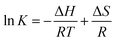Open Access Article
Open Access ArticleEffect of alkyl distribution in pyrazine on pyrazine flavor release in bovine serum albumin solution
Yun-Jiao Ma†
,
Jian-Hai Wu†,
Xiang Li,
Xian-Bing Xu *,
Zhen-Yu Wang,
Chao Wu,
Ming Du and
Liang Song
*,
Zhen-Yu Wang,
Chao Wu,
Ming Du and
Liang Song *
*
National Engineering Research Center of Seafood, School of Food Science and Technology, Dalian Polytechnic University, Dalian 116034, China. E-mail: xianbingxu@gmail.com; ryo.song@foxmail.com; Fax: +86 411 86323262; Fax: +86 411 86323453; Tel: +86 411 86332275 Tel: +86 15942804820
First published on 13th November 2019
Abstract
The flavor release mechanism related to the interaction of aroma compounds with proteins is still unclear. In this study, the interaction of protein with pyrazine homologues, such as 2-methylpyrazine (MP), 2,5-dimethylpyrazine (DP), 2,3,5-trimethylpyrazine (TRP) and 2,3,5,6-tetramethylpyrazine (TEP), was investigated to elucidate the effect of alkyl distribution in a pyrazine ring on its flavor release in bovine serum albumin (BSA) solution (pH 7.4). The results of SPME-GC-MS indicated that methyl distribution in a pyrazine ring significantly affected its release from BSA solution. The pyrazines released from BSA solution with an increasing order of MP, DP, TRP and TEP. The inhibition mechanism of alkyl-pyrazine release was further elucidated by the interaction between alkyl-pyrazines and BSA using multiple spectroscopic methods. The non-covalent interaction between alkyl-pyrazines and BSA was confirmed as the main interaction force by the value of the bimolecular quenching constant (Kq > 2 × 1010 L mol−1 s−1). A decrease in the hydrophobicity of the microenvironment between the alkyl-pyrazine and BSA was detected by synchronous fluorescence spectra, which revealed that alkyl-pyrazines were mainly bound on the sites of tyrosine and tryptophan in BSA. The UV-vis absorption spectra and circular dichromatic (CD) spectrum revealed that alkyl-pyrazines could induce polarity and conformation change of BSA. The above results indicated that the structure of the flavor homologues can affect their release in food matrices.
Introduction
The organoleptic property of food is highly related to the property of flavor release and perception, which finally affects the acceptability of food.1 In addition, flavor release could reduce flavor intensity during manufacture or long-term storage, which consequently causes flavor deterioration of food products.2 So a thorough understanding of flavor compound release is required to obtain a high quality of food flavor products. Flavor release is affected by the volatility of flavor compounds and the resistance to mass transfer from the matrix to the air.3 The food matrix such as proteins, fats and carbohydrates also had an important effect on the flavor release because of a strong interaction between flavor compounds and the food matrix.4–6 On the basis of this interaction, some studies have investigated the interfacial obstruction mechanism7–9 for controlling flavor release to preserve food flavor compounds, such as flavor encapsulation technology10–13 and emulsion delivery flavor system.14,15 The binding property of flavor compounds with food matrix could be evaluated by headspace analysis.16,17 Protein, an important food ingredient, can absorb or release flavor compounds due to the environment changes. It is necessary to understand the binding or releasing mechanism of flavor compounds in food because it is not only possible to modulate flavors, but also to improve the sensory properties of food.18 Therefore, the interaction between protein and flavor compounds was commonly investigated. The binding of proteins with flavor compounds not only depended on their intrinsic properties and conformational state of proteins,19,20 but also on the chemical property of flavor compounds. Numerous studies were carried out to elucidate the binding mechanisms of proteins with flavor compounds such as aldehydes, alcohols, ketones and esters.21–23 However, the interaction between flavor homologues and proteins has rarely been reported.Pyrazines were important heterocyclic flavor compounds, mainly formed during Maillard reaction24,25 or fermentation processing.26,27 Commonly, the produced pyrazines presented the roasted flavor or the baked flavor,28 which were important for evaluating the flavor quality of foodstuff, such as vinegar, meat production, flavoring, and so on. The pyrazine structures generally contained a heterocyclic structure with two nitrogen atoms (Fig. 1A). The functional groups (–R1, –R2, –R3 and –R4) like alkyl-, oxygenated- and acyl-groups could distribute at four carbon atom of heterocyclic structure. Recently, more than 70 different alkyl-pyrazines had been identified in nature.29 To some extent, the alkyl-pyrazines homologues exhibited the similar chemical properties, but it was still unknown that the different distributions of functional groups on a pyrazine ring could induce the difference in flavor release. Although the nature of interactions between pyrazines and β-lactoglobulin were theoretically calculated by 3D-QSAR,30 an experiment study was still not performed. In this study, alkyl-pyrazines homologues with different alkyl distributions including 2-methylpyrazine (MP), 2,5-dimethylpyrazine (DP), 2,3,5-trimethylpyrazine (TRP) and 2,3,5,6-tetramethylpyrazine (TEP) were selected to illustrate the effect of pyrazines homologues compounds on flavor release in food matrix.
