Open Access Article
Open Access ArticleA supramolecular hydrophobic guest transport system based on a biological macrocycle†
Yan Gea,
Xin Shena,
Hongqian Caoa,
Yuchong Haoa,
Lin Jina,
Jie Shanga,
Yangxin Wanga,
Tiezheng Pan*a and
Zhenhui Qi*ab
aSino-German Joint Research Lab for Space Biomaterials and Translational Technology, School of Life Sciences, Northwestern Polytechnical University, Xi'an, Shaanxi 710072, China. E-mail: pantiezheng@nwpu.edu.cn; qi@nwpu.edu.cn
bInstitute of Biomedical Materials & Engineering (IBME), Northwestern Polytechincial University, Xi'an, Shaanxi 710072, P. R. China
First published on 22nd November 2019
Abstract
A protein-based macrocyclic bioactive guest loading system has been developed, which not only provides a stable 10 nm scale lipophilic environment, but also increases the solubility of potent anticancer agent SN38 in its active lactone form in aqueous medium.
Macrocycle based supramolecular structures have been an intense area of research.1 In contrast to the vast number of fully organic synthetic macrocycles such as cyclodextrins,2 cucurbit[n]urils,3 pillar[n]arenes,4 calixarenes,5 and their derivatives,6 however, designing protein-based macrocycles and their corresponding guest carrier systems has largely lagged behind. Recently, protein-based macrocycles have attracted great attention owing not only to their biocompatible and biodegradable properties, but also to their precise macromolecular composition, conformational heterogeneity and abundant chemical/biological modification opportunities.7 Pioneering work from Aida,8 Liu,9 and others10 have utilized specific types of proteins (e.g. GroEL, SP1, GST) as building blocks to fabricate novel protein assemblies and functional entities. However, the construction of functional protein-based guest carrier systems by self-assembly is still a challenge for researchers. But, the design and preparation of a highly ordered protein assembly can not only help to understand the mechanism of protein nanostructures, but also provide a new way to construct functional biomaterials.9a
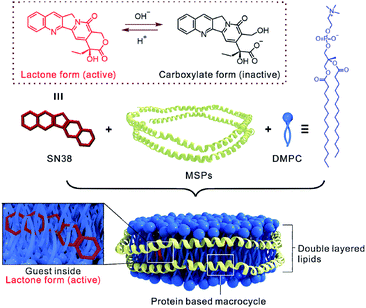
Nanodiscs (NDs) are biogenic macrocycles which are composed of phospholipids and encircling amphipathic helical belt proteins, termed membrane scaffold proteins (MSPs; Scheme 1).11 The self-assembly of NDs is originated from the robust tendency of phospholipids to form bilayers and on the enhanced stability of amphipathic helix structure of MSP due to the strong interaction with lipid acyl chains. Therefore, the internal space of NDs is filled by double-layered lipids (Scheme 1), which is dramatically different from current most water-soluble macrocycles (either organic synthetic or biogenic ones) by using water as internal medium.9a,12 Moreover, in contrast to liposome systems,13 the size of NDs is constantly monodisperse and stable at 10 nm scale, attributing to the constrained effect from coated MSPs. Consequently, NDs are found particularly useful in structural studies of membrane proteins previously, since different insoluble membrane proteins can be simultaneously monomerized, solubilized and incorporated into the well-defined membrane environment provided by NDs.14 More recently, by specific modifications of functional lipids on surface, NDs emerge as new protein-based building blocks in supramolecular polymerization,15 magnetic resonance imaging16 and light harvest device.17 Owing to the biogenicity of MSPs derives from human ApoA-I protein component of high-density lipoproteins (HDLs) particles, NDs were further found to effectively avoid systemic toxicity and autoimmunity.11b With all these abundant studies that have solidly proven that NDs represent a class of unique biological macrocycles and have immeasurable and valuable potential in biotechnological areas. Herein, we wonder whether the NDs macrocycles can be used as drug guest carrier systems to trap high-energy forms of bioactive ingredients to enhance their water-solubility and bioactivity.
