroxylation steps at C-6, C-8, and C-17, followed by oxidation to yield the anthraquinone structure. The thioesterase (cit 31) which plays as significant role in removal of acyl carrier protein (ACP) domain in type II PKS gene clusters34 would release the molecules from the ACP.
The assembly of a cyclic amide and formation of complicated heterocyclic structures in citreamicins might catalyzed by the asparagine synthetase homolog (cit 50). The asparagine synthetase that catalyzes the conversion of aspartic acid to asparagine in an ATP dependent fashion contains an N-terminal nucleophile cysteine that hydrolyzes the amine of the nitrogen donor glutamine.35 Cit 50 was 53% identical to the type II asparagine synthetases in pradimicin biosynthesis gene cluster (PdmN). In the case of PdmN, L-Ala was used as nitrogen source instead of free NH3.36 However, in the biosynthesis of citreamicin, serine might be nitrogen source to offer the fragment assembled to the polyketone backbone. The formation of the oxazolidinone ring was achieved by the dehydroxylation between C-25 and C-29, finally providing citreamicin ε. The UPLC-MS analysis of extract of the strain revealed an intermediate compound with the molecular weight at 594.1691 (Fig. 1), which provide the evidence to confirmed that the ring closing reaction of oxazolidinone might be the final step in the biosynthesis of citreamicin ε. The compounds identified in this study are boxed in red in Fig. 4.
3. Materials and methods
3.1. Bacteria strains and culture conditions
The Streptomyces sp. was isolated from the Red Sea coastal water side of fish market near Jeddah (21′29.622N 39′09.617E). The culture was isolated by Bd Difco 212168 Actinomycete Isolation Agar. The 16S rDNA (1575 base pairs long) similarity of this strain was assessed using the BLAST program in the NCBI Database. The strain was classified as Streptomyces caelestis according to the 16S rDNA sequence (99% similarity), which was submitted to GenBank under accession number JX204833. The strain was cultured in 250 mL flasks at 25 °C for 5 days and each flask contained 100 mL SPY media (10 g L−1 of starch, 2 g L−1 of peptone, 4 g L−1 of yeast extract and 20 g L−1 sea salt) and glass beads (3 mm in diameter). The culture broth (20 L) was extracted with ethyl acetate and dried under vacuum.3.2. Compounds isolation
The ethyl acetate extract (560 mg) was submitted to reversed-phase C18 flash chromatography, eluting with solvent mixtures of H2O–MeOH (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Fr. A), H2O–MeOH (7
1, Fr. A), H2O–MeOH (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, Fr. B), H2O–MeOH (5
3, Fr. B), H2O–MeOH (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, Fr. C), H2O–MeOH (3
5, Fr. C), H2O–MeOH (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7, Fr. D), H2O–MeOH (1
7, Fr. D), H2O–MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, Fr. E), and 100% MeOH (Fr. F). Each fraction was evaporated yielding residue and submitted to test the antifungal activity. The Fr. D (152 mg) which showed potential antifungal activity was then subjected to Sephadex LH-20 eluted with MeOH to yield 14 sub-fractions (named as Fr. D-1 to Fr. D-14). Fr. D-6 (59.8 mg), which showed significant antifungal activity, was submitted to a semi-preparative RP-HPLC, eluted with MeOH/H2O, using an increasing gradient of MeOH from 50% to 100% over 20 min to afford citreamicin ε (25.6 mg) and citreamicin θ (8.4 mg).
9, Fr. E), and 100% MeOH (Fr. F). Each fraction was evaporated yielding residue and submitted to test the antifungal activity. The Fr. D (152 mg) which showed potential antifungal activity was then subjected to Sephadex LH-20 eluted with MeOH to yield 14 sub-fractions (named as Fr. D-1 to Fr. D-14). Fr. D-6 (59.8 mg), which showed significant antifungal activity, was submitted to a semi-preparative RP-HPLC, eluted with MeOH/H2O, using an increasing gradient of MeOH from 50% to 100% over 20 min to afford citreamicin ε (25.6 mg) and citreamicin θ (8.4 mg).