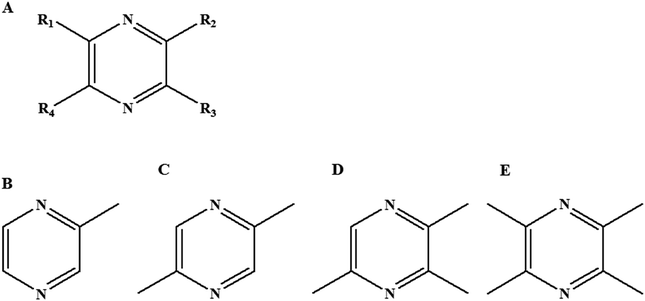 | ||
| Fig. 1 Molecular structure of pyrazines family(A), 2-methylpyrazine (B), 2,5-dimethylpyrazine (C), 2,3,5-trimethylpyrazine (D) and 2,3,5,6-tetramethylpyrazine (E). | ||
Protein, as an essential food component, was commonly used in food industry due to its emulsification,31 foamability32 and gelation property. Protein was widely used to control flavor release or deliver flavor by bonding hydrophobic or hydrogen interacted with flavor compounds or forming the weak and unspecific binding forces.33,34 Among the food proteins, bovine serum albumin (BSA),21 α-lactalbumin,35 β-lactoglobulin,36 and myofibrillar protein20,22 were usually used as food matrix to investigate the interaction mechanism between flavor compounds and protein. In the food systems, BSA has been universally applied in gel,37 delivery,38 nanoparticle39 and emulsion.40 Compared with soy protein, ovalbumin and β-casein, BSA has been used as emulsifier for food flavor delivery.41 In addition, BSA was frequently employed as model protein for the chemical mechanism study in food system.42 Generally, BSA has domains that can specifically bind to small molecules, such as metal ions and nucleotides.43,44 In this study, BSA as the ideal model protein was selected to reveal the flavor release mechanism between pyrazines and protein in food matrix.
The aim of this study was to investigate the effect of alkyl distribution of pyrazines on flavor release from protein matrix. The flavor release rate of pyrazines was detected by SPME-GC-MS. The interaction mechanism of pyrazines with BSA was evaluated by multispectroscopic methodologies, including fluorescence quenching analysis, ultraviolet visible (UV-vis) spectrophotometry and circular dichromatic (CD) spectrum.
Materials and methods
Materials
BSA was purchased from Sangon Biotech (Shanghai, China). MP, DP, TRP and TEP were provided by Sigma-Aldrich (St. Louis, MO, U.S.A). The BSA solution (15 × 10−4 mol L−1) was prepared in phosphate buffer solution (PBS, the concentration was 1 × 10−2 mol L−1 and pH = 7.4) and stored at 277 K for experiment. The solutions (1 × 10−2 mol L−1) of MP, DP, TRP and TEP were prepared in PBS and stored at 277 K for further experiments. The ultrapure water was used throughout the experiment.Solid phase micro-extraction gas chromatography mass spectrometry (SPME-GC-MS) analysis
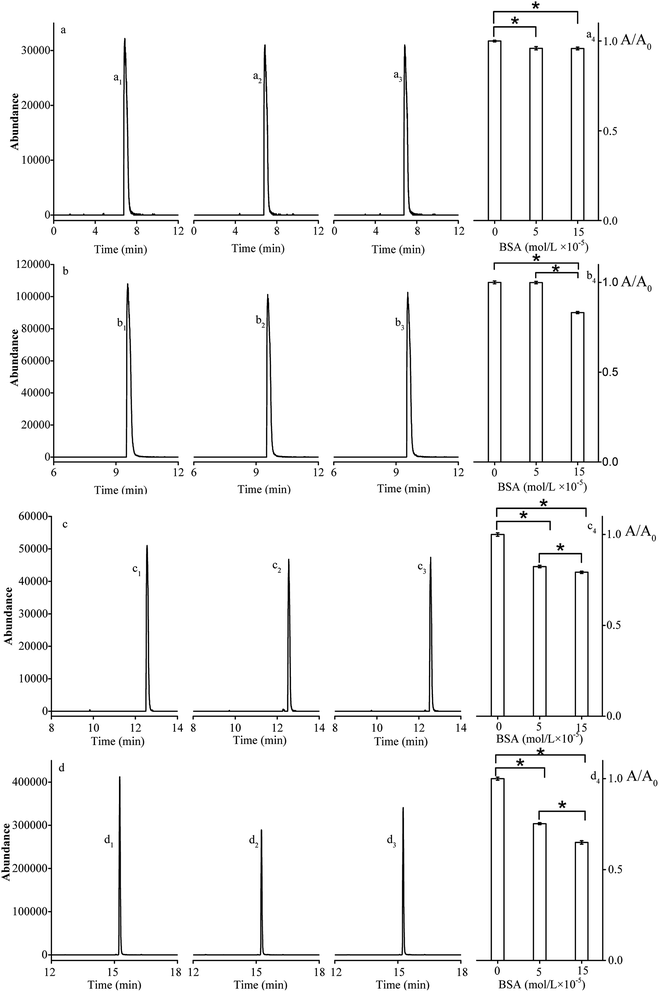
In this study, the manual SPME was used for collecting the volatile pyrazines. The method of GC-MS was established for the separation and identification of different kinds of pyrazines. For the SPME, the vials (20 mL) were filled with 1 mL solutions composed of a mixture of pyrazine solution (1 × 10−5 mol L−1) and BSA solution (0, 5, 15 × 10−4 mol L−1, respectively). The vials were heated at 323 K under stirring. The extraction fiber of SPME coatings with 50/30 μm DVB/CAR/PDMS was inserted into the vials and extracting the head space compounds for 30 minutes. When the extraction was completed, the fiber was placed in the injection port of the GC-MS system (Agilent 7890B-5977A, Agilent, U.S.A) with GC column (HP-5MS, 30 m × 250 μm ID). The time of desorption and the injection temperature were 5 minutes and 523 K, respectively. The oven heat program began at 308 K (3 minutes) and then was increased to 553 K at 283 K min−1, and maintained at 553 K for 5 min. The MS with electronic impact ionization source (ionization voltage: 70 eV) operated in scan mode and the mass detection range was from 40 to 400 m/z. In addition, the ability of pyrazine to release from BSA is reflected by the peak area ratios (area of pyrazine with BSA/area of pyrazine without BSA).Fluorescence spectrometry and synchronous fluorescence spectrometry analysis