SN38 (7-ethyl-10-hydroxy camptothecin, Scheme 1) is a potent antineoplastic drug belonging to the camptothecin family. Isolated from the Chinese tree Camptotheca acuminata, SN38 is up to 1000-fold more cytotoxic than the prodrug drug irinotecan. However, inherent poor water solubility and stability pose a great challenge to develop an optimal formulation of SN38.18 The lactone form of SN38 is active for its antineoplastic behavior, while its carboxylate form is inactive and favored at physiological pH, thus resulting in a loss of antineoplastic efficacy. By far, much effort has been made including using macrocycles to overcome the delivery issues of SN38 mostly through chemical modification, which transferred SN38 into water phase with multiple tedious steps for the preparation of the SN38 chemical conjugate at very low yield.19 In this study, SN38 was formulated in NDs macrocycle and characterized with respect to structural properties and biological activity without any chemical modification of SN38 or the nanodisc itself (Scheme 1).
Basically, the MSP, recombinant apolipoprotein A-I (apoA-I), was expressed in Escherichia coli and purified according to Sligar's and our own method.11a,20 SDS-PAGE showed the presence of only one protein band at ∼26 kDa, indicating that MSP was successfully purified (Fig. S1, ESI†). In the assembly of nanodisc, the SN38 DMSO solution was directly added into the lipid DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine) in NaOAc buffer (pH = 5.0), and exposed to short ultrasonication bath with 5 mg mL−1 MSP (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). The solution quickly became clear within minutes. After the dialysis of protein, DMPC and SN38 mixture overnight, the supernatant containing pure SN38-ND complex was ready for use. The transmission electron microscopy (TEM), dynamic light scattering (DLS) and size exclusion chromatography (SEC) characterizations consistently showed that the SN38-ND was successfully constructed with a diameter of 11 nm (Fig. 1).
2, v/v). The solution quickly became clear within minutes. After the dialysis of protein, DMPC and SN38 mixture overnight, the supernatant containing pure SN38-ND complex was ready for use. The transmission electron microscopy (TEM), dynamic light scattering (DLS) and size exclusion chromatography (SEC) characterizations consistently showed that the SN38-ND was successfully constructed with a diameter of 11 nm (Fig. 1).
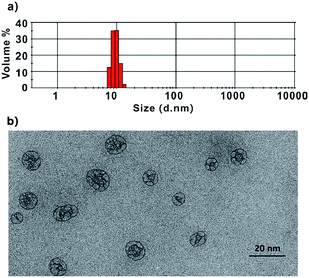 | ||
| Fig. 1 Dynamic light scattering (a) and transmission electron microscopy (TEM) image (b) of SN38-ND. | ||
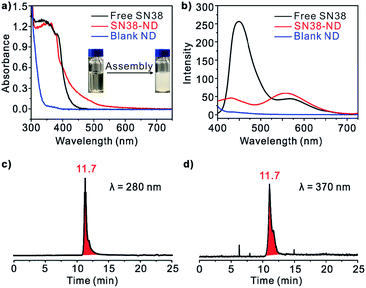
A UV-vis absorbance spectrum of SN38 gave rise to a single major peak centered at 370 nm. In comparison, the spectrum of SN38-NDs was of similar intensity to that of free SN38 in DMSO, and the SN38-ND sample became crystal clear after the assembly process, indicating the solubilization of SN38 in aqueous solution (Fig. 2a). In addition, excitation of free SN38 in DMSO gave rise to two emission peaks centered at 450 nm and 570 nm, respectively (excitation at 370 nm). SN38-NDs gave rise to an emission spectrum whose intensity was significantly attenuated and blue shifted at 450 nm compared with SN38 in DMSO, but the emission peak at 570 nm was enhanced (Fig. 2b). Moreover, the size exclusion chromatography (SEC) characterization of SN38-ND at 280 and 370 nm received the same retention time, showing that SN38 had been integrated into the NDs (Fig. 2c and d).