3.3. DNA isolation and sequencing
Isolation of genome DNA from S. caelestis Aw99c, was performed and the genome DNA was purified by precipitation in 70% EtOH. DNA sequencing was performed at the Chinese National Human Genome Center at Shanghai using the Illumina Hiseq2000 platform with 170 bp and 500 bp insert pair-end library. The 7 Mb draft genome was assembled into 297 contigs with the coverage approximately >200. The G + C content was up to 74%. The genome data was submitted to GenBank with the Bioproject ID PRJNA542982.3.4. Bioinformatics analysis of genome sequencing
Nucleotide and amino acid sequence similarity comparisons were carried out in public databases using the BLAST program. All contigs were subjected to antiSMASH (Version 4.0) (http://antismash.secondarymetabolites.org/) for the antibiotics and secondary metabolite analysis.37 The 48 kb type II PKS gene cluster cit was further analyzed using FramePlot 3.0. To understand the functions of each ORFs deduced by DNA sequencing, each ORFs was translated to its products and then submitted for BLAST search against the database to confirm the annotation.3.5. The inhibitory activity against mycelial growth of plant pathogenic fungi
Six plant pathogenic fungi Fusarium solani, Magnaporthe grisea, Cytospora sp., Colletotrichum gloeosporioides, Fusarium graminearum, Botrytis cinerea were used for bioassay. All of the fungi were provided by College of agriculture, Northwest A&F University. The growth inhibition of the test plant pathogenic fungi treated by compounds was studied. The fungus was firstly cultured on potato dextrose broth (PDB) in flask at 28 °C for 3 days. For determination of inhibitory activities, the fungal cells were smashed with vortex and filtrated with four layer gauze to afford the homogenous cell suspension. The cell density of the stock suspension was adjusted to 1 × 106 cells per mL by measuring the turbidity with a spectrophotometer (Bio-Tek, USA). The working suspension is made by a dilution of the stock suspension with PDB, which results in 5.0 × 102 cells per mL and then inoculated into 96-well flat-microtiter plates at a volume of 0.1 mL per well. The required concentrations of compounds dissolved in DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water
water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Triton X-100 (1
Triton X-100 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9
9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.01, v/v/v) and added into each well by serially diluted with PDB. The incubation of the microtiter plates was at 28 °C for 48 h and the results were measured with a spectrophotometer.38 The minimum inhibitory concentration (MIC) indicated the lowest concentrations in which no fungal growth could be observed. Each treatment was repeated for three times.
0.01, v/v/v) and added into each well by serially diluted with PDB. The incubation of the microtiter plates was at 28 °C for 48 h and the results were measured with a spectrophotometer.38 The minimum inhibitory concentration (MIC) indicated the lowest concentrations in which no fungal growth could be observed. Each treatment was repeated for three times.
3.6. The Cryo-SEM observation
The M. grisea was firstly cultured on potato dextrose agar (PDA) medium in Petri dishes for 3 days and then one discs of agar medium with fungal cultures was inoculate on PDA with or without citreamicin ε. After 3 days incubation, the PDA medium with fungal cells was cut into small pieces (1.0 cm × 1.0 cm) and transfer in a cryo-preparation chamber (PP3010T, Quorum, UK) for freezing dry. The freeze-fracture technique consists of physically breaking the frozen hydrated samples. The freezed discs were smeared on a silver stub and gold-coated by cathodic spraying. The coated samples were observed under a scanning electron microscope and digital images captured using a Nova Nona Scanning Electron Microscopic (SEM) 450 (FEI, USA) at an accelerating voltage of 5 kV.4. Conclusion
The antifungal activity of citreamicins was firstly reported based on in vitro assay and SEM observation. The biosynthetic gene cluster, which possesses a type II PKS and an asparagine synthetase, has been identified. The proposed biosynthesis pathway of citreamicins was consistent with the physical cit gene map developed from the draft genome of S. caelestis Aw99c. Characterization of the biosynthetic pathway of citreamicins provides the opportunity to understand unusual modifications involved in the biosynthesis of polycyclic xanthone antibiotics.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was co-financed by the grants from National Natural Science Foundation (21602177) and the Fundamental Research Funds for the Central Universities (Z109021508 and 2452017176). We also thank Mr Jia-Zhou Li from the Life Science Research Core Services of Northwest A&F University for providing scanning electron microscope.References
- M. W. Taylor, R. Radax, D. Steger and M. Wagner, Sponge-Associated Microorganisms: Evolution, Ecology, and Biotechnological Potential, Microbiol. Mol. Biol. Rev., 2007, 71(2), 295–347 CrossRef CAS PubMed.
- A. L. Demain and S. F. Sanchez, Microbial drug discovery: 80 years of progress, J. Antibiot., 2009, 62(1), 5–16 CrossRef CAS PubMed.
- J. Bérdy, Bioactive Microbial Metabolites, J. Antibiot., 2005, 58, 1 CrossRef PubMed.
- D. Tang, L. L. Liu, Q. R. He, W. Yan, D. Li and J. M. Gao, Ansamycins with Antiproliferative and Antineuroinflammatory Activity from Moss-Soil-Derived Streptomyces cacaoi subsp. asoensis H2S5, J. Nat. Prod., 2018, 81(9), 1984–1991 CrossRef CAS PubMed.
- N. Nakaew, S. Lumyong, W. T. Sloan and R. Sungthong, Bioactivities and genome insights of a thermotolerant antibiotics-producing Streptomyces sp. TM32 reveal its potentials for novel drug discovery, MicrobiologyOpen, 2019, e842 Search PubMed.