The fluorescence spectra of BSA in the presence of various concentrations of MP, DP, TRP and TEP were recorded by F-2700 (Hitachi, Japan) at 300, 310 and 323 K, the temperature was maintained by the thermostatic bath bought from (Nan Jing Xin Chen Biotechnology Co., Ltd). The excitation (EX) wavelength was 290 nm and the emission (EM) wavelength was recorded from 300 to 500 nm. The synchronous fluorescence spectra were measured with the D-value (Δλ) between excitation wavelength and emission wavelength at 15 or 60 nm. The slit widths of EX and EM were set at 5 nm. The quartz cell (1.0 cm) was used in this experiment. The concentration of BSA was diluted into 5 × 10−4 mol L−1 for this study. The concentration of MP, DP, TRP and TEP was 1 × 10−2 mol L−1. The selected pyrazine solutions (6 μL of each time, 6 times in total) were successively added into the BSA solution (2 mL in volume) and tested by 10 minutes interval. The fluorescence quenching was conducted in this study by adding different concentrations (0, 3, 6, 9, 12, 15 and 18 × 10−5 mol L−1) of DP, MP, TRP and TEP into BSA solution at 300, 310 or 323 K, respectively.Ultraviolet visible (UV-vis) spectrophotometry analysis
The UV-vis absorption spectra were recorded from 200 to 500 nm by Lambda 35 (PerkinElmer, USA). The UV absorption spectra of BSA solution (2 mL, 2 × 10−6 mol L−1) with pyrazines (5 × 10−5 mol L−1) or without pyrazines were measured. In addition, the UV spectra of pyrazines solution (2 mL, 5 × 10−5 mol L−1) were also recorded. The entire experiment was conducted at room temperature (300 K) and the quartz cell (1.0 cm) was used for trials.Circular dichromatic (CD) spectrum analysis
The CD spectra were recorded at room temperature (300 K) under constant nitrogen flush using a CD spectropolarimeter (J-1500, Jassco, Japan), and the range of wavelength was set from 200 to 280 nm. The scan speed was set at 100 nm min−1. The bandwidth was set at 1 nm, and the path length was 0.1 cm quartz cell. The concentration of BSA was 5 × 10−6 mol L−1. The volume (350 μL) of BSA was tested. The volume (10 μL) of pyrazines (1 × 10−2 mol L−1) was added into the BSA solution (5 × 10−6 mol L−1) for measurement.Statistical analysis
The experimental data were presented in form of mean and standard deviation. Analysis of variance was used to determine any significant differences (p < 0.05) among the applied treatments by the SPSS software package (SPSS 10.0 for Windows).Results and discussion
Release of pyrazines in BSA solution
The SPME-GC-MS and the headspace GC were applied to clarify the flavor release mechanisms.45 As shown in Fig. 2, the peak area ratios (area of pyrazine with BSA/area of pyrazine without BSA) significantly decreased with the increasing concentrations of BSA. This decrease in peak area ratio could be explained by a decrease in the amount of free pyrazine in solution after interaction with BSA. This result was consistent with the previous studies.46,47 Interestingly, the area ratio of DP (from 1 to 0.83), TRP (from 1 to 0.79) and TEP (from 1 to 0.65) decreased significantly, while the area ratio of MP (from 1 to 0.95) decreased slightly with the increasing concentration of BSA (the concentration ranging from 0 to 15 × 10−5 mol L−1). The results indicated that the distribution and the number of functional groups may have an important effect on release of pyrazines in food matrix.Fluorescence quenching analysis and binding mode