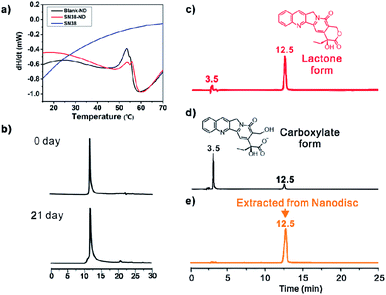
Upon formation of SN38-ND, the solubility of SN38 increased to 69.6 μg mL−1 or 177.4 μM, which was 14-fold higher than that of the native SN38 (<5 μg mL−1).21 The reason was that the phospholipid in the center of NDs offered a lipophilic environment for SN38. At the same time, the concentration of ND was 76.6 μM, indicating that there were ca. 2.3 molecules of SN38 in a single ND. Compared with liposomes, the smaller size and monodisperse characteristic of SN38-ND are beneficial for cell internalization. More importantly, SN38-ND was proven to be very stable and to maintain its structural integrity for as long as 3 weeks at 4 °C (Fig. 3a and S2†). Given the remarkable stability upon storage in the refrigerator of the HB-containing NDs, further evaluation of their thermal stability was achieved by differential scanning calorimetry (DSC). Heating curves of the empty ND (blank ND) and the HB-ND complex were recorded at a scan rate of 10 °C min−1 (Fig. 3b). The phase-transition temperature (Tm) of SN38-ND appeared at a similar position (53.5 °C) as that of the empty ND (Tm ≈ 53 °C), demonstrating that the SN38 loading did not interface the stability of the ND to a large extent (Fig. 3b).
Apart from the enhancement in solubility of SN38, NDs also significantly improved the stability of SN38 from hydrolysis in physiological environment. For free SN38, most of SN38 in the lactone form was transformed into the carboxylate form within 2 h in 20 mM PBS buffer at pH = 7.4. However, the hydrolysis rate greatly slowed upon formation of SN38-ND (less than 5% SN38 was hydrolyzed into carboxylate form, Fig. 3c–e), because SN38 deeply penetrated into the hydrophobic cavity of NDs, inhibiting the lactone ring opening. Because of the noncovalent complexation, the lactone form of SN38 was greatly maintained. Negligible carboxylate forms of SN38 were detected in the same period of time. This observation confirmed that the antineoplastic efficacy could be effectively preserved by the formation of SN38-ND. The 31P NMR spectra further indicated that the encapsulation of SN38 did not affect the polar region of bilayer lipids in NDs, since no significant chemical shifts were observed (Fig. S3, ESI†).
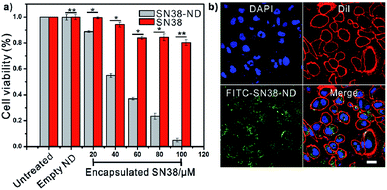
Cell viability assay (CCK-8) was used to assess the antineoplastic efficacy of the SN38-NDs (Fig. 4a). MCF-7 cells were treated with the solutions of free SN38 and SN38-ND at concentrations from 20 to 100.0 μM and the incubation time was 24 hours. As minimal changes of cell viability and cell proliferation occurred in the empty ND group, NDs showed great biocompatibility to the cell lines. On the other hand, the viability of cells incubated with free SN38/SN38-ND decreased with the increasing concentration. However, free SN38 showed limited antineoplastic efficacy because the cultivation pH was 7.4 when SN38 would transform into carboxylate form. By contrast, SN38-ND exhibited obvious antineoplastic efficacy with an IC50 value of 48 μM, this could be attributed to that SN38 remained lactone form inside NDs. These results demonstrated SN38-ND not only maintained the bioactivity of the antineoplastic drug but also enhanced the pH-stability of SN38.
