Open Access Article
Open Access ArticleClinical training and validation of the LeukoScope: a low-cost, point-of-care device to perform white blood cell and neutrophil counts
Catherine E. Majors a,
Michal E. Pawlowskia,
Daniel C. Burke
a,
Michal E. Pawlowskia,
Daniel C. Burke a,
Tomasz S. Tkaczyk
a,
Tomasz S. Tkaczyk a,
Alyssa Rieberb and
Rebecca Richards-Kortum
a,
Alyssa Rieberb and
Rebecca Richards-Kortum *a
*a
aDepartment of Bioengineering, Rice University, 6100 Main Street, MS-142, Houston, TX 77005, USA. E-mail: rkortum@rice.edu; Tel: +1 713 348 3823
bDepartment of General Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA
First published on 30th August 2019
Abstract
A white blood cell (WBC) count with partial differential is an important clinical laboratory test. However, current methods to perform a WBC count and differential are difficult to use at the point of care or too expensive for use in low-resource settings. To meet this need, we developed the LeukoScope: a low-cost system to measure a WBC and neutrophil count from a single drop of blood at the point of care. The LeukoScope is battery powered and has a sample-to-answer time of <5 minutes. A drop of blood from a finger stick is added to a LeukoScope sample cartridge where pre-dried acridine orange fluorescently stains WBCs. The cartridge is then inserted into the LeukoScope reader where a portable fluorescence microscope captures a color image of the sample, which is analyzed to report results to the user. The LeukoScope system was tested at the point of care using fingerprick samples collected from 105 general oncology patients in Houston, TX. Performance of the LeukoScope was compared to that of a HemoCue WBC DIFF performed using the same fingerprick sample; clinical laboratory analysis of a venous blood draw was used as the gold standard in all cases. Bland–Altman analysis showed that the LeukoScope and HemoCue WBC DIFF had similar accuracy for measurement of WBC and neutrophil counts as compared to the gold standard. Seven out of eight patients with abnormal WBC count values were correctly identified using the LeukoScope, while six out of eight were correctly identified using the HemoCue WBC DIFF. Five out of six patients with abnormal neutrophil counts were correctly identified using the LeukoScope, while six of six were correctly identified using the HemoCue WBC DIFF.
Introduction
White blood cells (WBCs or leukocytes) are a central component of the immune system. There are five subtypes of WBCs (lymphocytes, monocytes, neutrophils, eosinophils, and basophils); because neutrophils, eosinophils, and basophils have a granular appearance, they are collectively known as granulocytes, while lymphocytes and monocytes are collectively known as agranulocytes.1 A WBC count and differential is an important clinical laboratory test; together with clinical signs and symptoms, it can be used to help differentiate between viral and bacterial infections, to assess the effectiveness of an antibiotic treatment, and to predict clinical outcomes.2–4 Bacterial infections are associated with an elevated WBC count and an elevated neutrophil count.5 During treatments that may reduce the WBC count, such as chemotherapy, WBC and neutrophil counts are routinely performed to assess whether additional treatment could place a patient at too great a risk of infection.1 It has been previously demonstrated that patient satisfaction could be improved if monitoring of these counts could be performed in a home setting in order to avoid trips to a treatment center if counts are too low for treatment to proceed, especially in patients traveling to a centralized center from a rural setting.6Historically, to obtain a WBC count, a trained user dilutes whole blood, inserts the sample into a hemocytometer, and counts the number of WBCs visible under microscopic examination of a pre-specified area. To perform a differential, whole blood is smeared on a glass slide, fixed and stained, and a trained user differentiates the five subtypes based on the staining pattern. This technique is time consuming, labor intensive, and requires a trained technician.2
Automatic hematology analyzers are used to perform WBC counts and differentials in most high-resource clinical settings today. These analyzers are flow-based systems that use a combination of electrical impedance and light scattering to count and differentiate stained WBCs, as well as to count red blood cells and platelets. However, these devices are large, expensive, need specialized reagents, and require high levels of infrastructure.7
HemoCue developed a method to perform a WBC count at the point of care using a small, disposable cuvette which is preloaded with methylene blue; the HemoCue WBC device captures several images of the stained blood and uses image analysis to calculate the WBC count in under 3 minutes. The HemoCue WBC system has shown high levels of accuracy, with 96% of tested samples having a less than 10% difference from reference values.8 However, the per-test cost of the HemoCue WBC ($3.52 per test, purchased in US in December 2018) is too high for routine use in low-resource areas, and it does not provide information about the WBC differential. HemoCue recently released the HemoCue WBC DIFF, which can perform a WBC count and five-part differential. Preliminary validation of this device shows accurate performance for WBC counts and neutrophil counts, adequate performance for lymphocyte counts, and weak performance for monocyte and eosinophil counts.9–12 Despite promising results, this device is also too costly for routine use in many low-resource areas ($5.58 per test, purchased in Malawi in November 2018). While other groups have developed similar imaging-based systems to perform a WBC count and differential, these systems lack the portability and/or simple sample preparation schemes to be truly implemented in at the point of care.13,14
Here we describe the LeukoScope, a low-cost system to perform a WBC and neutrophil count using small volumes of blood at the point-of-care. The system consists of a disposable cartridge and portable imaging system. A drop of blood is collected into the cartridge which is preloaded with acridine orange to fluorescently stain WBCs. The disposable cartridge is inserted into the LeukoScope reader where a fluorescence image is captured using a portable microscope; the resulting image is analyzed by an integrated single board computer, and the WBC and neutrophil counts are reported to the user in under five minutes.