Fluorescence quenching was used to assess the decrease in quantum yield of fluorophore fluorescence induced by interactions with a variety of molecular of quenchers.48 The quenching mechanisms could be categorized as dynamic and static quenching. In general, the different temperature dependence of fluorescence quenching behavior was applied to distinguish dynamic and static quenching.49 Thus, the fluorescence quenching was investigated at different temperatures (300, 310 or 323 K) in this study.To avoid the inner filter effects, fluorescence quenching experiments were performed in the linear range of Stern–Volmer curve.50 The Stern–Volmer equation was used to analyze the fluorescence quenching.51
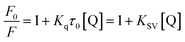 | (1) |
| Complex | T (K) | Ksv (L mol−1) | Kq (L mol−1 s−1) | Ra | SDb | c |
|---|---|---|---|---|---|---|
a The correlation coefficient.b The standard deviation for the Ksv, n = 3.c y represents ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K, x represents 1/T. K, x represents 1/T. |
||||||
| 2-Methylpyrazine | 300 | 0.61 × 103 | 0.61 × 1011 | 0.9548 | 0.0573 | y = 1378.3x + 1.8219 |
| 310 | 0.53 × 103 | 0.53 × 1011 | 0.9837 | 0.0427 | ||
| 323 | 0.44 × 103 | 0.44 × 1011 | 0.9192 | 0.0607 | ||
| 2,5-Dimethylpyrazine | 300 | 1.24 × 103 | 1.24 × 1011 | 0.9927 | 0.0429 | y = 244.81x + 6.3068 |
| 310 | 1.17 × 103 | 1.17 × 1011 | 0.9934 | 0.0235 | ||
| 323 | 1.17 × 103 | 1.17 × 1011 | 0.9918 | 0.0699 | ||
| 2,3,5-Trimethylpyrazine | 300 | 1.67 × 103 | 1.67 × 1011 | 0.9935 | 0.0140 | y = 101.86x + 7.0807 |
| 310 | 1.65 × 103 | 1.65 × 1011 | 0.9909 | 0.0185 | ||
| 323 | 1.63 × 103 | 1.63 × 1011 | 0.9919 | 0.0199 | ||
| 2,3,5,6-Tetramethylpyrazine | 300 | 6.18 × 103 | 6.18 × 1011 | 0.9932 | 0.0044 | y = 1296.6x + 4.4289 |
| 310 | 5.72 × 103 | 5.72 × 1011 | 0.9918 | 0.0114 | ||
| 323 | 4.56 × 103 | 4.56 × 1011 | 0.9841 | 0.0205 | ||
Significantly, the fluorescence intensity of BSA decreased with addition of pyrazine into BSA solution (Fig. 3A–D). The values of KSV in the pyrazine–BSA interaction systems were decreased with the increasing temperature and Kq > 2 × 1010 L mol−1 s−1 (Table 1). The results indicated that the interaction between pyrazines and BSA was attributed to the static quenching effect. In addition, as shown in Table 1, TRP and TEP exhibited strong interaction with BSA compared to DP, MP at same temperature. The interaction intensity of BSA with pyrazines was listed in order: MP < DP < TRP < TEP, which was consistent with the GC-MS results.
The interaction forces may be the weak interaction force, such as hydrogen bonding, van der Waals force, hydrophobic effect and electrostatic interaction, which are the main interactions between a small molecule and biological macromolecule.52,53 The specific interactions can be determined based on the enthalpy change (ΔH) and entropy change (ΔS) in the interaction process. The energy change in quenching process can be calculated by the slope and intercept of the ratio graph of the quench constant and 1/T. In the studied temperature range if the enthalpy change (ΔH) no significant changes, then its value and that of entropy change (ΔS) can be determined from van't Hoff equation:54
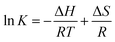 | (2) |
G = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = ΔH − TΔS K = ΔH − TΔS
| (3) |
The thermodynamic parameter of the pyrazine–BSA system was listed in Table 1. The negative enthalpy (ΔH), positive entropy (ΔS), and negative free energy (ΔG) values can be obtained by the slope and intercept of the equations (Table 1). The negative sign for ΔG meant that the interaction process was spontaneous. The negative ΔH and positive ΔS values of the interaction of pyrazine and BSA indicate that electrostatic interactions played a major role in the reaction. In addition, the positive ΔS was taken as evidence for hydrophobic interactions.55 Therefore, the major intermolecular interactions between BSA and pyrazines may be electrostatic interactions and hydrophobic interactions.