- B. O. Bachmann, S. G. Van Lanen and R. H. Baltz, Microbial genome mining for accelerated natural products discovery: is a renaissance in the making?, J. Ind. Microbiol. Biotechnol., 2014, 41(2), 175–184 CrossRef CAS PubMed.
- J. P. Gomez-Escribano, S. Alt and M. J. Bibb, Next Generation Sequencing of Actinobacteria for the Discovery of Novel Natural Products, Mar. Drugs, 2016, 14(4), 78 CrossRef PubMed.
- H. Komaki, N. Ichikawa, A. Oguchi, M. Hamada, T. Tamura and N. Fujita, Genome-based analysis of non-ribosomal peptide synthetase and type-I polyketide synthase gene clusters in all type strains of the genus Herbidospora, BMC Res. Notes, 2015, 8, 548 CrossRef PubMed.
- M. Nett, H. Ikeda and B. S. Moore, Genomic basis for natural product biosynthetic diversity in the actinomycetes, Nat. Prod. Rep., 2009, 26(11), 1362–1384 RSC.
- L. Liao, S. Su, B. Zhao, C. Fan, J. Zhang, H. Li and B. Chen, Biosynthetic Potential of a Novel Antarctic Actinobacterium Marisediminicola antarctica ZS314T Revealed by Genomic Data Mining and Pigment Characterization, Mar. Drugs, 2019, 17(7), 388 CrossRef PubMed.
- K. S. Masters and S. Bräse, Xanthones from Fungi, Lichens, and Bacteria: The Natural Products and Their Synthesis, Chem. Rev., 2012, 112(7), 3717–3776 CrossRef CAS PubMed.
- C. Risdian, T. Mozef and J. Wink, Biosynthesis of Polyketides in Streptomyces, Microorganisms, 2019, 7(5), 124 CrossRef PubMed.
- J. F. Sanchez, R. Entwistle, J. H. Hung, J. Yaegashi, S. Jain, Y. M. Chiang, C. C. C. Wang and B. R. Oakley, Genome-based deletion analysis reveals the prenyl xanthone biosynthesis pathway in Aspergillus nidulans, J. Am. Chem. Soc., 2011, 133(11), 4010–4017 CrossRef CAS PubMed.
- G. Carter, J. A. Nietsche, D. R. Williams and D. B. Borders, Citreamicins, novel antibiotics from Micromonospora citrea: isolation, characterization, and structure determination, J. Antibiot., 1990, 43(5), 504–512 CrossRef CAS PubMed.
- G. T. Carter, D. B. Borders, J. J. Goodman, J. Ashcroft, M. Greenstein, W. M. Maiese and C. J. Pearce, Biosynthetic origins of the polycyclic xanthone antibiotic, citreamicin, J. Chem. Soc., Perkin Trans. 1, 1991,(9), 2215–2219 RSC.
- D. C. Hopp, D. J. Milanowski, J. Rhea, D. Jacobsen, J. Rabenstein, C. Smith, K. Romari, M. Clarke, L. Francis and M. Irigoyen, Citreamicins with potent Gram-positive activity, J. Nat. Prod., 2008, 71(12), 2032–2035 CrossRef CAS PubMed.
- A. J. Peoples, Q. Zhang, W. P. Millett, M. T. Rothfeder, B. C. Pescatore, A. A. Madden, L. L. Ling and C. M. Moore, Neocitreamicins I and II, Novel Antibiotics with Activity Against Methicillin Resistant Staphylococcus aureus and Vancomycin-Resistant Enterococci, J. Antibiot., 2008, 61(7), 457 CrossRef CAS PubMed.
- L. L. Liu, Y. Xu, Z. Han, Y. X. Li, L. Lu, P. Y. Lai, J. L. Zhong, X. R. Guo, X. X. Zhang and P. Y. Qian, Four New Antibacterial Xanthones from the Marine-Derived Actinomycetes Streptomyces caelestis, Mar. Drugs, 2012, 10(11), 2571–2583 CrossRef CAS PubMed.
- J. Xiao, Q. Zhang, Y. Q. Gao, J. J. Tang, A. L. Zhang and J. M. Gao, Secondary metabolites from the endophytic Botryosphaeria dothidea of Melia azedarach and their antifungal, antibacterial, antioxidant, and cytotoxic activities, J. Agric. Food Chem., 2014, 62(16), 3584–3590 CrossRef CAS PubMed.
- H. Li, J. Wei, S.-Y. Pan, J. M. Gao and J. M. Tian, Antifungal, phytotoxic and toxic metabolites produced by Penicillium purpurogenum, Nat. Prod. Res., 2014, 28, 1–4 CrossRef PubMed.