Materials & methods
Disposable cartridge design
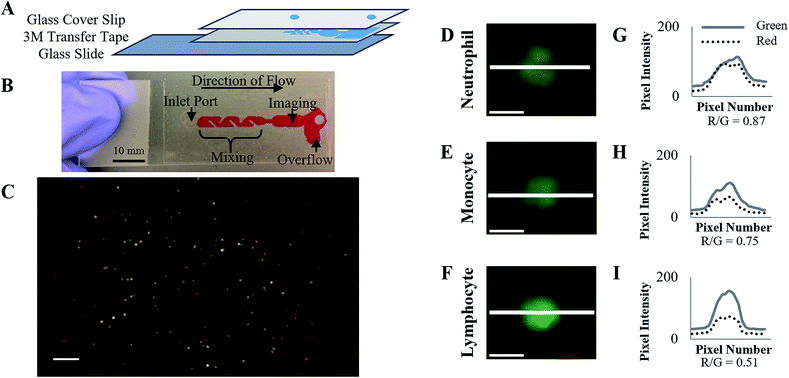
The disposable cartridge (Fig. 1) is composed of three layers: a glass microscope slide (46004-368, VWR Funding Inc, West Chester, PA, USA), a layer of transfer tape (3M, 8153LE, Maplewood, MN, USA) and a no. 1.5 glass cover slip (48393-241, VWR Funding Inc, West Chester, PA, USA). The transfer tape and glass cover slip were cut using a VLS3.60 laser cutter (Universal Laser Systems, Scottsdale, AZ, USA) to create a microfluidic channel and inlet/outlet ports on top of the slide, as shown in Fig. 1. The cut tape was adhered to the slide, and the coverslip was affixed atop the tape, creating a microfluidic channel with a depth of 88 μm. The channel consists of a passive mixing segment, an imaging chamber, and an overflow chamber. When a drop of blood is applied to the inlet, approximately 12–15 μL of blood is drawn into the cartridge by capillary action, filling the imaging chamber; excess blood is contained in the overflow chamber.During cartridge fabrication, acridine orange (Life Technologies, Carlsbad, CA, USA) was deposited onto the slide surface just after the inlet port and allowed to dry. Previous work has demonstrated 10 μg mL−1 gives the optimal signal-to-noise ratio to image WBCs;15 as such 0.4 μL of acridine orange (400 μg mL−1 in water) was deposited on the slide surface to achieve an acridine orange concentration of approximately 10 μg mL−1 in whole blood when imaged. Acridine orange was deposited onto the slide surface using a sciFLEXARRAYER S3 (Scienion, Berlin, Germany) and allowed to dry for 1 hour before placing the cover slip enclosing the cartridge. Cartridges were stored at room temperature in foil bags with desiccant until used.
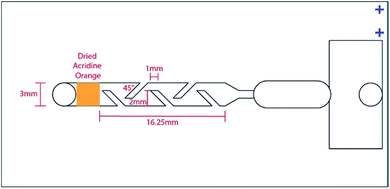
A passive microfluidic mixing design was incorporated into the microfluidic channel to ensure uniform distribution of acridine orange in the blood sample within the imaging chamber. Passive microfluidic mixers rely on chaotic advection effects of disrupting the laminar flow in a microfluidic chamber.16 To achieve this disruption of laminar flow, parallelogram barriers were incorporated into the channel. The mixing region was 16.25 mm in length and 3 mm in width. Five parallelogram mixing barriers were incorporated along the length of the mixing region, with a width of 1 mm and angle of 45°. These barriers obstructed 2/3 of the channel width, as shown in Fig. 2.
When a whole blood sample is introduced at the inlet port, acridine orange dissolves and stains the white blood cells. Acridine orange differentially stains double stranded-DNA and single stranded-RNA. When bound to DNA, the dye fluoresces green (excitation maximum = 502 nm, emission maximum = 525 nm). Alternatively, when bound to RNA it fluoresces orange (excitation maximum = 460 nm, emission maximum = 650 nm); the dye also fluoresces orange in highly acidic environments, such as lysosomes.15 As such, agranulocytes exhibit primarily green fluorescence due to the lack of cytoplasmic granules, while granulocytes exhibit a mixture of green and red fluorescence. Red blood cells do not contain nucleic acids and therefore do not fluoresce.
Development of image analysis method
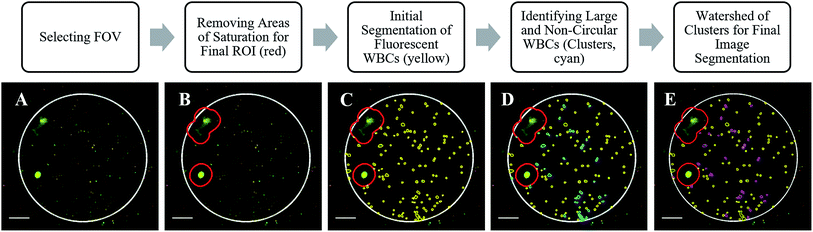
Venous whole blood samples from fifteen normal volunteers were added to the disposable cartridges and imaged on a Zeiss AxioImager Z1 microscope (Oberkochen, Germany). The resulting images were used to develop an image analysis algorithm to segment and count fluorescent WBCs and differentiate subtypes of WBCs. The resulting values were compared to reference values as determined by a Beckman Coulter AcT Diff2 hematology analyzer (Lab Resources Inc, Tomball, TX, USA).17 The blood collection protocol followed the United States Department of Health & Human Services (HHS) guidelines for the protection of human subjects (45 CFR part 46); the protocol was reviewed and approved by the Rice University Institutional Review Board (IRB). All volunteers provided written informed consent before participating.Fig. 3 illustrates the steps in the algorithm developed to analyze images of stained WBCs. First, the image is cropped to remove regions that are saturated, dimly fluorescent, or out of focus. The remaining region of interest is segmented to identify fluorescent WBCs that meet pre-specified intensity, size, and shape criteria. Any abnormally large or non-circular fluorescent areas are analyzed using a watershed algorithm to distinguish individual cells that may be present in clusters, resulting in the final segmentation of WBCs. The average WBC size is calculated in pixels and used to determine a size correction factor to calculate the size of the field of view (FOV), correcting for minor changes in magnification associated with adjustments in focus between cartridges. The number of WBCs per mm2 in the image is then calculated. This value is used to calculate the WBC count using clinical training data, as described in detail later.