Microenvironment change
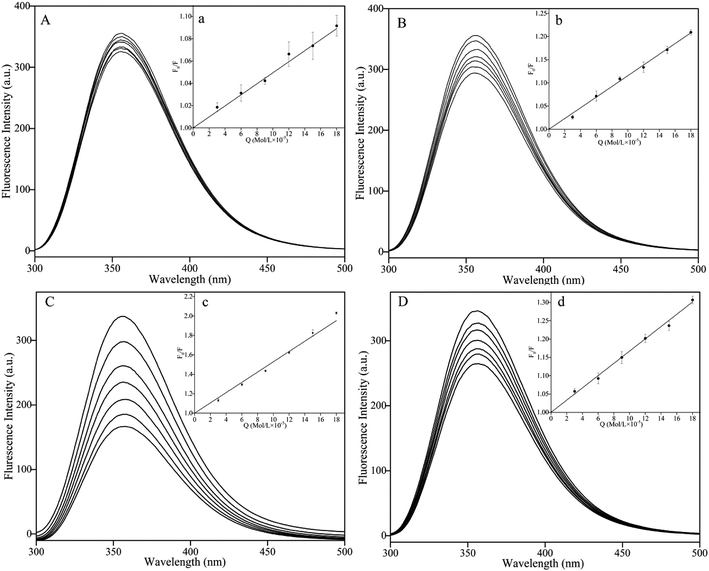
The change in the synchronous fluorescence spectra was relative to the microenvironment change in fluorophore of protein interacting with ligand,56 especially the polarity change of tyrosine (Δλ = 15 nm) and tryptophan (Δλ = 60 nm) in protein.57 As shown in Fig. 4, the fluorescence intensities of BSA solution at Δλ = 15 nm and Δλ = 60 nm were decreased with the increasing concentration of pyrazines. Addition of TRP and TEP induced a significant quenching at Δλ = 15 nm and Δλ = 60 nm compared to the addition of MP and DP. The results confirmed that tyrosine and tryptophan of BSA were contained in the binding sites of BSA and pyrazines. In addition, significant red shifts were observed in Fig. 4(C, D, G and H). The results indicated that BSA conformation had been altered and the hydrophobicity of microenvironment around tyrosine and tryptophan residues both decreased with the increase of concentration of pyrazine,58,59 which meant that the hydrophilicity and polarity of the microenvironment increased. The phenomenon indicated that these amino acid residues were exposed to the solvent at a higher level due to the addition of pyrazine.60Conformational changes
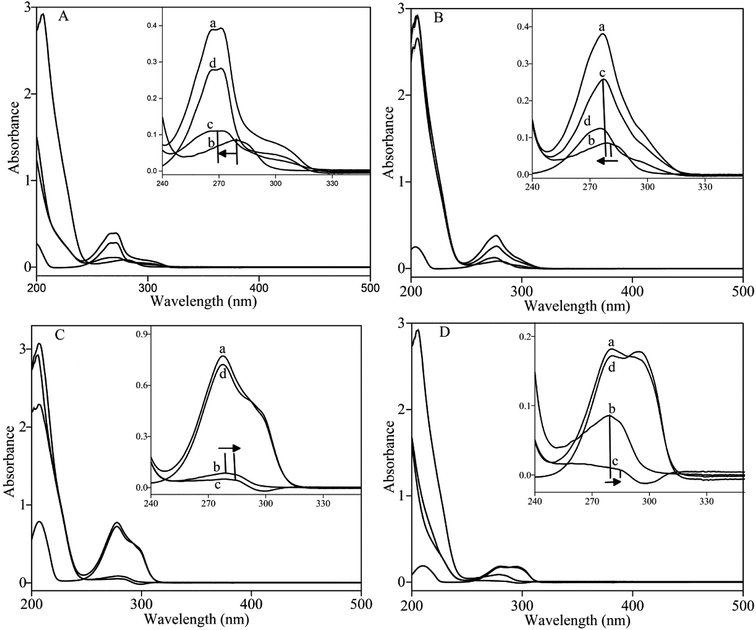
The UV-vis absorption spectra were widely used to evaluate the structure changes of protein.61 As shown in Fig. 5, a strong absorption of BSA at 210 nm and a weak absorption at 278 nm were observed. Generally, absorption in the region of 190–240 nm was induced by n–π* and π–π* transition of the amide group, which could be applied to indicate the conformational change of protein.62 A significant decrease at A210 nm indicated that pyrazines may affect the steric structure of BSA when pyrazines were added into BSA solution. The result was consistent with that of synchronous fluorescence.The value of A278 nm indicated the polarity of the microenvironment of tryptophan and tyrosine resides could be used to reflect the change in the tertiary structure of protein.63,64 As shown in Fig. 5 A and B, the A278 nm value of BSA increased with addition of MP and DP and a blue shift of the UV spectra was observed. However, the A278 nm value of BSA decreased with addition of TRP and TEP and a red shift of the UV spectra was detected (Fig. 5C and D). The results demonstrated that the addition of the pyrazines induced the change in polarity microenvironment of BSA.65
Secondary structure changes
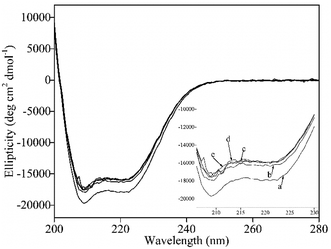
The CD spectra could directly reflect the secondary structure change of protein.66 In this study, CD spectra were carried out at room temperature (T = 300 K) to further reveal the changes of secondary structure of BSA interacted with pyrazines. The CD spectra presented two negative bands at around 208 and 220 nm (Fig. 6), which were the characteristic of helix and beta structure in the second structure of BSA according to the previous literatures.67 As shown in Table 2, the helix of BSA with the addition of pyrazines decreased from 59.03 to 53.17%. A possible reason could be explained that pyrazines attacked amino acid residues in the main polypeptide chain of BSA and destroyed their hydrogen bond network.60 Interestingly, the random of BSA with the addition of pyrazines had no significant changes. However, the turns of BSA presented a significant increase (from 2.97 to 9.27%) with addition of MP, DP, TRP and TEP. Obviously, the addition of pyrazines could induce the secondary structure change of BSA, which could be related to the change of pyrazines release in BSA solution.| Complex | Helix (%) | Beta (%) | Turn (%) | Random (%) |