Furthermore, confocal laser scanning microscopy (CLSM) was used to assess the internalization ability of the SN38-NDs. Free particles outside the plasma membrane were removed and the live cells were imaged by one-photon fluorescence at multitrack mode. Green fluorescence signal arising from FITC labelled SN38-NDs was observed in cytoplasm after culturing the MCF-7 cells with SN38-NDs for 6 h. The fluorescence and luminescence signals in Fig. 4b appear to be strongly localized at the area of treated cells, thus suggesting a substantial degree of interactions between cells and these particles. The green dots scattered in the cytoplasm and on the membrane indicated that the SN38-NDs were effectively internalized. Also, three-dimensional image (Fig. S4 ESI†) provides additional evidence for endocytosis of SN38-NDs according to the distribution of the green and red channels.
In summary, we fabricated a 10 nm scale biological macrocycle (nanodisc) equipped with antineoplastic agent through a supramolecular strategy. Differing from previous reported organic macrocycles systems, nanodisc macrocycle is absolutely based on protein and natural lipids. Taking advantage of the unique bilayer lipid environment in nanodisc macrocycle, the solubility enhancement ability of nanodisc for hydrophobic anticancer agent SN38 does not require any chemical modification, thus effectively avoiding multiple tedious steps and antineoplastic activity loss of SN38. Considering the increasing interest in functional biological macrocyclic self-assemblies, our results would be inspirational for designing intelligent biological nanocarrier system.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge financial support from Postdoctoral Innovative Talents Supporting Project (BX20180255) of China, Shaanxi Province (2019KW-031, 2019KW-038, 2018JQ2017), Fundamental Research Funds for the Central Universities (3102018jcc007, 3102017OQD044, 3102017OQD040, 3102017OQD115, 3102019ghxm005). We thank the Analytical & Testing Center of NPU for the characterization of materials.Notes and references
- (a) Z. Laughrey and B. C. Gibb, Chem. Soc. Rev., 2011, 40, 363 RSC; (b) K. I. Assaf and W. M. Nau, Chem. Soc. Rev., 2015, 44, 394 RSC; (c) J. A. Foster and J. W. Steed, Angew. Chem., Int. Ed., 2010, 49, 6718–6724 CrossRef CAS PubMed; (d) H. Wang, X. Ji, Z. Li and F. Huang, Adv. Mater., 2017, 29, 1606117 CrossRef; (e) X. Wang, Y. Han, Y. Liu, G. Zou, Z. Gao and F. Wang, Angew. Chem., Int. Ed., 2017, 56, 12466 CrossRef CAS; (f) H. Wang, H. Zhu, W. Fu, Y. Zhang, B. Xu, F. Gao, Z. Cao and W. Liu, Macromol. Rapid Commun., 2017, 38, 1600695 CrossRef; (g) X. Ji, R. T. Wu, L. Long, C. Guo, N. M. Khashab, F. Huang and J. L. Sessler, J. Am. Chem. Soc., 2018, 140, 2777 CrossRef CAS; (h) E. A. Appel, X. J. Loh, S. T. Jones, F. Biedermann, C. A. Dreiss and O. A. Scherman, J. Am. Chem. Soc., 2012, 134, 11767 CrossRef CAS; (i) X. Ma and H. Tian, Acc. Chem. Res., 2014, 47, 1971 CrossRef CAS PubMed; (j) M. Ni, N. Zhang, W. Xia, X. Wu, C. Yao, X. Liu, X. Hu, C. Lin and L. Wang, J. Am. Chem. Soc., 2016, 138, 6643 CrossRef CAS PubMed; (k) C. Zhang, B. Ou, S. Jiang, G. Yin, L. Chen, L. Xu, X. Li and H. Yang, Polym. Chem., 2018, 9, 2021 RSC; (l) Y. Zheng, D. Wang, J. Cui, M. Mezger, G. K. Auernhammer, K. Koynov, H. J. Butt and T. Ikeda, Macromol. Rapid Commun., 2018, 39, 1800282 CrossRef PubMed; (m) N. Song, D. Chen, M. Xia, X. Qiu, K. Ma, B. Xu, W. Tian and Y. Yang, Chem. Commun., 2015, 51, 5526 RSC; (n) C. Sun, H. Peng, L. Niu, Y. Chen, L. Wu, C. Tung and Q. Yang, Chem. Commun., 2018, 54, 1117 RSC; (o) J. Zhang, J. Zhu, C. Lu, Z. Gu, T. He, A. Yang, H. Qiu, M. Zhang and S. Yin, Polym. Chem., 2016, 7, 4317 RSC; (p) X. Yan, T. R. Cook, J. B. Pollock, P. Wei, Y. Zhang, Y. Yu, F. Huang and P. J. Stang, J. Am. Chem. Soc., 2014, 136, 4460 CrossRef CAS; (q) X. Hou, C. Ke, C. J. Bruns, P. R. McGonigal, R. B. Pettman and J. F. Stoddart, Nat. Commun., 2015, 6, 6884 CrossRef PubMed; (r) L. Ma, S. Wang, C. Li, D. Cao, T. Li and X. Ma, Chem. Commun., 2018, 54, 2405 RSC; (s) H. Chen, J. Fan, X. Hu, J. Ma, S. Wang, J. Li, Y. Yu, X. Jia and C. Li, Chem. Sci., 2015, 6, 197 RSC.
- (a) Q.-D. Hu, G.-P. Tang and P. K. Chu, Acc. Chem. Res., 2014, 47, 2017 CrossRef CAS; (b) C. O. Mellet, J. M. G. Fernández and J. M. Benito, Chem. Soc. Rev., 2011, 40, 1586 RSC; (c) Y. Liu, Z.-L. Yu, Y.-M. Zhang, D.-S. Guo and Y.-P. Liu, J. Am. Chem. Soc., 2008, 130, 10431 CrossRef CAS PubMed.
- (a) K. Kim, Chem. Soc. Rev., 2002, 31, 96 RSC; (b) B. Vinciguerra, L. Cao, J. R. Cannon, P. Y. Zavalij, C. Fenselau and L. Isaacs, J. Am. Chem. Soc., 2012, 134, 13133 CrossRef CAS PubMed; (c) D. Ma, G. Hettiarachchi, D. Nguyen, B. Zhang, J. B. Wittenberg, P. Y. Zavalij, V. Briken and L. Isaacs, Nat. Chem., 2012, 4, 503 CrossRef CAS PubMed.
- (a) X.-B. Hu, Z. Chen, G. Tang, J.-L. Hou and Z.-T. Li, J. Am. Chem. Soc., 2012, 134, 8384 CrossRef CAS; (b) Y. Yao, M. Xue, J. Chen, M. Zhang and F. Huang, J. Am. Chem. Soc., 2012, 134, 15712 CrossRef CAS PubMed; (c) H. Zhang, X. Ma, K. T. Nguyen and Y. Zhao, ACS Nano, 2013, 7, 7853 CrossRef CAS PubMed; (d) W. Si, Z. Li and J. Hou, Angew. Chem., Int. Ed., 2014, 53, 4578 CrossRef CAS PubMed; (e) X. Hu, K. Jia, Y. Cao, Y. Li, S. Qin, F. Zhou, C. Lin, D. Zhang and L. Wang, Chem.–Eur. J., 2015, 21, 1208 CrossRef CAS PubMed; (f) Z. Qi, K. Achazi, R. Haag, S. Dong and C. Schalley, Chem. Commun., 2015, 51, 10326 RSC; (g) L. Gao, B. Zheng, W. Chen and C. A. Schalley, Chem. Commun., 2015, 51, 14901 RSC; (h) K. Yang, Y. Chang, J. Wen, Y. Lu, Y. Pei, S. Cao, F. Wang and Z. Pei, Chem. Mater., 2016, 28, 1990 CrossRef CAS; (i) L. Rui, Y. Xue, Y. Wang, Y. Gao and W. Zhang, Chem. Commun., 2017, 53, 5126 RSC; (j) L. Shuangguan, Q. Chen, B. Shi and F. Huang, Chem. Commun., 2017, 53, 9749 RSC.
- (a) J. S. Kim and D. T. Quang, Chem. Rev., 2007, 107, 3780 CrossRef CAS PubMed; (b) D.-S. Guo and Y. Liu, Chem. Soc. Rev., 2012, 41, 5907 RSC.
- (a) H. Ke, L.-P. Yang, M. Xie, Z. Chen, H. Yao and W. Jiang, Nat. Chem., 2019, 11, 470 CrossRef CAS PubMed; (b) X. Chi, G. M. Peters, C. Brockman, V. M. Lynch and J. L. Sessler, J. Am. Chem. Soc., 2018, 140, 13219 CrossRef CAS PubMed; (c) X. Chi, W. Cen, J. A. Queenan, L. Long, V. M. Lynch, N. M. Khashab and J. L. Sessler, J. Am. Chem. Soc., 2019, 141, 6468 CrossRef CAS PubMed.
- (a) E. J. Lee, N. K. Lee and I. S. Kim, Adv. Drug Delivery Rev., 2016, 106, 157 CrossRef CAS PubMed; (b) B. J. G. E. Pieters, M. B. Van Eldijk, R. J. M. Nolte and J. Mecinović, Chem. Soc. Rev., 2016, 45, 24 RSC; (c) N. F. Steinmetz, V. Hong, E. D. Spoerke, P. Lu, K. Breitenkamp, M. G. Finn and M. Manchester, J. Am. Chem. Soc., 2009, 131, 17093 CrossRef CAS PubMed.
- (a) S. Sim and T. Aida, Acc. Chem. Res., 2017, 50, 492 CrossRef CAS PubMed; (b) S. Biswas, K. Kinbara, T. Niwa, H. Taguchi, N. Ishii, S. Watanabe, K. Miyata, K. Kataoka and T. Aida, Nat. Chem., 2013, 5, 613 CrossRef CAS PubMed.
- (a) Q. Luo, C. Hou, Y. Bai, R. Wang and J. Liu, Chem. Rev., 2016, 116, 13571 CrossRef CAS PubMed; (b) Y. Bai, Q. Luo and J. Liu, Chem. Soc. Rev., 2016, 45, 2756 RSC; (c) Q. Luo, Z. Dong, C. Hou and J. Liu, Chem. Commun., 2014, 50, 9997 RSC; (d) L. Miao, J. Han, H. Zhang, L. Zhao, C. Si, X. Zhang, C. Hou, Q. Luo, J. Xu and J. Liu, ACS Nano, 2014, 8, 3743 CrossRef CAS PubMed; (e) Y. Bai, Q. Luo, W. Zhang, L. Miao, J. Xu, H. Li and J. Liu, J. Am. Chem. Soc., 2013, 135, 10966–10969 CrossRef CAS PubMed.
- (a) J. R. Petersburg, J. Shen, C. M. Csizmar, K. A. Murphy, J. Spanier, K. Gabrielse, T. S. Griffith, B. Fife and C. R. Wagner, ACS Nano, 2018, 12, 6563 CrossRef CAS PubMed; (b) W. Wang, L. Wang, H. Chen, J. Zang, X. Zhao, G. Zhao and H. Wang, J. Am. Chem. Soc., 2018, 140, 14078 CrossRef CAS PubMed; (c) X. Gao, S. Yang, C. Zhao, Y. Ren and D. Wei, Angew. Chem., Int. Ed., 2014, 53, 14027 CrossRef CAS PubMed.
- (a) I. G. Denisov and S. G. Sligar, Chem. Rev., 2017, 117, 4669 CrossRef CAS PubMed; (b) R. Kuai, X. Sun, W. Yuan, Y. Xu, A. Schwendeman and J. J. Moon, Bioconjugate Chem., 2018, 29, 771 CrossRef CAS PubMed.