To classify whether each segmented WBC is a neutrophil, the mean red and mean green pixel intensities are calculated for each WBC, and the median background levels of red and green pixel intensity from that image are subtracted from the respective channels. From the background corrected intensities, a red-to-green ratio is calculated for each WBC. This ratio of red-to-green fluorescence is used to classify each cell as a neutrophil or other WBC subtype, as described in detail later.
The image analysis algorithm was initially coded in MATLAB, before being translated to OCTAVE for implementation on the single board computer within the LeukoScope device.
LeukoScope reader design
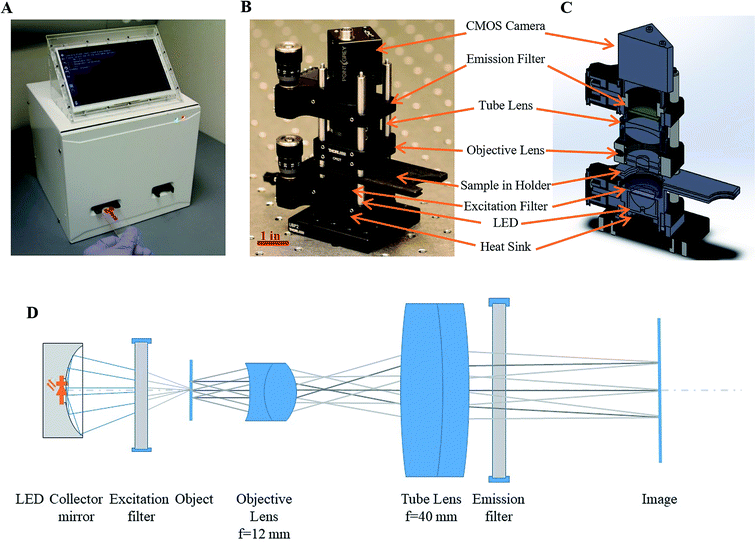
The LeukoScope reader consists of a custom portable microscope to capture fluorescence images, a single board computer (NP93-2930, Jetway, Newark, CA, USA), an Arduino Nano 3.0 for device control, a screen to display the user interface, and a Rii Mini X1 Bluetooth enabled keyboard for user interaction; all parts are housed in a custom housing (Fig. 4A).LeukoScope optical design
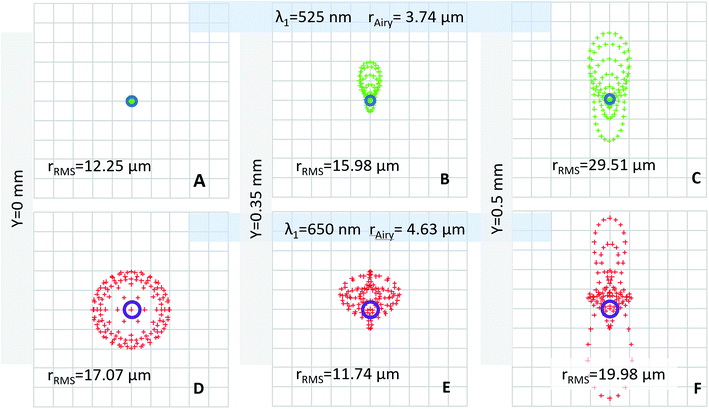
Fig. 4B shows a photograph of the portable microscope, and Fig. 4C shows a CAD drawing with the internal components. The optical schematic of the LeukoScope is presented in Fig. 4D, and its essential optical parameters are gathered in Table 1. The optical system of the LeukoScope was constructed from two off-the-shelf lenses. An achromatic doublet with focal length of 9 mm (EO 49-656, Edmund Optics, Barrington, NJ, USA) was placed 7.8 mm from the surface of the cartridge to serve as the objective lens. A techspec lens with focal length of f = 40 mm (EO49-664, Edmund Optics, Barrington, NJ, USA) was placed 10 mm from the objective lens and was followed by long-pass filter with central wavelength of 532 nm (BLP01-532R-25, Semrock, Rochester, NY, USA). Together both lenses formed the optical train of the LeukoScope. Lens placement and orientation were optimized to provide the best performance with a numerical aperture (NA) of 0.3 in the object space and −3.5× magnification required by the application; these parameters were set by the necessity to resolve individual WBCs with diameters ranging from 7 to 20 μm![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 during image segmentation for the purpose of counting and analysis. Position and orientation of both lenses were numerically optimized using OpticStudio (Zemax LLC, Kirkland, USA). Based on the experimental procedure described above, an average of 90 cells per mm2 was observed using a 5× magnification; thus in order to guarantee at least 100 cells were captured in an image, the field of view was set to 1 mm in diameter. This parameter is important in order to mimic the performance of a characteristic manual WBC differential, in which at least 100 WBCs are classified from a peripheral blood smear and reported as percentages.18 Nominal spot diagrams for the complete LeukoScope optical train, including 100 μm of fluid within cartridge, the cover slip thickness, and the emission filter, are given in Fig. 5 for axial, mid-field, and marginal field points at each of acridine orange's emission wavelengths. Nominally, the system resolution in the object space is limited to 205, 298, and 175 lp per mm for λ1 = 650 nm and 286, 219 and 119 lp per mm for λ2 = 525 nm for axial, mid-field, and marginal field points, respectively. The above performance, while on average 4-times larger than diffraction limited performance, was achieved using only off-the-shelf components. It should be noted that further system improvements could be made by restricting the numerical aperture of the system, such as through the modification of the illumination system or the installation of an adjustable aperture stop. However, while system performance would improve with the decrease of numerical aperture, the system's light throughput would decrease proportionally and image exposure time would need to be increased to compensate for this loss. In order to balance resolution with image acquisition time, we decided to use the system at 0.3 NA with resultant exposure time of 200 ms.