|---|---|---|---|---|
| a Average ± standard deviations, n = 3. Different letters in the same column indicate significant differences (p < 0.05). | ||||
| Bovine serum albumin | 59.03 ± 0.04a | 12.03 ± 0.07a | 2.97 ± 0.49a | 25.97 ± 0.03a |
| 2-Methylpyrazine | 53.20 ± 0.03b | 11.20 ± 0.19a | 8.03 ± 0.29b | 27.53 ± 0.03a |
| 2,5-Dimethylpyrazine | 53.27 ± 0.05b | 12.80 ± 0.08a | 7.30 ± 0.29b | 26.63 ± 0.06a |
| 2,3,5-Trimethylpyrazine | 53.73 ± 0.01b | 9.67 ± 0.18a | 9.27 ± 0.09b | 26.33 ± 0.07a |
| 2,3,5,6-Tetramethylpyrazine | 53.17 ± 0.04b | 13.70 ± 0.17a | 7.17 ± 0.42b | 26.00 ± 0.04a |
Conclusion
The methyl distribution of pyrazines had a significant effect on the pyrazines release from BSA solution, which was mainly due to electrostatic interaction and hydrophobic interaction. The interaction between pyrazines homologues (MP, DP, TRP, and TEP) and BSA revealed that non-covalent interaction presented in BSA-pyrazine systems, which induced the difference release pattern of pyrazines in BSA solution. The disclosed mechanism of pyrazines release was helpful for the flavor compounds analysis and the control of flavor release in food matrix.Conflicts of interest
There are no conflicts to declare.Abbreviations
| MP | 2-Methylpyrazine |
| DP | 2,5-Dimethylpyrazine |
| TRP | 2,3,5-Trimethylpyrazine |
| TEP | 2,3,5,6-Tetramethylpyrazine |
| BSA | Bovine serum albumin |
| CD | Circular dichromatic |
| UV-vis | Ultraviolet visible |
| PBS | Phosphate buffer solution |
| SPME-GC-MS | Solid phase micro-extraction gas chromatography mass spectrometry |
| EX | Excitation |
| EM | Emission |
Acknowledgements
This work was supported by the 13th Five Years Key Programs for Science and Technology Development of China (Grant No. 2017YFD0400103) and the National Natural Science Foundation of China (Grant No. 31871795; 31601467).References
- K. Wang and S. D. Arntfield, Flavour Fragrance J., 2017, 32, 92–101 CrossRef CAS.
- S. Lubbers, P. Landy and A. Voilley, Food Technol., 1998, 52, 68–74 CAS.
- K. B. D. Roos, in Physiochemical Models of Flavor Release from Foods, ed. D. D. Roberts and A. J. Taylor, ACS Publications, 2000, p. 126 Search PubMed.
- P. Relkin, M. Fabre and E. Guichard, J. Agric. Food Chem., 2004, 52, 6257–6263 CrossRef CAS.
- X. Liu, J. R. Powers, B. G. Swanson, H. H. Hill and S. Clark, J. Food Sci., 2010, 70, C581–C585 CrossRef.
- E. Guichard, Food Rev. Int., 2002, 18, 49–70 CrossRef CAS.
- S. Ghosh, D. G. Peterson and J. N. Coupland, J. Agric. Food Chem., 2006, 54, 1829–1837 CrossRef CAS PubMed.
- S. Karaiskou, G. Blekas and A. Paraskevopoulou, Food Res. Int., 2008, 41, 637–645 CrossRef CAS.
- A. Meynier, C. Lecoq and C. Genot, Food Chem., 2005, 93, 153–159 CrossRef CAS.
