Open Access Article
Open Access ArticleCurcumin alleviates LPS-induced inflammation and oxidative stress in mouse microglial BV2 cells by targeting miR-137-3p/NeuroD1†
Feng Gaoab,
Jing Leia,
Zhaowei Zhanga,
Yanling Yangb and
Haojun You *a
*a
aCenter for Biomedical Research on Pain (CBRP), Xi'an Jiaotong University Health Science Center, No. 76, Yantaxi Road, Xi'an, Shaanxi Province, P. R. China 710061. E-mail: yanjilinlbehl@163.com; Tel: +86-82657041
bDepartment of Physiology, School of Medicine, Yan'an University, Yan'an, Shaanxi Province, P. R. China 710061
First published on 25th November 2019
Abstract
Curcumin has been reported to exert protective effects on inflammation-related diseases, including spinal cord injury (SCI). Numerous evidence have suggested miRNAs are one of the important targets for curcumin during its anti-inflammatory function. However, little is known about the contribution of miRNAs on the role of curcumin in SCI. Thus, the objective of this study is to determine the role of miRNA (miR)-137-3p during curcumin treatment after SCI. Expression of miR-137-3p and NeuroD1 was detected using RT-qPCR and western blot assay. Inflammation and oxidative stress were measured with the protein expression levels of tumor necrosis factor (TNF)-α, interleukin (IL)-1β, and inducible nitric oxide synthase (iNOS). The target binding between miR-137-3p and NeuroD1 was confirmed via the luciferase reporter assay and RNA immunoprecipitation. LPS induced a higher expression of TNF-α, IL-1β, and iNOS in mouse microglia BV2 cells, which was attenuated by curcumin. miR-137-3p was downregulated and NeuroD1 was upregulated under LPS challenge. Curcumin also alleviated LPS-induced regulation on miR-137-3p and NeuroD1. The knockdown of miR-137-3p and ectopic expression of NeuroD1 could individually abolish the curcumin-mediated downregulation of TNF-α, IL-1β, and iNOS in LPS-challenged BV2 cells. Besides, NeuroD1 was inversely regulated by miR-137-3p via direct binding. Silencing of NeuroD1 reversed the miR-137-3p downregulation-mediated promoting effect on inflammation and oxidative stress in the presence of LPS and curcumin. Downregulation of miR-137-3p abolishes curcumin-mediated protection on LPS-induced inflammation and oxidative stress in mouse microglial BV2 cells depending on the direct upregulation of NeuroD1.
Introduction
Spinal cord injury (SCI) is a devastating condition that often affects young and healthy individuals worldwide. Most of these injuries are caused by trauma, which accounts for approximately 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 of the new cases annually.1 More than half of all SCI patients suffer cervical spine injury.2 The incidence of SCI is relatively low compared with other types of injuries or major debilitating diseases; however, the current incidence of SCI has been increasing each year.3 Moreover, SCI always results in disability, and places a huge burden on human society and a tremendous impact to the mind and body of patients.
000 of the new cases annually.1 More than half of all SCI patients suffer cervical spine injury.2 The incidence of SCI is relatively low compared with other types of injuries or major debilitating diseases; however, the current incidence of SCI has been increasing each year.3 Moreover, SCI always results in disability, and places a huge burden on human society and a tremendous impact to the mind and body of patients.
SCI consists of primary injury and secondary injury. The primary injury immediately destroys the cell membrane, myelin and axon, and microvessels, thus triggering secondary injuries, inflammation response and oxidative stress.4 Recent research in models of SCI has shown that spinal cord microglia becomes activated and underlie the pathological pain.5 Microglial cells derived from tissue monocytes differentiate into macrophages.6 Activated microglia begin infiltrating the injury site around 24 h after SCI and rapidly release cytokines, chemokines, nitric oxide (NO) and reactive oxygen species (ROS), which stimulate the inflammation cascade.1 Many studies have focused on the acute response to SCI, such as biochemical cascades that contribute to secondary damage. However, the prognosis of SCI patients is greatly determined by the inflammation response, which plays an important role in regulating the pathogenesis of SCI.7,8
Multiple neuroprotective agents have been found for SCI therapy, including curcumin. Curcumin [1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione] is a natural polyphenolic compound extracted from the rhizome of Curcuma longa L. Studies indicate that it has potent anti-cancer, anti-arthritic, and anti-diabetic activities. Recently, curcumin has emerged as a potential therapeutic drug in SCI treatment.6 In SCI, curcumin exerts a treatment effect to protect neurons and to inhibit oxidant and inflammatory reactions.9 The relevant mechanisms underlying curcumin protecting SCI include the promotion of superoxide dismutase (SOD) and GSH, and the suppression of inflammation-related factors (e.g. NF-κB), pro-inflammatory cytokines (e.g. TNF-α, IL-1β, IL-6 and RANTES), apoptosis-related genes (e.g. caspase-3/7), and oxidation-associated factors (e.g. MDA and CAT),9 as well as signaling pathways.10–13
MicroRNAs (miRNAs) are endogenous small non-coding RNAs consisting of about 22 nucleotides. It is noted that miRNAs can serve as diagnostic, prognostic and therapeutic biomarkers in diseases.14 Altered miRNA expression is closely correlated with many pathological processes, including cell proliferation, apoptosis, carcinogenesis, neuroinflammation and traumatic SCI.15,16 The imbalance of miRNA is related to a variety of central nervous system diseases, and additionally, it is also associated with glial differentiation.17,18 miRNA (miR)-137-3p, a well-documented tumor repressor, has been reported to function in SCI mice/rat models19,20 induced by spinal cord contusion, as well as in SCI cell models induced by high glucose and H2O2.21,22 Besides, the down-regulation of miR-137-3p is invariably observed in the serum from SCI patients, rat SCI models, and H2O2-induced astrocytes in vitro.19,20 However, the role of miR-137-3p and its detailed working mechanism in SCI has not been fully elucidated to date. Although evidence has suggested miRNAs as one of the important targets for curcumin when exhibiting its anti-cancer properties,23,24 little is known about the contribution of miRNAs on the role of curcumin in SCI.