1 during image segmentation for the purpose of counting and analysis. Position and orientation of both lenses were numerically optimized using OpticStudio (Zemax LLC, Kirkland, USA). Based on the experimental procedure described above, an average of 90 cells per mm2 was observed using a 5× magnification; thus in order to guarantee at least 100 cells were captured in an image, the field of view was set to 1 mm in diameter. This parameter is important in order to mimic the performance of a characteristic manual WBC differential, in which at least 100 WBCs are classified from a peripheral blood smear and reported as percentages.18 Nominal spot diagrams for the complete LeukoScope optical train, including 100 μm of fluid within cartridge, the cover slip thickness, and the emission filter, are given in Fig. 5 for axial, mid-field, and marginal field points at each of acridine orange's emission wavelengths. Nominally, the system resolution in the object space is limited to 205, 298, and 175 lp per mm for λ1 = 650 nm and 286, 219 and 119 lp per mm for λ2 = 525 nm for axial, mid-field, and marginal field points, respectively. The above performance, while on average 4-times larger than diffraction limited performance, was achieved using only off-the-shelf components. It should be noted that further system improvements could be made by restricting the numerical aperture of the system, such as through the modification of the illumination system or the installation of an adjustable aperture stop. However, while system performance would improve with the decrease of numerical aperture, the system's light throughput would decrease proportionally and image exposure time would need to be increased to compensate for this loss. In order to balance resolution with image acquisition time, we decided to use the system at 0.3 NA with resultant exposure time of 200 ms.
| Parameter | Value | Unit | Remarks |
|---|---|---|---|
| Numerical aperture | 0.3 | Object side | |
| Design wavelength | λ1 = 525, λ2 = 650 | nm | Main emission lines of AO bound to DNA (λ1) and RNA (λ2) |
| Field of view | 1 | mm | Diameter |
| Magnification | −3.5 | × | |
| Total length | 72.5 | mm |
While performance of the LeukoScope optical system is far from diffraction limited, it is sufficient to identify individual non-tightly clustered cells in fluorescently stained images. Further, because our image analysis program relies on red and green pixel intensity of individual cells rather than sub-cellular morphology as in typical microscopy, a partial differential of WBC subtypes is possible at this system's level of performance.
The illumination system of the LeukoScope was constructed from a 470 nm LED (SR-01-B0040, Luxeon Star, Lethbridge, Alberta, Canada) mounted with a matching collector mirror (Dialite OPC-1-COL, Luxeon Star, Lethbridge, Alberta, Canada). A 470 nm bandpass filter (FF01-470-28-25, Semrock, Rochester, NY, USA) was placed directly behind collector unit; its spectral band-pass was experimentally optimized for balanced radiant response of both emission lines of acridine orange. The measured power output of the illumination system at the sample plane was 63.2 mW.
All parts were mounted in an optical cage mount (Thor Labs, Ann Arbor, MI, USA). The disposable cartridge was inserted into a custom 3D-printed sample holder that also holds the illumination system; the sample holder aligns the imaging window with the illumination and optical pathways for image capture. The sample holder was initially mounted in a z-axis translation mount (SM1Z, Thor Labs, Ann Arbor, MI, USA) to enable rough focus adjustments; this mount was later replaced with a fixed mount (CP08, Thor Labs, Ann Arbor, MI, USA). Images are captured by a Flea3 8.8 MP Color Vision CMOS camera (Point Gray, Wilsonville, OR, USA); the camera was mounted in a precision focusing mount (SM1Z, Thor Labs, Ann Arbor, MI, USA) for fine focus adjustments.
LeukoScope device control
Image capture is controlled by a custom software interface installed on an enclosed single board computer. The user initiates the program using a Bluetooth enabled keyboard. Once the program is initiated, a sample cartridge is inserted and the user enters a sample identification number. The sample is illuminated, and an image is captured; the program prompts the user to reposition the cartridge to collect two additional images within the cartridge imaging window. To reposition the cartridge, the user pulls the cartridge out of the sample holder in 1.5 mm steps by aligning the outside edge with two 3D printed alignment keys. The resulting images are then analyzed, and results are reported to the user within five minutes; additionally, the images are saved within the device for further analysis if needed.Laboratory testing with spiked normal volunteer blood
To verify the incorporation of dried acridine orange within the cartridge, five whole blood samples which were stained within the cartridge were compared to five in which acridine orange was added to whole blood to a final concentration of 10 μg mL−1, mixed by gentle pipetting, and added to a cartridge without dried acridine orange. Both samples were imaged on a Zeiss AxioImager Z1 microscope (Oberkochen, Germany), and the fluorescence of WBCs and background were compared.A whole blood sample from a volunteer was manipulated to vary the concentration of WBCs in order to validate the portable imaging system in a laboratory setting. Fresh blood was obtained from a normal volunteer via venous draw into EDTA-coated vacutainer tubes. The sample was aliquoted into 23 sample containers and centrifuged at 100 RCF for 15 minutes to separate red blood cells, buffy coat (which include the WBCs), and plasma. The WBC count was then artificially manipulated by adding or removing red blood cells and plasma without disturbing the buffy coat, resulting in WBC counts that span the entire normal range from 3000 cells per μL to 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 cells per μL. Each sample was measured using the LeukoScope and results were compared to reference values measured with a Beckman Coulter AcT Diff2 hematology analyzer (Lab Resources Inc, Tomball, TX, USA). The normal blood collection protocol followed United States HHS guidelines for the protection of human subjects (45 CFR part 46); the protocol was reviewed and approved by the Rice University IRB. Volunteers provided written informed consent before participating.