- F. Biedermann, W. M. Nau and H.-J. Schneider, Angew. Chem., Int. Ed., 2014, 53, 11158 CrossRef CAS PubMed.
- (a) I. G. Denisov, Y. V. Grinkova, A. A. Lazarides and S. G. Sligar, J. Am. Chem. Soc., 2004, 126, 3477 CrossRef CAS PubMed; (b) N. Skar-Gislinge, N. T. Johansen, R. Hoiberg-Nielsen and L. Arleth, Langmuir, 2018, 34, 12569 CrossRef CAS PubMed.
- F. Hagn, M. L. Nasr and G. Wagner, Nat. Protoc., 2018, 13, 79 CrossRef CAS PubMed.
- P. A. Beales, N. Geerts, K. K. Inampudi, H. Shigematsu, C. J. Wilson and T. K. Vanderlick, J. Am. Chem. Soc., 2013, 135, 3335 CrossRef CAS PubMed.
- C. E. Carney, I. L. Lenov, C. J. Baker, K. W. Macrenaris, A. L. Eckermann, S. G. Sligar and T. J. Meade, Bioconjugate Chem., 2015, 26, 899 CrossRef CAS PubMed.
- M. H. Ham, J. H. Choi, A. A. Boghossian, E. S. Jeng, R. A. Graff, D. A. Heller, A. C. Chang, A. Mattis, T. H. Bayburt, Y. V. Grinkova, A. S. Zeiger, K. J. Van Vliet, E. K. Hobbie, S. G. Sligar, C. A. Wraight and M. S. Strano, Nat. Chem., 2010, 2, 929 CrossRef CAS PubMed.
- (a) J. Cheng, K. T. Khin, G. S. Jensen, A. Liu and M. E. Davis, Bioconjugate Chem., 2003, 14, 1007 CrossRef CAS PubMed; (b) A. G. Cheetham, P. Zhang, Y. Lin, L. L. Lock and H. Cui, J. Am. Chem. Soc., 2013, 135, 2907 CrossRef CAS PubMed; (c) J. Kang, V. Kumara, D. Yang, P. R. Chowdhury and R. Hohl, Eur. J. Pharm. Sci., 2002, 15, 163 CrossRef CAS PubMed; (d) T. Schluep, J. Hwang, I. J. Hildebrandt, J. Czernin, C. H. J. Choi, C. A. Alabi, B. C. Mack and M. E. Davis, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 11394 CrossRef CAS PubMed.
- (a) H. Zhao, B. Rubio, P. Sapra, D. Wu, P. Reddy, P. Sai, A. Martinez, Y. Gao, Y. Lozanguiez, C. Longley, L. M. Greenberger and I. D. Horak, Bioconjugate Chem., 2008, 19, 849 CrossRef CAS PubMed; (b) Y. Yang, Y.-M. Zhang, D. Li, H.-L. Sun, H.-X. Fan and Y. Liu, Bioconjugate Chem., 2016, 27, 2834 CrossRef CAS PubMed; (c) G. Yu, X. Zhao, J. Zhou, Z. Mao, X. Huang, Z. Wang, B. Hua, Y. Liu, F. Zhang, Z. He, O. Jacobson, C. Gao, W. Wang, C. Yu, X. Zhu, F. Huang and X. Chen, J. Am. Chem. Soc., 2018, 140, 8005 CrossRef CAS PubMed; (d) U. Y. Lau, L. T. Benoit, N. S. Stevens, K. K. Emmerton, M. Zaval, J. H. Cochran and P. D. Senter, Mol. Pharm., 2018, 15, 4063 CrossRef CAS PubMed.
- Y. Ge, A. Draycheva, T. Bornemann, M. V. Rodnina and W. Wintermeyer, Nat. Commun., 2014, 5, 5263 CrossRef CAS PubMed.
- S. Palakurthi, Expert Opin. Drug Delivery, 2015, 12, 1911 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed experimental methods and additional data. See DOI: 10.1039/c9ra07054k |
| This journal is © The Royal Society of Chemistry 2019 |