Lipopolysaccharide (LPS) is the most abundant component within the cell wall of Gram-negative bacteria, which has been extensively used in models studying inflammation.25 Cell-based SCI models induced by LPS have been widely used for exploring the pathogenesis of SCI and testing new therapeutic medicine for SCI. Therefore, we herein, constructed an SCI cell injury model in mouse microglia BV2 cells challenged by LPS. The protective effect of curcumin in LPS-induced inflammation and oxidative stress was also verified. Finally, the role of miR-137-3p in the process of curcumin-mediated protection in LPS-challenged BV2 cells was investigated.
Materials and methods
Cells and cell culture
Mouse microglial BV2 cells (ATL03001) were purchased from Interlab Cell Line Collection (ICLC; Genova, Italy) and cultured in cell growth medium containing 90% high glucose Dulbecco's Modified Eagle Medium (DMEM; Gibco, Grand Island, NY, USA), 10% (v/v) fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) and 1% penicillin–streptomycin in a humidified incubator at 37 °C in 5% CO2. All BV2 cells were passaged with fresh growth medium every 3 days.Chemicals
Lipopolysaccharide (LPS; L4391) from Escherichia coli 0111:B4 and curcumin (C1386) were purchased from Sigma Aldrich (St. Louis, MO, USA). Stock solutions of 100 μg mL−1 LPS and 100 mM curcumin were prepared in cell culture medium and DMSO, respectively.LPS stimulation of microglia
For LPS stimulation, BV2 cells were cultured in 6-well plates (Corning, NY, USA) prior to different processing. BV2 cells (90% confluence) were incubated in cell growth medium and LPS was added to obtain a final concentration of 1 μg mL−1 for 48 h. The control group was cells without any treatment.Treatment of curcumin and experimental groups
BV2 cells were exposed to curcumin at concentration of 10 μM for 48 h. In the preliminary experiments, BV2 cells were divided into three groups, control (without any treatment), LPS (with 1 μg mL−1 of LPS challenge) and LPS + curcumin (with simultaneous treatment of 1 μg mL−1 of LPS and 10 μM of curcumin). For the loss-of function assays, BV2 cells were transfected with anti-miR-137-3p or anti-miR-NC, followed by treatment with LPS and curcumin. Cells in gain-of function analysis were grouped similar to that in the loss-of function assays. In the rescue experiments, BV2 cells were pretreated with co-transfection of anti-miR-137-3p and siRNA against NeuroD1 (siNeuroD1) or non-special genes (scramble), and then co-transfected cells were subjected to LPS and curcumin for 48 h. Notably, all the groups involved curcumin treatment contained less than 0.1% DMSO.Methyl thiazolyl tetrazolium (MTT) assay
The MTT assay was performed to evaluate the cell viability of BV2 cells after treatment with LPS and curcumin or not for 48 h. Subsequently, the cells were incubated with 5 mg mL−1 of MTT (Sigma Aldrich; 20 μL in FBS-free medium) for 4 h, then 100 μL of dimethyl sulfoxide (Sigma Aldrich) was added with vigorous shaking for 5 min. The optimal density (OD) at 490 nm was read on a Benchmark Plus™ microplate spectrometer (Bio-Rad, Hercules, CA, USA). The measurement for each group was repeated 4 times.Cell transfection
BV2 cells were seeded into a 6-well plate (Corning) and incubated overnight. When the cells reached 80% confluence, transient transfection was carried out with Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's instruction. The pcDNA4.1 vector was purchased from Thermo Fisher Scientific (Waltham, MA, USA). The recombinant vector pcDNA4.1-NeuroD1 was constructed. Special siRNAs against mouse NeuroD1 (siNeuroD1), mmu-miR-137-3p mimic, and anti-miR-137-3p were obtained from GenePharma (Shanghai, China), as well as their negative controls (scramble, miR-NC mimic and anti-NC). Uniformly, 30 nM of miRNA mimics, 50 nM siRNAs or 2 μg eukaryotic vectors was used for transfection. For the rescue assays, miRNA (20 nM) and vectors (1 μg) were co-transfected into BV2 cells. The transfected cells were incubated for 24 h for further study.RNA extraction and real-time quantitative PCR (RT-qPCR)
Total RNA in cultured BV2 cells was isolated in TRIzol (Beyotime, Shanghai, China). 200 ng total RNA was used as the template for reverse transcription using the Fastking RT kit (KR116; TIANGEN, Beijing, China) (for mRNA) and miRcute miRNA cDNA first strand synthesis kit (KR201; TIANGEN) (for miRNA). Quantitative PCR was performed with the Quant One-Step qRT-PCR kit (FP303; TIANGEN) and miRcute enhanced miRNA qPCR kit (FP411; TIANGEN) on an ABI 7900 real-time PCR system (Applied Biosystems, Foster City, CA, USA). GAPDH mRNA and U6 small nuclear RNA (U6) were used as the internal control to NeuroD1 mRNA and mature miR-137-3p, separately. The reactions were performed in triplicate for each sample in at least three independent runs, and the primers involved were as follows: mmu-miR-137-3p, 5′-TGACAGCGGTAGCAGAGGCAGAG-3′ (sense) and 5′-CCGCTG CCCGCCTGCCGCTGGTA-3′ (anti-sense); NeuroD1, 5′-CGAAUUUGGUGUGGCUGUA-3′ (sense) and 5′-UACAGCCACACCAAAUUCG-3′ (anti-sense); U6, 5′-CGCTTCGGCAGCACATATAC-3′ (sense) and 5′-TTCACGAATTTGCGTGTCAT-3′ (anti-sense). GAPDH, 5′-AGAACATCATCCCAGCGT-3′ (sense) and 5′-AGCCTTCACTACCCTCTTG-3′ (anti-sense). The expression levels of NeuroD1 mRNA and miR-137-3p were analyzed using the threshold cycle 2−ΔΔCt method. Results are presented as fold change normalized to the control group.Western blot assay
Total protein from cultured BV2 cells was isolated in RIPA lysis buffer (Beyotime) supplemented with phenylmethyl sulfonyl fluoride (PMSF; Sangon, Shanghai, China). The protein concentrations were determined using the Bradford protein assay reagent (Sangon). Equal amounts of protein (20 μg) from each sample were loaded for the standard procedures of the western blot assay. GAPDH on the same membrane was an internal standard to normalize the protein levels. The primary antibodies were purchased from Cell Signaling Technology (CST; Danvers, Massachusetts, USA) and as follows: TNF-α (#3707, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), IL-1β (#31202, 1
1000), IL-1β (#31202, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), iNOS (#2982, 1
1000), iNOS (#2982, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), and GAPDH (#97166, 1
1000), and GAPDH (#97166, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000). Anti-NeuroD1 (#213725, 1
1000). Anti-NeuroD1 (#213725, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) was obtained from Abcam (Cambridge, UK). Quantification of the western blot bands was performed on Image J, and the results are presented as fold change normalized to the control group.