700 cells per μL. Each sample was measured using the LeukoScope and results were compared to reference values measured with a Beckman Coulter AcT Diff2 hematology analyzer (Lab Resources Inc, Tomball, TX, USA). The normal blood collection protocol followed United States HHS guidelines for the protection of human subjects (45 CFR part 46); the protocol was reviewed and approved by the Rice University IRB. Volunteers provided written informed consent before participating.
Training and validation with patient samples
Venous and capillary blood samples were collected from 105 general oncology patients at the Lyndon B. Johnson Hospital Outpatient Center in Houston, TX. Patients were eligible to participate if they were 18 years of age or older and their treating physician ordered a complete blood count (CBC). Patients were excluded if having a fingerprick blood sample collection was contraindicated by their treating physician, if they were unwilling or unable to provide informed consent, or if they were below 18 years of age. The protocol followed United States HHS guidelines for the protection of human subjects (45 CFR part 46) and was reviewed and approved by the IRBs at the MD Anderson Cancer Center, the Harris Health System, and Rice University. Patients provided written informed consent prior to participation.Venous blood samples were submitted to the hospital laboratory for CBC measurement using a Sysmex XN-3000 hematology analyzer, following the Lyndon B. Johnson Hospital clinical standard of care. Fingerprick samples were collected for measurement with the LeukoScope as well as with the HemoCue WBC DIFF. Because the WBC count in fingerprick blood samples can vary significantly from one drop to another,19 fingerprick samples were collected into EDTA coated microtainer tubes (Fisher Scientific Company, Pittsburgh, PA, USA); these samples were well mixed before being split and introduced to the cartridges for each POC device.
Up to three LeukoScope cartridges were filled and imaged per patient, based on the amount of blood available. Three images were collected for each cartridge. A user reviewed all available images to identify images which were out of focus, dim, saturated, or contained high background fluorescence. Images that did not pass quality control were not used for further analysis. For patients with images from multiple cartridges passing quality control, the average result was used for data analysis.
The remaining samples were randomly split into training and validation sets of equal size. The training set was used to develop a linear fit to convert WBC per mm2 measured with the LeukoScope to a WBC count (WBC per μL), using the venous results as the gold standard. The resulting fit was applied to the validation set to convert the LeukoScope output (WBC per mm2 in the FOV) to the WBC count.
To perform a differential, the ratio of red-to-green fluorescence was calculated for each cell. When these values were plotted as a histogram for individual sample images, a characteristic bimodal distribution was observed, as shown in Fig. 6. The two peaks visible in these plots correspond to neutrophils, which have the highest red-to-green ratios due to characteristic ratios of nuclear-to-cytoplasmic material, and other agranulocyte WBC subtypes. Therefore, cells with a red-to-green ratio above a threshold value were classified as neutrophils. Data in the training set were used to determine the optimal threshold value as that which gave best agreement to the venous standard of care neutrophil percentage measurements. Visual inspection of training set red-to-green ratio histogram plots was used to identify a range of potential threshold values, which were then applied to training set samples. The resulting neutrophil percentages for each threshold value was compared to venous gold standard neutrophil percentages, and the threshold resulting in the minimum mean squared error for the training set was then applied to the validation set. Bland–Altman analyses were performed to compare results measured with the LeukoScope and the HemoCue WBC DIFF to the clinical standard of care measured from venous blood.20
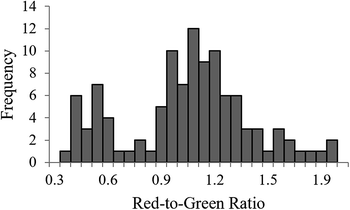 | ||
| Fig. 6 Histogram of red-to-green ratio values for all WBCs in a single sample, illustrating the bimodal distribution of measured values. | ||
Results & discussion
Laboratory based validation of the LeukoScope
Acridine orange staining of whole blood within the cartridge was verified by comparing samples that were well-mixed with acridine orange before addition to the cartridge with those that were stained with dried acridine orange in cartridge. Fluorescence intensity levels observed in both samples were comparable within WBCs and background levels of the FOV (data not shown).The number of WBCs detected per unit area in fluorescence LeukoScope images of donor blood spiked with increasing numbers of WBCs were compared to reference values measured with the AcT Diff2 hematology analyzer. Images of manipulated blood exhibited a higher background level than observed with unprocessed blood; this was attributed to lysing of some cells and release of cellular material during sample dilution. Despite higher background levels, a linear correlation was observed between the WBC per mm2 in the FOV of the LeukoScope and the WBC count as determined by the AcT Diff2 hematology analyzer, with a slope of 0.237 and an R2 of 0.68 (data not shown).
Clinical sample validation of the LeukoScope
Of the 105 patients enrolled, 42 were male and 63 were female. The average age was 52 years (range = 20 to 75 years). Capillary patient samples could not be measured from six patients due to an insufficient volume of fingerprick blood. Venous blood results were not returned from the hematology lab for three patients.Of the 96 remaining patient samples measured with the LeukoScope, 38 samples were removed due to quality control. The majority of samples failing quality control were measured during the first four days of patient enrollment when it was noted that the LeukoScope imaging system did not remain in focus during transportation and only 12 of 37 samples (32%) measured passed quality control. After this problem was identified, the mounting was reinforced, and 46 of 59 samples (78%) passed quality control. The 58 samples remaining after quality control were randomly split into training (n = 29) and validation sets (n = 29). Table 2 summarizes results of the venous WBC count and differential for all 58 patient samples. Images had an average of 59.6 ± 29.3 cells per FOV. This limited accuracy of differential counts reported in cases with a small number of cells per FOV.
| Venous results | |
|---|---|
| WBC count | 6.2 × 103 cell per μL (1.2, 16.9) |
| Neutrophil percentage | 64% (40, 86) |
| Lymphocyte percentage | 24% (3, 48) |
| Monocyte percentage | 9% (2, 21) |
| Eosinophil percentage | 2% (0, 10) |
| Basophil percentage | 1% (0, 4) |
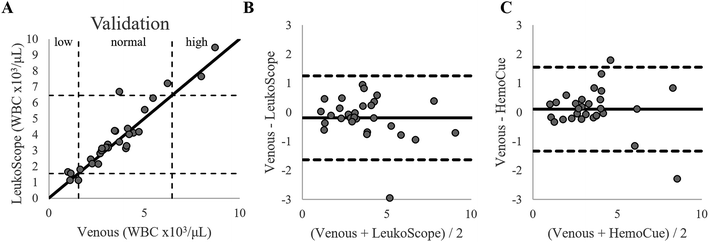
Fig. 7A shows the relationship between the number of WBC per mm2 measured with the LeukoScope to the clinically reported WBC count for all samples in the training set. As expected, the linear relationship between the number of WBC per mm2 measured with the LeukoScope and the WBC count is similar to that measured for spiked normal volunteer blood in the laboratory. The fit determined from the training set was used to calculate the WBC count for all 29 samples in the validation set. Fig. 7B compares WBC counts from the LeukoScope to reference venous measurements.
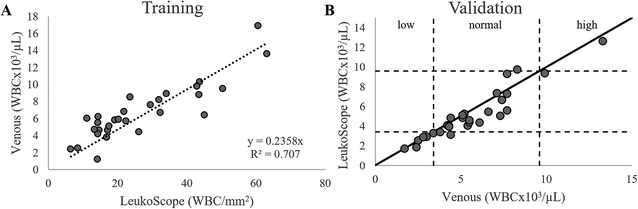 | ||
| Fig. 7 (A) Number of WBCs per unit area measured by the LeukoScope vs. reference venous WBC counts for the training set (n = 29). The resulting linear fit was used to calculate the WBC concentration for samples in the validation set. (B) WBC count measured using LeukoScope vs. reference venous WBC counts for the validation set (n = 29). The solid line represents perfect agreement, and the dashed lines show the cutoffs for low, normal, and high WBC counts.21 | ||
As shown in Fig. 7B, all six samples with low WBC counts (<3400 cells per μL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21) were correctly identified as low by the LeukoScope, and one of two samples with a high WBC count (>9600 cells per μL
21) were correctly identified as low by the LeukoScope, and one of two samples with a high WBC count (>9600 cells per μL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21) were correctly identified by the LeukoScope. The one incorrectly classified sample was measured as 9900 cells per μL by venous measurement and 9400 cells per μL on the LeukoScope. Nineteen of the 21 samples falling in the normal range were correctly identified by the LeukoScope. One sample was measured as 4400 cells per μL by venous measurement and as 3100 cells per μL on the LeukoScope, and one was measured as 8300 cells per μL by venous measurement and as 9700 cells per μL on the LeukoScope. In comparison, the HemoCue WBC DIFF correctly identified four out of six samples with a low WBC count, two out of two samples with a high WBC count, and 20 of 21 samples with a normal WBC count.
21) were correctly identified by the LeukoScope. The one incorrectly classified sample was measured as 9900 cells per μL by venous measurement and 9400 cells per μL on the LeukoScope. Nineteen of the 21 samples falling in the normal range were correctly identified by the LeukoScope. One sample was measured as 4400 cells per μL by venous measurement and as 3100 cells per μL on the LeukoScope, and one was measured as 8300 cells per μL by venous measurement and as 9700 cells per μL on the LeukoScope. In comparison, the HemoCue WBC DIFF correctly identified four out of six samples with a low WBC count, two out of two samples with a high WBC count, and 20 of 21 samples with a normal WBC count.
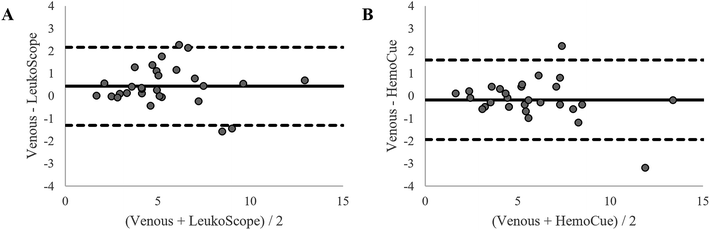
Fig. 8 shows Bland–Altman plots comparing the performance of the LeukoScope and the HemoCue WBC DIFF to that of the clinical standard of care. The 95% limits of agreement for the two point of care systems are similar: the limits for the LeukoScope ranged from −1300 cells per μL to 2200 cells per μL (Fig. 8A), and those for the HemoCue WBC DIFF ranged from −1900 cells per μL to 1600 cells per μL (Fig. 8B).
WBC differential results: neutrophil count
A red-to-green ratio of 0.76 was determined to be the optimal threshold to differentiate neutrophils from other types of WBCs. This threshold was then used to calculate the neutrophil count for all samples in the validation set. Fig. 9A shows the relationship between the neutrophil count measured using the LeukoScope and the venous clinical standard of care for all samples in the validation set. As shown in Fig. 9A, three out of four samples with low neutrophil counts (<1560 cells per μL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21) were correctly identified as low using the LeukoScope, and two of two samples with a high neutrophil count (>6450 cells per μL
21) were correctly identified as low using the LeukoScope, and two of two samples with a high neutrophil count (>6450 cells per μL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21) were correctly identified using the LeukoScope. The incorrectly classified sample was measured as 1000 cells per μL by venous measurement and 1600 cells per μL using the LeukoScope. Twenty-one of the 23 samples falling in the normal range were correctly identified with the LeukoScope. One sample was measured as 6200 cells per μL by venous measurement and as 7200 cells per μL using the LeukoScope, and one was measured as 3700 cells per μL by venous measurement and as 6700 cells per μL with the LeukoScope. In comparison, the HemoCue WBC DIFF correctly identified four out of four samples with a low neutrophil count, two out of two samples with a high neutrophil count, and 22 of 23 samples with a normal neutrophil count.
21) were correctly identified using the LeukoScope. The incorrectly classified sample was measured as 1000 cells per μL by venous measurement and 1600 cells per μL using the LeukoScope. Twenty-one of the 23 samples falling in the normal range were correctly identified with the LeukoScope. One sample was measured as 6200 cells per μL by venous measurement and as 7200 cells per μL using the LeukoScope, and one was measured as 3700 cells per μL by venous measurement and as 6700 cells per μL with the LeukoScope. In comparison, the HemoCue WBC DIFF correctly identified four out of four samples with a low neutrophil count, two out of two samples with a high neutrophil count, and 22 of 23 samples with a normal neutrophil count.
 | ||
| Fig. 9 (A) The neutrophil count measured using the LeukoScope vs. reference venous neutrophil counts for the validation set (n = 29). The solid line represents perfect agreement, and the dashed lines show the cutoffs for low, normal, and high neutrophil counts.21 Bland–Altman plots comparing the neutrophil counts in the validation set measured by (B) the LeukoScope to venous measurement and (C) the HemoCue WBC DIFF to venous measurement. The solid line shows the mean offset, and the dotted lines show the 95% limits of agreement. All units are in WBC × 103/μL. | ||
Fig. 9B and C show Bland–Altman plots comparing the LeukoScope and HemoCue WBC DIFF measurements of the neutrophil count to that of the clinical gold standard. The 95% limits of agreement for the LeukoScope are −1600 cells per μL to 1300 cells per μL (Fig. 9B), similar to the range measured for the HemoCue WBC DIFF of −1300 cells per μL to 1600 cells per μL (Fig. 9C).
Per test cost analysis
The total cost of goods to produce a LeukoScope cartridge is approximately $0.31 when materials are purchased at small volume (∼1000 pieces). These estimates do not include the cost to manufacture, assemble, or package cartridges. The cost of parts for the prototype a single LeukoScope reader was approximately $1800.Conclusions
This study evaluates the performance of the LeukoScope, a new portable, integrated, low-cost system designed to measure WBC and neutrophil counts at the point of care. A disposable cartridge accepts whole blood from a fingerprick sample, and the system requires minimal user steps and reports results within five minutes. After initial modification, 77% of samples measured with LeukoScope at the point of care passed quality control. For samples passing quality control, the accuracy of LeukoScope is comparable to that of the HemoCue WBC DIFF as judged by Bland–Altman analysis and using clinical measurement of venous blood as the reference standard.Despite promising results, performance of the LeukoScope must be improved to ensure that quality images are obtained from all samples and to increase the number of WBCs analyzed per test to meet the 100 cells per FOV criterion for a statistically accurate differential.18 In this system's design, we wanted to provide the most compact, portable system possible; as such the optical track of the system was minimized. To achieve this, some design compromises were made, such as the simple illumination system design consisting of only a LED and catoptric collector, the decreased optical path, and the use of low-cost mechanical mounting for the positioning of components. Additionally, the system presented here had a limited depth of field (5.8 μm), which was less than the expected variability in cartridge thickness due to manufacturing tolerances of component pieces. As such, variation in cartridge thickness that fell within manufacturing tolerances could cause images to be out of focus. In response to the quality of images seen in this study, we have re-designed the portable microscope to a telecentric system with an increased field of view and increased the depth of focus to improve image quality. Additional clinical studies are planned to evaluate the accuracy of the revised LeukoScope.
Our system is also limited by its inability to perform a three-part differential; similar limitations have been reported in the HemoCue WBC DIFF when measuring WBC subtypes other than neutrophils.12 While previous studies have shown that differentiating lymphocytes, monocytes, and granulocytes in an imaging-based POC system is theoretically possible,14,17 the current spatial resolution and FOV of our system limit this application. However, promising results have been reported in the development of POC blood analysis systems with large FOVs, improved spatial resolution, and accurate results.22–24 Techniques such as these could be implemented to further improve the proposed system.
The ability to perform an accurate, low-cost WBC and neutrophil count at the point of care can improve clinical care in many settings. In this study population of general oncology patients, routine WBC and neutrophil counts are performed to determine if a patient is able to undergo chemotherapy treatment. However, patients must travel to the hospital in order to have these levels determined and undergo treatment, and if the counts are too low the trip will have been made in vain. The ability to screen for low counts prior to making a trip to the hospital for treatment has been shown to increase patient satisfaction by saving patients time and expense.6 The ability to monitor WBC and neutrophil counts would also improve clinical care in many low-resource settings. Patients with bacterial infections are more likely to have elevated WBC and neutrophil counts; conversely, severely decreased neutrophil counts (neutropenia) indicate a patient is at higher risk for developing a severe infection.1 Thus, a WBC and neutrophil count could help reduce overuse of antibiotics by informing clinicians when the use of antibiotics is appropriate. In the absence of such information, overuse of antibiotics has led to an increase in antibiotic resistance globally.25
Conflicts of interest
The authors have no conflicts to declare.Acknowledgements
This work was supported by Grant #2013138 from the Doris Duke Charitable Foundation. This work was funded by Grant #OPP1028843 from the Bill and Melinda Gates Foundation through the Grand Challenges in Global Health Initiative. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. 1450681. We would like to acknowledge Kristi Villalta, RN, and her team for her assistance in enrolling study participants at Lyndon B. Johnson Hospital.References
- R. J. Laudicina and Y. Simonian, in Clinical Laboratory Hematology, ed. S. B. McKenzie and J. L. Williams, Pearson, Boston, 2nd edn, 2010, pp. 104–145 Search PubMed.
- B. Houwen, Lab. Hematol., 2001, 7, 89–100 Search PubMed.
- D. J. Williams, M. Hall, K. A. Auger, J. S. Tieder, K. Jerardi, M. A. Queen, A. Statile, A. Myers, S. S. Shah and C. Author, Pediatr. Infect. Dis. J., 2015, 34, 792–793 CrossRef PubMed.
- C. Fleming, H. Russcher, J. Lindemans and R. de Jonge, Clin. Chem. Lab. Med., 2015, 53, 1689–1706 CAS.
- A. J. Kapasi, S. Dittrich, I. J. González and T. C. Rodwell, PLoS One, 2016, 11, e0160278 CrossRef PubMed.
- A. C. Lohman, I. Van Rijn, C. L. Lindhardt, R. Vonthein, D. Rades and N. H. Hollander, In Vivo, 2018, 32, 1283–1288 CrossRef PubMed.
- K. Fujimoto, Sysmex Journal International, 1999, 9, 31–44 Search PubMed.
- A. Osei-Bimpong, C. Jury, R. Mclean and S. M. Lewis, Int. J. Lab. Hematol., 2009, 31, 657–664 CrossRef CAS PubMed.
- H. Russcher, N. Van Deursen, T. Ermens and R. De Jonge, Ned. Tijdschr. Klin. Chem. Laboratoriumgeneeskd., 2013, 38, 140–141 Search PubMed.
- B. A. Spaeth, M. D. S. Shephard, B. McCormack and G. Sinclair, Pathology, 2015, 47, 91–95 CrossRef PubMed.
- M. Karawajczyk, S. Haile, M. Grabski and A. Larsson, Acta Paediatr., 2017, 106, 974–978 CrossRef CAS PubMed.
- H. N. Bui, J. P. A. M. Bogers, D. Cohen, T. Njo and M. H. Herruer, Int. J. Lab. Hematol., 2016, 38, 703–709 CrossRef CAS PubMed.
- Z. J. Smith, T. Gao, K. Chu, S. M. Lane, D. L. Matthews, D. M. Dwyre, J. Hood, K. Tatsukawa, L. Heifetz and S. Wachsmann-Hogiu, Lab Chip, 2014, 14, 3029–3036 RSC.
- A. J. Powless, R. J. Conley, K. A. Freeman and T. J. Muldoon, J. Biomed. Opt., 2017, 22, 035001 CrossRef PubMed.
- S. Zheng, J. Lin, H. Kasdan and Y. Tai, Sens. Actuators, B, 2008, 132, 558–567 CrossRef CAS.
- C.-Y. Lee, C.-L. Chang, Y.-N. Wang and L.-M. Fu, Int. J. Mol. Sci., 2011, 12, 3263–3287 CrossRef CAS PubMed.
- C. E. Majors, M. E. Pawlowski, T. Tkaczyk and R. R. Richards-Kortum, Presented in Part at Health Innovations and Point-of-Care Technologies Conference, Seattle, WA, 2014 Search PubMed.
- B. F. Rodak and J. H. Carr, Clinical Hematology Atlas, Elsevier Inc., St. Louis, 4th edn, 2013 Search PubMed.
- M. M. Bond and R. R. Richards-Kortum, Am. J. Clin. Pathol., 2015, 144, 885–894 CrossRef CAS PubMed.
- J. M. Bland and D. G. Altman, Lancet, 1986, 327, 307–310 CrossRef.
- Mayo Clinic Laboratories, CBC with Differential, Blood, https://www.mayocliniclabs.com/test-catalog/Clinical+and+Interpretive/9109, accessed February 2019 Search PubMed.
- A. Skandarajah, C. D. Reber, N. A. Switz and D. A. Fletcher, PLoS One, 2014, 9, e96906 CrossRef.
- S. Seo, S. O. Isikman, I. Sencan, O. Mudanyali, T.-W. Su, W. Bishara, A. Erlinger and A. Ozcan, Anal. Chem., 2010, 82, 4621–4627 CrossRef CAS PubMed.
- C. Wong, M. E. Pawlowski, A. Forcucci, C. E. Majors, R. Richards-Kortum and T. S. Tkaczyk, Biomed. Opt. Express, 2018, 9, 1041 CrossRef CAS PubMed.
- WHO, Antimicrobial Resistance: Global Report on Surveillance 2014, 2014 Search PubMed.
| This journal is © The Royal Society of Chemistry 2019 |