- A. H. King, in Encapsulation of food ingredients: a review of available technology, focusing on hydrocolloids, ed. S. J. Rish and G. A. Reineccius, ACS Publications, 1995, p. 26 Search PubMed.
- T. A. Tari and R. S. Singhal, Carbohydr. Polym., 2002, 50, 279–282 CrossRef CAS.
- A. Madene, M. Jacquot, J. l. Scher and S. Desobry, Int. J. Food Sci. Technol., 2010, 41, 1–21 CrossRef.
- B. Lin, F. Zhang, T. Chen, J. Cao, Y. Chen and Y. Bai, Flavour Fragrance J., 2015, 30, 459–466 CrossRef.
- L. Mao, Y. H. Roos, C. G. Biliaderis and S. Miao, Crit. Rev. Food Sci. Nutr., 2017, 57, 3173–3187 CrossRef CAS.
- Z. Xiao, T. Tian, J. Hu, M. Wang and R. Zhou, Flavour Fragrance J., 2014, 29, 22–34 CrossRef CAS.
- J. Kühn, T. Considine and H. Singh, J. Food Sci., 2006, 71, R72–R82 CrossRef.
- K. Wang and S. D. Arntfield, Food Chem., 2014, 157, 364–372 CrossRef PubMed.
- H. Shen, M. Zhao and W. Sun, Food Chem., 2019, 287, 93–99 CrossRef CAS PubMed.
- T. E. O'Neill and J. E. Kinsella, J. Food Sci., 1987, 52, 98–101 CrossRef.
- F. Zhou, M. Zhao, G. Su and W. Sun, J. Agric. Food Chem., 2014, 62, 9544–9552 CrossRef CAS PubMed.
- Y. Tan and K. J. Siebert, J. Food Sci., 2010, 73, S56–S63 CrossRef PubMed.
- M. PéRezjuan, M. Flores and F. Toldra, Food Chem., 2007, 105, 932–939 CrossRef.
- M. Pérez-Juan, M. Flores and F. Toldrá, J. Agric. Food Chem., 2006, 54, 4802–4808 CrossRef PubMed.
- H.-I. Hwang, T. G. Hartman and C.-T. Ho, J. Agric. Food Chem., 1995, 43, 179–184 CrossRef CAS.
- M. Amrani-Hemaimi, C. Cerny and L. B. Fay, J. Agric. Food Chem., 1995, 43, 2818–2822 CrossRef CAS.
- I. Besson, C. Creuly, J. B. Gros and C. Larroche, Appl. Microbiol. Biotechnol., 1997, 47, 489–495 CrossRef CAS.
- C. Larroche, I. Besson and J.-B. Gros, Process Biochem., 1998, 34, 667–674 CrossRef.
- T. Shibamoto, T. Akiyama, M. Sakaguchi, Y. Enomoto and H. Masuda, J. Agric. Food Chem., 1979, 27, 1027–1031 CrossRef CAS.
- R. Wagner, M. Czerny, J. Bielohradsky and W. Grosch, Z. Lebensm.-Unters. -Forsch. A, 1999, 208, 308–316 CrossRef CAS.
- A. Tromelin and E. Guichard, J. Agric. Food Chem., 2003, 51, 1977–1983 CrossRef CAS PubMed.
- E. A. Foegeding and J. P. Davis, Food Hydrocolloids, 2011, 25, 1853–1864 CrossRef CAS.
- L.-y. Yin, X.-g. Zhou, J.-s. Yu, H.-l. Wang, S. Zhao, Z. Luo and B. Yang, J. Eur. Ceram. Soc., 2013, 33, 1387–1392 CrossRef CAS.
- E. Guichard, Biotechnol. Adv., 2006, 24, 226–229 CrossRef CAS PubMed.
- J. Kühn, T. Considine and H. Singh, J. Agric. Food Chem., 2008, 56, 10218–10224 CrossRef PubMed.
- J. Kühn, X.-Q. Zhu, T. Considine and H. Singh, J. Agric. Food Chem., 2007, 55(9), 3599–3604 CrossRef PubMed.
- H. Jegasothy, L. Bennett and R. Stockmann, Int. Dairy J., 2015, 47, 46–51 CrossRef CAS.
- C. Semasaka, L. Katopo, R. Buckow and S. Kasapis, Food Hydrocolloids, 2018, 76, 141–149 CrossRef CAS.
- J. Zhu, X. Sun, S. Wang, Y. Xu and D. Wang, Food Hydrocolloids, 2017, 63, 391–403 CrossRef CAS.
- J. Y. Jun, H. H. Nguyen, S.-Y.-R. Paik, H. S. Chun, B.-C. Kang and S. Ko, Food Chem., 2011, 127, 1892–1898 CrossRef CAS.
- K.-Y. Park, D.-Y. Kim and W.-S. Shin, Food Sci. Biotechnol., 2015, 24, 1583–1589 CrossRef CAS.