1000) was obtained from Abcam (Cambridge, UK). Quantification of the western blot bands was performed on Image J, and the results are presented as fold change normalized to the control group.
Bioinformatics analysis
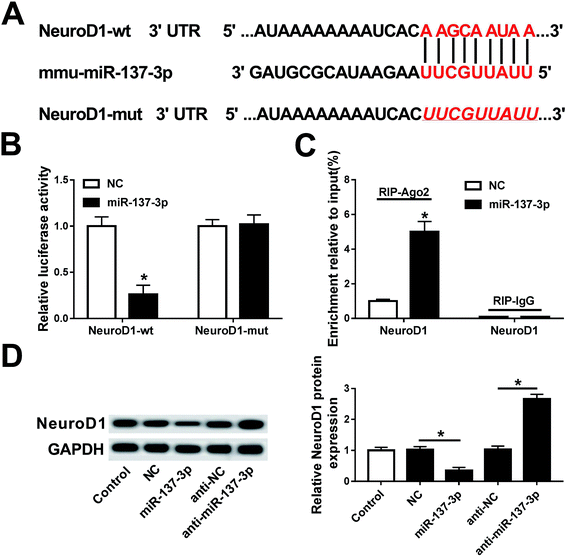
The identification of the putative miRNA target was performed using miRNA target analysis tools TargetScan Mouse Release 5.2 (http://www.targetscan.org/). With the key research word NeuroD1, several miRNAs were shown to have potential to bind to mouse NeuroD1 ENSMUST00000041099.4 3′ UTR.Luciferase reporter assay and RNA immunoprecipitation (RIP)
Considering the bioinformatics analysis, we hypothesized that NeuroD1 is a potential downstream target for miR-137-3p, and the luciferase reporter assay was adopted to verify this binding. Mouse NeuroD1 3′ UTR fragment (NeuroD1-wt) containing the potential binding sites of mmu-miR-137-3p was cloned by PCR methods into psi-CHECK vector (Invitrogen) and the mutated NeuroD1 3′ UTR sequence (NeuroD1-mut). BV2 cells were transfected according to the following groups: NeuroD1-wt + miR-NC mimic (miR-NC), NeuroD1-wt + miR-137-3p mimic (miR-137-3p), NeuroD1-mut + miR-NC, and NeuroD1-mut + miR-137-3p. The psi-CHECK vector itself provides a strong Renilla luciferase signal as normalization. After 24 h incubation, the cells were collected to measure the Firefly and Renilla luciferase activity using the dual-luciferase reporter assay system (Promega). All the data are the average of at least three independent transfections.RIP was performed in BV2 cell extract after transfection of miR-137-3p/NC. The Magna RIP™ RNA-binding protein immunoprecipitation kit (Millipore, Bradford, MA, USA) was chosen to detect the expression of NeuroD1 mRNA from the samples bound to Ago2 or IgG antibody with RT-qPCR. All operations obeyed the standard instructions.
Statistical analysis
Statistics were analyzed using SPSS 21.0 (SPSS Inc, IBM Corp. Armonk, NY, USA) and presented as the mean ± SD. The Student's t-test method was utilized for comparison between two groups, and one-way ANOVA was used for data comparison in multiple groups. P < 0.05 was considered as statistically significant.Results
Curcumin relieves LPS-induced inflammation and oxidative stress in mouse microglial BV2 cells
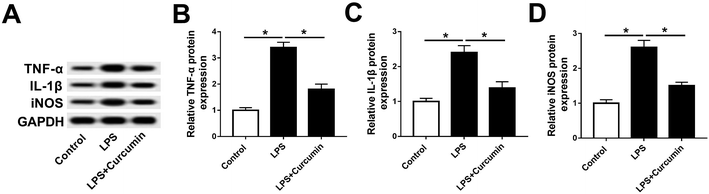
To study the effect of curcumin on the SCI-induced inflammation response and oxidative stress, we utilized mouse microglia BV2 cells stimulated by LPS to mimic SCI in vitro. According to the preliminary experiment, 1 μg mL−1 of LPS treatment for 48 h led to ∼50% cell viability inhibition, and 5–20 μM of curcumin significantly improved the LPS-induced cell injury. Moreover, 10 μM of curcumin showed the same strong protection as 20 μM (ESI Fig. 1A and B†). Thus, the concentration of LPS was fixed at 1 μg mL−1, and the concentration of curcumin was 10 μM. As shown in Fig. 1A, LPS stimulation greatly up-regulated the expression of TNF-α, IL-1β, and iNOS on protein level, which apparently decreased with the simultaneous treatment of LPS and curcumin. The quantification of western blot on Image J indicated that the alteration of the TNF-α, IL-1β, and iNOS levels was significantly different (Fig. 1B–D). This result supports the protective effect of curcumin on LPS-induced inflammation and oxidative stress in mouse microglia cells in vitro.Curcumin alleviates LPS-mediated regulation on miR-137-3p and NeuroD1 in mouse microglial BV2 cells in vitro
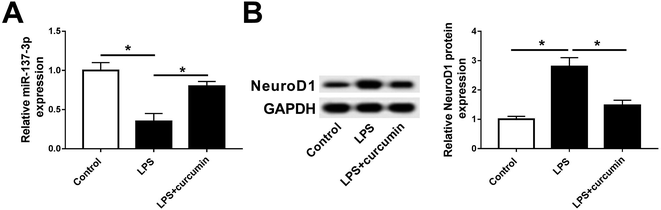
Considering the vital functions of miR-137-3p19,20,22,26 and NeuroD1 (ref. 14, 27 and 28) in the nervous system, we investigated their role in the SCI cell model. Especially, their expression levels were measured in BV2 cells in response to LPS. miR-137-3p was less expressed under LPS stimulation, which was elevated in the presence of curcumin (Fig. 2A). In the case of NeuroD1, it was up-regulated in the stress of LPS compared to the control cells. In addition, curcumin down-regulated the NeuroD1 protein level in LPS-challenged BV2 cells (Fig. 2B). These results demonstrate the aberrant expression of miR-137-3p and NeuroD1 in LPS-induced BV2 cells, suggesting their potential role in curcumin-mediated protection in microglia after SCI.Down-regulation of miR-137-3p abolishes curcumin-mediated protection on LPS-induced inflammation and oxidative stress in mouse microglial BV2 cells
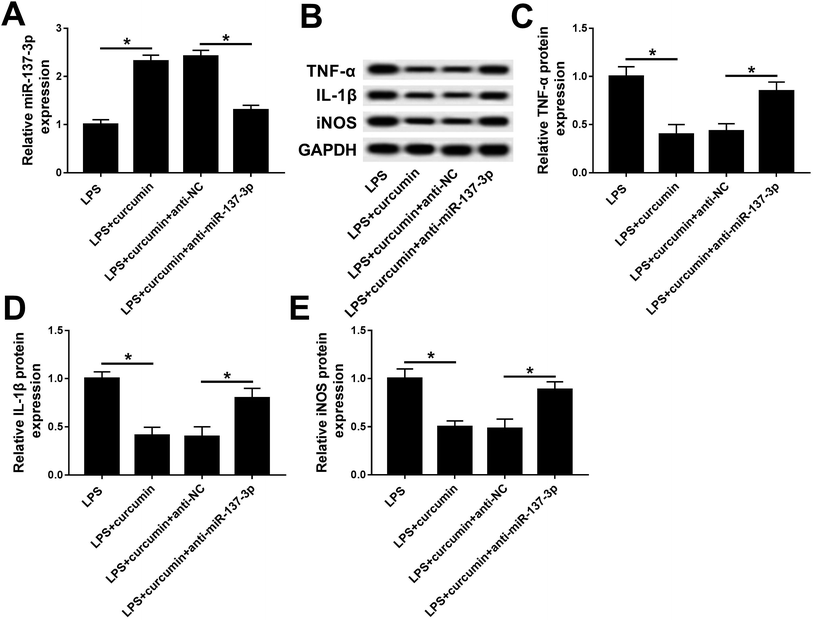
Based on the present study, we hypothesized curcumin can up-regulate miR-137-3p expression when exerting protective effects under LPS challenge. Then, consequently, we wondered whether miR-137-3p affects the role of curcumin in LPS-induced cell injury. Thus, a series of loss-of-function assays were carried out. miR-137-3p expression was knocked down in BV2 cells with transient transfection of anti-miR-137-3p. The transfection efficiency was determined by RT-qPCR (Fig. 3A). When simultaneously treated with LPS and curcumin, the anti-miR-137-3p-transfected cells expressed higher levels of TNF-α, IL-1β, and iNOS versus the anti-NC-transfected cells (Fig. 3B–E). These outcomes indicate that miR-137-3p down-regulation abolished the curcumin-mediated negative regulatory effect on TNF-α, IL-1β, and iNOS expression, implying curcumin protects LPS-induced inflammation and oxidative stress depending on the up-regulation of miR-137-3p.Ectopic expression of NeuroD1 suppresses curcumin-mediated protection on LPS-induced inflammation and oxidative stress in mouse microglial BV2 cells
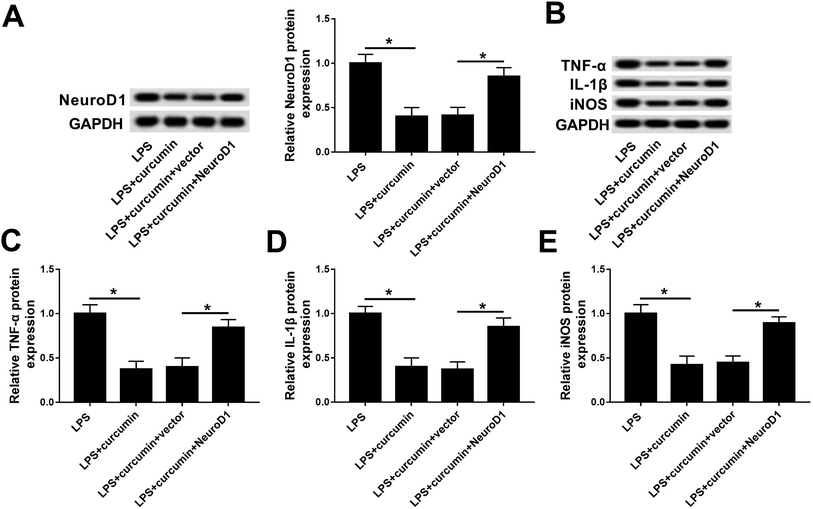
Similarly, we wondered if NeuroD1 can affect the role of curcumin in LPS-induced cell injury. Gain-of-function analyses were conducted in BV2 cells transfected with pcDNA4.1-NeuroD1 (NeuroD1) or pcDNA4.1-empty (vector). As depicted, NeuroD1 was forcedly expressed during the treatment with LPS and curcumin (Fig. 4A). Ectopic NeuroD1 up-regulated the expression of TNF-α, IL-1β, and iNOS in BV2 cells exposed to LPS and curcumin simultaneously (Fig. 4B–E). These results show that up-regulated NeuroD1 relieved the curcumin-mediated inhibition on LPS-induced inflammation and oxidative stress, namely, the protection effect of curcumin on LPS-induced inflammation and oxidative stress relies on the down-regulation of NeuroD1.miR-137-3p regulates NeuroD1 expression by target binding in mouse microglial BV2 cells
Next, the regulatory relationship between miR-137-3p and NeuroD1 was investigated in this study. Algorithm analysis by the publicly available database TargetScan Mouse was used to identify the targets of miR-137-3p. The analysis suggested that mmu-miR-137-3p had 2 potential binding sites on NeuroD1 3′ UTR: a highly conserved binding site at position 324–331 (Fig. 5A) and a poorly conserved binding site at position 1300–1306 of NeuroD1 3′ UTR (not shown). To determine whether miR-137-3p regulates NeuroD1 by binding to the 3′ UTR, we performed the luciferase reporter assay integrating sequences of the NeuroD1 3′ UTR containing the binding sites for miR-137-3p or the sequences whose binding site was mutated into a luciferase reporter vector (named NeuroD1-wt and NeuroD1-mut). BV2 cells were co-transfected with NeuroD1-wt/mut and miR-137-3p/NC. The luciferase activity was remarkably reduced when miR-137-3p and NeuroD1-wt were overexpressed; however, there was no difference in the NeuroD1-mut groups (Fig. 5B). The RNA immunoprecipitation (RIP) assay further identified the target binding of miR-137-3p and NeuroD1 (Fig. 5C). The western blot assay showed that NeuroD1 expression was inhibited by the miR-137-3p mimic and promoted by anti-miR-137-3p in the LPS-induced BV2 cells (Fig. 5D). Thus, these results support that NeuroD1 is a direct target of miR-137-3p.NeuroD1 silencing reverses miR-137-3p down-regulation effect on inflammation and oxidative stress
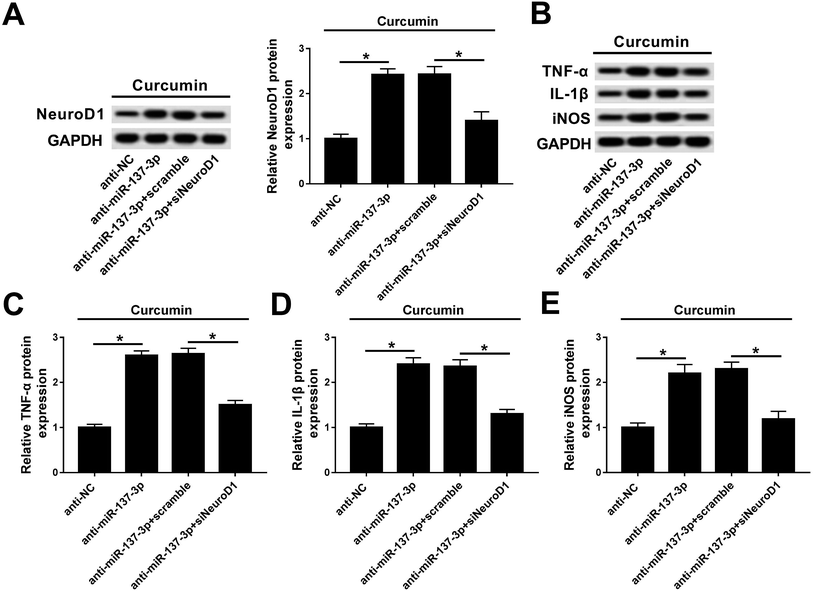
Rescue experiments were performed to clarify the activity of NeuroD1 in mediating the biological action of miR-137-3p under treatment of LPS and curcumin. BV2 cells were co-transfected with anti-miR-137-3p/NC and siNeuroD1/scramble. As shown in Fig. 6A, the NeuroD1 up-regulation induced by anti-miR-137-3p was impaired by siNeuroD1. Under the condition of miR-137-3p knockdown, TNF-α, IL-1β, and iNOS synthesis was promoted, which was blocked by silencing of NeuroD1 (Fig. 6B). The quantification of western blot was performed using Image J (Fig. 6C–E). These results indicate that the down-regulation of NeuroD1 can reverse the promotion effect of miR-137-3p knockdown on LPS-induced inflammation and oxidative stress in mouse microglia cells in vitro.Discussion
Treatment of SCI is currently a significant challenge in clinic and in research worldwide. Plant-derived medicines have gained increasing attention worldwide due to their safety, high efficiency and minimum side effects.9 Curcumin is a yellow pigment that exerts powerful anti-inflammation and anti-oxidant potential.29,30 It has been documented that curcumin protects animals and cell lines against acute toxicity. LPS mediates its toxic effects through the activation of glial cells and the generation of ROS and RNS species via NADPH oxidase activation.31 In the present study, we used LPS to induce SCI cell injury in BV2 cells. Simultaneous treatment of curcumin alleviated LPS-induced elevation of inflammatory factors synthesis and oxidative-related gene expression (TNF-α, IL-1β, and iNOS). Mechanistically, curcumin treatment rescued LPS-induced miR-137-3p expression loss in BV2 cells. Knockdown of miR-137-3p deregulated the inhibitory effect of curcumin on TNF-α, IL-1β, and iNOS expression. Simultaneously, we identified that NeuroD1 was negatively regulated by miR-137-3p through target binding. Ectopic expression of NeuroD1 abolished curcumin-mediated protection in LPS-induced BV2 cells, and silencing the expression of NeuroD1 reversed the pro-inflammation effect of miR-137-3p downregulation.In recent years, curcumin has emerged as a potential therapeutic drug in SCI treatment. After SCI, curcumin protects neurons and inhibits the inflammation response and oxidative stress.32,33 For example, Zaky et al.25 screened an overall altered expression profile in five types of let-7 miRNAs in LPS-induced and curcumin- and/or valproic acid-rat, including in the self-recovery after SCI. The anti-inflammatory effects of curcumin were uncovered in the research by Ma et al.34 to be associated with the down-regulation of miR-155 in LPS-treated macrophages and mice. Another study from Hong et al.35 discovered that the treatment of curcumin could down-regulate the expression of NF-κB/miR-155, thus inhibiting the NF-κB signal pathway and the apoptosis of extravillus trophoblast cells. To the best of our knowledge, these are the only reports descripting the role of miRNAs during curcumin functions before the present study. Here, we found that curcumin induced a higher expression of miR-137-3p during LPS-induced neuroinflammation in mouse microglia BV2 cells; moreover, the anti-inflammatory effect of curcumin relied on the miR-137-3p/NeuroD1 axis. Even though numerous articles have reported that curcumin promotes the degradation of TNF-α and IL-6 stimulated by LPS, there is scarce literature on the involvement of miRNAs when curcumin displays protective roles among diseases, including SCI and LPS-induced inflammation. Therefore, the mechanism underlying the protective activity of curcumin, especially through regulating miRNAs needs to be extensively and urgently elaborated.
The exact significance of the deregulated miRNAs after SCI is still obscure. Nevertheless, miR-137-3p was declared to be down-regulated after SCI, together with other miRNAs such as miR-138 and miR-124, and some other miRNAs were up-regulated.36 It is well accepted that miRNAs exert their functions by targeting their downstream genes by binding to some given area of the 3′ UTRs. miR-137-3p functions after SCI through direct negative regulation of several downstream target genes. For example, Wang et al.22 firstly demonstrated that heme oxygenase-1 decreased H2O2-induced spinal cord neuron injury (primary neuronal apoptosis and necrosis) through MLK3/MKK7/JNK3 signaling by downregulating Cdc42, during which miR-137-3p was the essential factor. However, in their research, they did not identify whether miR-137-3p targeted Cdc42 in primary neurons, even though the target relationship between them was widely found in H2O2-induced cardiomyocyte apoptosis37 and cancer cells.38–41 Although miR-137-3p targeting Cdc42 is likely conserved across different cell types, other target genes for this miRNA have also been reported, such as CDK6 and MITF.39,42 In SCI, miR-137-3p attenuated rat spinal cord tissue inflammation (TNF-α and IL-1) and oxidative stress (SEPN1, GPX1, iNOS, and eNOS) by targeting and modulating NeuroD4.20 Additionally, H2O2-induced astrocytes inflammation (TNF-α and IL-6) and apoptosis in C8-D1A and C8-B4 cells were inhibited by miR-137-3p/MAPK-activated protein kinase 2 (MK2).19 Tang et al.43 reported that the ventral horn of the spinal cord is associated with motoneurons degeneration and upregulation of miR-137-3p in spinal cord reduced nNOS expression and motoneurons death through inhibiting its target calpain-2 levels. In the present study, we identified a new downstream target gene, NeuroD1, in mouse microglia cells. Functionally and mechanically, miR-137-3p down-regulation promoted the synthesis of TNF-α, IL-1β, and iNOS in BV2 cells subjected to LPS and curcumin. Therefore, we believe that miR-137 targeting NeuroD1 mediates the curcumin-mediated protective effect on inflammation and oxidative stress in mouse microglia in vitro.
Neurogenic differentiation factor (Neuro) is a family of basic helix-loop-helix (bHLH) transcription factors that plays an important role in the development of neurons and reprogramming other cell types into neurons.28 After SCI, NeuroD1 was distinctively increased in the microglia.14 As a target of miR-30a-5p, silencing NeuroD1 expression can block the increase in inflammatory cytokine product and oxygen removal-related gene expression. NeuroD1 has been proposed to be essential for the survival and maturation of adult-born neurons.44 For instance, Guo et al. demonstrated that reactive glial cells in a stab-injured cortex can be directly reprogrammed into functional neurons in vivo using retroviral expression of NeuroD1.45 Recently, Chen et al. concluded that lentivirus carrying the NeuroD1 gene promoted the conversion from glial cells into neurons in a spinal cord injury model.28 In their study, NeuroD1 up-regulated cells could be reprogrammed into neural stem cells (nestin-positive), and immature (DCX-positive) and functional neurons (NeuN-positive) on days 7, 14 and 21 after SCI. In our study, NeuroD1 expression was proven to be up-regulated in LPS-induced microglia BV2 cells, and was sensitive to curcumin treatment. A higher expression of NeuroD1 eliminated the protective effect of curcumin on the synthesis of TNF-α, IL-1β, and iNOS in LPS-induced injury. Thus, all these results demonstrate that NeuroD1 may have crucial effects on spinal cord injury recovery.
Extensive pharmacological effects, low toxicity and good tolerance make curcumin a hot spot for exploration in fundamental research and clinical application. However, some studies indicate that curcumin is associated with some limitations such as low oral absorption, bio-distribution, and systemic bioavailability.46,47 These limitations have led to curcumin not being approved as a drug in clinic. It has been shown that curcumin exerts its effects by targeting a wide sequence of cellular and molecular pathways10–13 including the NF-κB, MAPK, Nrf2/ARE, and mTOR signaling pathways. However, in SCI, much more experiments should be performed to discuss the precise molecular mechanism of the “curcumin protective effect”, especially through regulating miRNAs. Therefore, the effect of curcumin on the expression of various miRNAs is an important area of scientific investigation. Based on our findings in this study, the signaling pathways underlying the curcumin/miR-137-3p/NeuroD1 axis should be revealed in inflammation response and oxidative stress after SCI, even though miR-137 and NeuroD1 take part in the MLK3/MKK7/JNK3 and MAPK/ERK signaling pathway.14,22
In summary, our research verified the protective activity of curcumin on an LPS-induced SCI cell injury model in mouse microglia BV2 cells. Curcumin treatment decreased LPS-induced inflammatory injury by up-regulating miR-137-3p and down-regulating NeuroD1. Moreover, miR-137-3p can directly negatively modulate NeuroD1 expression in LPS-induced BV2 cells. This work provides novel evidence for further understanding the anti-inflammation mechanism of curcumin and affords a theoretical basis for the development of miR-137-3p as a new biomarker for the molecular treatment of SCI.
Funding
This work was supported by the National Natural Science Foundation of China (81772451), National Natural Science Foundation of China (81760235) and Health Commission of Shaanxi Provincial (2018D080).Conflicts of interest
The authors declare that they have no conflicts of interests.References
- C. C. Lin, T. H. Chiang, W. J. Chen, Y. Y. Sun, Y. H. Lee and M. S. Lin, Injury, 2015, 46, 2341–2350 CrossRef PubMed.
- K. Stephan, S. Huber, S. Häberle, K. G. Kanz, V. Bühren, V. M. Griensven, B. Meyer, P. Biberthaler and R. Lefering, The Spine Journal, 2015, 15, 1994–2001 CrossRef PubMed.
- S. Selvarajah, E. R. Hammond, A. H. Haider, C. J. Abularrage, D. Becker, N. Dhiman, O. Hyder, D. Gupta, J. H. Black 3rd and E. B. Schneider, J. Neurotrauma, 2014, 31, 228–238 CrossRef PubMed.
- J. Chen, Z. Wang, Z. Zheng, Y. Chen, S. Khor, K. Shi, Z. He, Q. Wang, Y. Zhao, H. Zhang, X. Li, J. Li, J. Yin, X. Wang and J. Xiao, Cell Death Dis., 2017, 8, e3090 CrossRef PubMed.
- P. Zhao, S. G. Waxman and B. C. Hains, J. Neurosci., 2007, 27, 8893–8902 CrossRef CAS PubMed.
- R. Sanivarapu, V. Vallabhaneni and V. Verma, Neurology Research International, 2016, 2016, 9468193 CrossRef PubMed.
- D. J. Donnelly and P. G. Popovich, Exp. Neurol., 2008, 209, 378–388 CrossRef CAS PubMed.
- L. V. Blomster, F. H. Brennan, H. W. Lao, D. W. Harle, A. R. Harvey and M. J. Ruitenberg, Exp. Neurol., 2013, 247, 226–240 CrossRef CAS PubMed.
- Q. Zhang, H. Yang, J. An, R. Zhang, B. Chen and D. J. Hao, Evid. Based Complement Altern. Med., 2016, 2016, 8958721 Search PubMed.
- N. Zhang, G. Wei, J. Ye, L. Yang, Y. Hong, G. Liu, H. Zhong and X. Cai, Pharmacol. Rep., 2017, 69, 1001–1006 CrossRef CAS PubMed.
- J. Yuan, W. Liu, H. Zhu, Y. Chen, X. Zhang, L. Li, W. Chu, Z. Wen, H. Feng and J. Lin, Brain Res., 2017, 1655, 90–103 CrossRef CAS PubMed.
- J. Lin, X. Huo and X. Liu, BioMed Res. Int., 2017, 2017, 1634801 Search PubMed.
- E. Balogun, M. Hoque, P. Gong, E. Killeen, C. J. Green, R. Foresti, J. Alam and R. Motterlini, Biochem. J., 2003, 371, 887–895 CrossRef CAS PubMed.
- X. Fu, Y. Shen, W. Wang and X. Li, Clin. Exp. Pharmacol. Physiol., 2018, 45, 68–74 CrossRef CAS PubMed.
- Z. Shi, H. Zhou, L. Lu, X. Li, Z. Fu, J. Liu, Y. Kang, Z. Wei, B. Pan, L. Liu, X. Kong and S. Feng, Int. J. Neurosci., 2017, 127, 1104–1115 CrossRef CAS PubMed.
- B. Ning, L. Gao, R. H. Liu, Y. Liu, N. S. Zhang and Z. Y. Chen, Int. J. Biol. Sci., 2014, 10, 997–1006 CrossRef PubMed.
- L. H. Nguyen, H. J. Diao and S. Y. Chew, Adv. Drug Delivery Rev., 2015, 88, 53–66 CrossRef CAS PubMed.
- E. D. Ponomarev, T. Veremeyko and H. L. Weiner, Glia, 2013, 61, 91–103 CrossRef PubMed.
- L. Gao, C. Dai, Z. Feng, L. Zhang and Z. Zhang, J. Cell. Biochem., 2018, 119, 3280–3292 CrossRef CAS PubMed.
- J. Dai, L. J. Xu, G. D. Han, H. L. Sun, G. T. Zhu, H. T. Jiang, G. Y. Yu and X. M. Tang, Eur. Rev. Med. Pharmacol. Sci., 2018, 22, 1884–1890 CAS.
- J. Li, J. Li, T. Wei and J. Li, Cell. Physiol. Biochem., 2016, 39, 847–859 CrossRef CAS PubMed.
- S. Wang, T. Zhang, Z. Yang, J. Lin, B. Cai, Q. Ke, W. Lan, J. Shi, S. Wu and W. Lin, Apoptosis, 2017, 22, 449–462 CrossRef CAS PubMed.
- J. A. McCubrey, K. Lertpiriyapong, L. S. Steelman, S. L. Abrams, L. V. Yang, R. M. Murata, P. L. Rosalen, A. Scalisi, L. M. Neri, L. Cocco, S. Ratti, A. M. Martelli, P. Laidler, J. Dulińska-Litewka, D. Rakus, A. Gizak, P. Lombardi, F. Nicoletti, S. Candido, M. Libra, G. Montalto and M. Cervello, Aging, 2017, 9, 1477–1536 CAS.
- H. Mirzaei, A. Masoudifar, A. Sahebkar, N. Zare, J. SadriNahand, B. Rashidi, E. Mehrabian, M. Mohammadi, H. R. Mirzaei and M. R. Jaafari, J. Cell. Physiol., 2018, 233, 3004–3015 CrossRef CAS PubMed.
- A. Zaky, M. Mahmoud, D. Awad, B. M. El Sabaa, K. M. Kandeel and A. R. Bassiouny, Front. Cell. Neurosci., 2014, 8, 337 Search PubMed.
- Y. Tang, R. Fu, Z. M. Ling, L. L. Liu, G. Y. Yu, W. Li, X. Y. Fang, Z. Zhu, W. T. Wu and L. H. Zhou, Nitric Oxide, 2018, 74, 74–85 CrossRef CAS PubMed.
- M. B. Breslin, M. Zhu and M. S. Lan, J. Biol. Chem., 2003, 278, 38991–38997 CrossRef CAS PubMed.
- W. Chen, B. Zhang, S. Xu, R. Lin and W. Wang, Brain Res. Bull., 2017, 135, 143–148 CrossRef CAS PubMed.
- B. Park, S. You, W. C. S. Cho, J. Y. Choi and M. S. Lee, J. Zhejiang Univ., Sci., B, 2019, 20, 9–22 CrossRef CAS PubMed.
- L. Machova Urdzikova, K. Karova, J. Ruzicka, A. Kloudova, C. Shannon, J. Dubisova, R. Murali, S. Kubinova, E. Sykova, M. Jhanwar-Uniyal and P. Jendelova, Int. J. Mol. Sci., 2015, 17, E49 CrossRef PubMed.
- N. Sharma and B. Nehru, Neurochem. Int., 2015, 87, 92–105 CrossRef CAS PubMed.
- Q. Hao, H. W. Wang, Q. Yu, J. Shen, L. Zhao, F. F. Shi, M. M. Chen and Y. L. Yang, Zhongguo Yingyong Shenglixue Zazhi, 2017, 33, 441–444 Search PubMed.
- Y. F. Wang, J. N. Zu, J. Li, C. Chen, C. Y. Xi and J. L. Yan, Neurosci. Lett., 2014, 560, 51–56 CrossRef CAS PubMed.
- F. Ma, F. Liu, L. Ding, M. You, H. Yue, Y. Zhou and Y. Hou, Pharm. Biol., 2017, 55, 1263–1273 CrossRef CAS PubMed.
- G. M. Hong, P. Gong, M. Y. Yang, L. Shen, Z. Y. Diao and Y. L. Hu, Zhonghua Fuchanke Zazhi, 2016, 51, 859–864 CAS.
- N. K. Liu, X. F. Wang, Q. B. Lu and X. M. Xu, Exp. Neurol., 2009, 219, 424–429 CrossRef CAS PubMed.
- J. Wang, R. Xu, J. Wu and Z. Li, Med. Sci. Monit., 2015, 21, 3498–3504 CrossRef CAS PubMed.
- M. Gao, L. Liu, S. Li, X. Zhang, Z. Chang and M. Zhang, Oncol. Rep., 2015, 34, 2523–2532 CrossRef CAS PubMed.
- X. Zhu, Y. Li, H. Shen, H. Li, L. Long, L. Hui and W. Xu, FEBS Lett., 2013, 587, 73–81 CrossRef CAS PubMed.
- M. Liu, N. Lang, M. Qiu, F. Xu, Q. Li, Q. Tang, J. Chen, X. Chen, S. Zhang, Z. Liu, J. Zhou, Y. Zhu, Y. Deng, Y. Zheng and F. Bi, Int. J. Cancer, 2011, 128, 1269–1279 CrossRef CAS PubMed.
- Q. Chen, X. Chen, M. Zhang, Q. Fan, S. Luo and X. Cao, Dig. Dis. Sci., 2011, 56, 2009–2016 CrossRef CAS PubMed.
- B. Zhang, L. Ma, J. Wei, J. Hu, Z. Zhao, Y. Wang, Y. Chen and F. Zhao, Curr. Cancer Drug Targets, 2016, 16, 807–817 CrossRef CAS PubMed.
- Y. Tang, R. Fu, Z. M. Ling, L. L. Liu, G. Y. Yu, W. Li, X. Y. Fang, Z. Zhu, W. T. Wu and L. H. Zhou, Nitric Oxide, 2018, 74, 74–85 CrossRef CAS PubMed.
- Z. Gao, K. Ure, J. L. Ables, D. C. Lagace, K. A. Nave, S. Goebbels, A. J. Eisch and J. Hsieh, Nat. Neurosci., 2009, 12, 1090–1092 CrossRef CAS PubMed.
- Z. Guo, L. Zhang, Z. Wu, Y. Chen, F. Wang and G. Chen, Cell Stem Cell, 2014, 14, 188–202 CrossRef CAS PubMed.
- P. S. Lu, B. S. Inbaraj and B. H. Chen, J. Sci. Food Agric., 2018, 98, 51–63 CrossRef CAS PubMed.
- H. Mirzaei, A. Shakeri, B. Rashidi, A. Jalili, Z. Banikazemi and A. Sahebkar, Biomed. Pharmacother., 2017, 85, 102–112 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra07266g |
| This journal is © The Royal Society of Chemistry 2019 |