- G. T. Carter, J. J. Goodman, M. J. Torrey, D. B. Borders and S. J. Gould, Biosynthetic origin of the carbon skeleton of simaomicin alpha., a hexacyclic xanthone antibiotic, J. Org. Chem., 1989, 54(18), 4321–4323 CrossRef CAS.
- H. Bockholt, G. Udvarnoki, J. Rohr, U. Mocek, J. M. Beale and H. G. Floss, Biosynthetic studies on the xanthone antibiotics lysolipins X and I, J. Org. Chem., 1994, 59(8), 2064–2069 CrossRef CAS.
- S. Bartnicki-Garcia and E. Lippman, Fungal morphogenesis: cell wall construction in Mucor rouxii, Science, 1969, 165(3890), 302–304 CrossRef CAS PubMed.
- N. Zhang, J. Luo and D. Bhattacharya, Chapter eight–advances in fungal phylogenomics and their impact on fungal systematics, in Advances in genetics, ed. J. P. Townsend and Z. Wang, Academic Press, 2017, vol. 100, pp. 309–328 Search PubMed.
- W. Zhang, L. Wang, L. Kong, T. Wang, Y. Chu, Z. Deng and D. You, Unveiling the Post-PKS Redox Tailoring Steps in Biosynthesis of the Type II Polyketide Antitumor Antibiotic Xantholipin, Chem. Biol., 2012, 19(3), 422–432 CrossRef CAS PubMed.
- C. Ratledge, Fatty acid biosynthesis in microorganisms being used for single cell oil production, Biochimie, 2004, 86(11), 807–815 CrossRef CAS PubMed.
- F. Kudo, T. Yonezawa, A. Komatsubara, K. Mizoue and T. Eguchi, Cloning of the biosynthetic gene cluster for naphthoxanthene antibiotic FD-594 from Streptomyces sp. TA-0256, J. Antibiot., 2011, 64(1), 123–132 CrossRef CAS PubMed.
- M. P. Beam, M. A. Bosserman, N. Noinaj, M. Wehenkel and J. r. Rohr, Crystal Structure of Baeyer–Villiger Monooxygenase MtmOIV, the Key Enzyme of the Mithramycin Biosynthetic Pathway, Biochemistry, 2009, 48(21), 4476–4487 CrossRef CAS PubMed.
- H. Kumar and M. W. Fraaije, Conversion of Furans by Baeyer–Villiger Monooxygenases, Catalysts, 2017, 7(6), 179 CrossRef.
- T. P. Korman, Y. H. Tan, J. Wong, R. Luo and S. C. Tsai, Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase, Biochemistry, 2008, 47(7), 1837–1847 CrossRef CAS PubMed.
- H. Fu, S. Ebert-Khosla, D. A. Hopwood and C. Khosla, Relaxed specificity of the oxytetracycline polyketide synthase for an acetate primer in the absence of a malonamyl primer, J. Am. Chem. Soc., 1994, 116(14), 6443–6444 CrossRef CAS.
- M. F. Reich, V. J. Lee, J. Ashcroft and G. O. Morton, An unusual cyclization product from the reaction of citreamicin η with sulfene, J. Org. Chem., 1993, 58(19), 5288–5290 CrossRef CAS.
- A. Li and J. Piel, A Gene Cluster from a Marine Streptomyces Encoding the Biosynthesis of the Aromatic Spiroketal Polyketide Griseorhodin A, Chem. Biol., 2002, 9(9), 1017–1026 CrossRef CAS PubMed.
- M. L. Heathcote, J. Staunton and P. F. Leadlay, Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units, Chem. Biol., 2001, 8(2), 207–220 CrossRef CAS PubMed.
- S. K. Boehlein, J. D. Stewart, E. S. Walworth, R. Thirumoorthy, N. G. Richards and S. M. Schuster, Kinetic Mechanism of Escherichia c oli Asparagine Synthetase B, Biochemistry, 1998, 37(38), 13230–13238 CrossRef CAS PubMed.
- B. C. Kim, J. M. Lee, J. S. Ahn and B. S. Kim, Cloning, sequencing, and characterization of the pradimicin biosynthetic gene cluster of Actinomadura hibisca P157-2, J. Microbiol. Biotechnol., 2007, 17(5), 830–835 CAS.
- K. Blin, M. H. Medema, D. Kazempour, M. A. Fischbach, R. Breitling, E. Takano and T. Weber, antiSMASH 2.0--a versatile platform for genome mining of secondary metabolite producers, Nucleic Acids Res., 2013, 41, W204–W212 CrossRef PubMed.
- Clinical and Laboratory Standards Institute, Reference Method for Broth dilution Antifungal Susceptibility Testing of Filamentous Fungi; Approved Standard, CLSI document 2008 M38-A2, Pennsylvania, 2nd edn Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra07345k |
| This journal is © The Royal Society of Chemistry 2019 |