- F. Liu, J. Zheng, C.-H. Huang, C.-H. Tang and S.-Y. Ou, Food Hydrocolloids, 2018, 82, 96–105 CrossRef CAS.
- O. Becconi, E. Ahlstrand, A. Salis and R. Friedman, Isr. J. Chem., 2017, 57, 403–412 CrossRef CAS.
- J. Li and P. Yao, Langmuir, 2009, 25, 6385 CrossRef CAS PubMed.
- Y. D. Livney, in Biopolymeric amphiphiles and their assemblies as functional food ingredients and nutraceutical delivery systems, ed. N. Garti and J. Mcclements, Woodhead Publishing Limited: 80 High Street, Cambridge, 2012, p. 252 Search PubMed.
- K. G. Weel, A. E. Boelrijk, A. C. Alting, P. J. Van Mil, J. J. Burger, H. Gruppen, A. G. Voragen and G. Smit, J. Agric. Food Chem., 2002, 50, 5149–5155 CrossRef CAS PubMed.
- S. Damodaran and J. E. Kinsella, J. Agric. Food Chem., 1980, 28, 567 CrossRef CAS PubMed.
- F. M. Gianelli M P and F. Toldrá, J. Agric. Food Chem., 2003, 51, 6828–6834 CrossRef PubMed.
- A. Papadopoulou, R. J. Green and R. A. Frazier, J. Agric. Food Chem., 2005, 58, 158–163 CrossRef PubMed.
- H. Mohammadzadeh-Aghdash, J. Ezzati Nazhad Dolatabadi, P. Dehghan, V. Panahi-Azar and A. Barzegar, Food Chem., 2017, 228, 265–269 CrossRef CAS PubMed.
- S. Huang, H. Qiu, J. Xie, C. Huang, W. Su, B. Hu and Q. Xiao, RSC Adv., 2016, 6, 44531–44542 RSC.
- J. R. Lakowicz, in Principles of fluorescence spectroscopy, Woodhead Publishing Limited: Springer Science+Business Media, New York, 1999 Search PubMed.
- D. Leckband, Annu. Rev. Biophys. Biomol. Struct., 2003, 29, 1–26 CrossRef PubMed.
- G. Li, B. S. Liu, Q. Zhang and R. Han, Luminescence, 2016, 31, 1054–1062 CrossRef CAS PubMed.
- Y. J. Hu, L. Yi, J. B. Wang, X. H. Xiao and S. S. Qu, J. Pharm. Biomed. Anal., 2005, 36, 915–919 CrossRef PubMed.
- B. Pang, S. Bi, Y. Wang, L. Yan and T. Wang, J. Lumin., 2012, 132, 895–900 CrossRef CAS.
- Y. J. Hu, Y. Liu, T. Q. Sun, A. M. Bai, J. Q. Lu and Z. B. Pi, Int. J. Biol. Macromol., 2006, 39, 280–285 CrossRef CAS PubMed.
- Y. Q. Wang, H. M. Zhang, G. C. Zhang, S. X. Liu, Q. H. Zhou, Z. H. Fei and Z. T. Liu, Int. J. Biol. Macromol., 2007, 41, 243–250 CrossRef CAS PubMed.
- N. Wang, L. Ye, B. Q. Zhao and J. X. Yu, Braz. J. Med. Biol. Res., 2008, 41, 55–60 Search PubMed.
- Z. Nan, C. Hao, X. Ye, Y. Feng and R. Sun, Spectrochim. Acta, Part A, 2019, 210, 348–354 CrossRef CAS PubMed.
- H. Shen, M. Huang, M. Zhao and W. Sun, Food Chem., 2019, 298, 125060 CrossRef CAS PubMed.
- K. Nienhaus and G. U. Nienhaus, in Probing Heme Protein-Ligand Interactions by UV/Visible Absorption Spectroscopy, ed. G. U. Nienhaus, Woodhead Publishing Limited: Humana Press, 2005, p. 251 Search PubMed.
- A. N. Glazer and E. L. Smith, J. Biol. Chem., 1961, 236, 2942–2947 CAS.
- K. Rosenheck and P. Doty, Proc. Natl. Acad. Sci. U. S. A., 1961, 47, 1775–1785 CrossRef CAS PubMed.
- D. Li, B. Ji and J. Jin, J. Lumin., 2008, 128, 1399–1406 CrossRef CAS.
- Y. Zhang and Q. Zhong, J. Agric. Food Chem., 2012, 60, 1880–1886 CrossRef CAS PubMed.
- S. M. Darwish, S. E. Abu sharkh, M. M. Abu Teir, S. A. Makharza and M. M. Abu-hadid, J. Mol. Struct., 2010, 963, 122–129 CrossRef CAS.
- J. Liu, J. Tian, X. Tian, Z. Hu and X. Chen, Bioorg. Med. Chem., 2004, 12, 469–474 CrossRef CAS.
Footnote |
| † Both authors have contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |