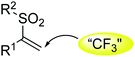Open Access Article
Open Access ArticleCombinatorial synthesis and biological evaluations of (E)-β-trifluoromethyl vinylsulfones as antitumor agents†
Haosha
Tang
a,
Yunyan
Kuang
b,
Julan
Zeng
c,
Xiaofang
Li
d,
Wei
Zhou
 *b and
Yuan
Lu
*a
*b and
Yuan
Lu
*a
aObstetrics and Gynecology Hospital, Fudan University, 419 Fangxie Road, Shanghai 200011, China
bDepartment of Chemistry, Fudan University, 2005 Songhu Road, Shanghai 200433, China. E-mail: zhouw@fudan.edu.cn
cDepartment of Chemistry, Changsha University of Science and Technology, Changsha 410114, China
dSchool of Chemistry and Chemical Engineering, Hunan University of Science and Technology, Xiangtan 411201, China
First published on 3rd October 2019
Abstract
Combinatorial synthesis of (E)-β-trifluoromethyl vinylsulfones is accomplished through a reaction of alkynes, Togni reagent, and sodium benzenesulfinates in DMSO under metal-free conditions at room temperature. These compounds are evaluated in several assays against different tumor cells. Some hits are identified against ES-2, HO-8910, and K562.
Introduction
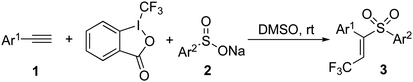
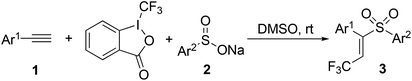
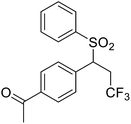
The chalcogen-containing skeleton has become a privileged and attractive scaffold in medicinal chemistry owing to its unique biological activities.1 In particular, more and more novel biological effects of vinyl sulfone compounds have been discovered recently. It has been reported that structures containing an α,β-unsaturated vinylsulfone moiety exhibit modest inhibitory potencies in inflammation as a novel class toward Parkinson's2,3 and arthritis4 disease therapy by depressing the expression of endothelial cells of adhesion molecules, including vascular cell adhesion molecule-1 (VCAM-1) and intercellular adhesion molecule-1 (ICAM-1), as well as inhibiting the nuclear factor E2-related factor 2 (Nrf2) pathway which is responsible for the cellular defense system against oxidative stress. They also possess other biological activities, such as anti-Gram-positive bacteria as SrtA transpeptidase inhibitors,5 anti-parasitic as cysteine protease inhibitors,6 and anti-virus as potent inhibitors of HIV-1 integrase.7Since compounds with substitution of fluorine might have a higher stability against metabolic enzymes and a better membranous permeability,8,9 we therefore considered to introduce a trans-trifluoromethyl group to the α,β-unsaturated vinylsulfone entity (Scheme 1). Thus, we initiated a program for the combinatorial synthesis of trifluoromethyl-substituted (E)-vinylsulfones and their biological evaluations as antitumor agents. Recently, we and others have involved in the synthesis of sulfonyl compounds,10,11 and we identified that (E)-β-trifluoromethyl vinylsulfones could be accessed through a three-component reaction of alkynes, Togni reagent, and sodium benzenesulfinates in DMSO under metal-free conditions at room temperature.12 We envisioned that the library of (E)-β-trifluoromethyl vinylsulfones could be constructed via this method through diversity-oriented synthesis.
Herein, we report the biological evaluation of a series of trans-trifluoromethyl vinylsulfone derivatives by aim to study the structure–activity relationship and identify a hit structure. By initial screening, we found that the introduction of β-trans-trifluoromethyl group led to a potent activity against tumor cells proliferation.
Results and discussion
Chemistry
The synthetic route for the target (E)-β-trifluoromethyl vinylsulfones was described in Scheme 2.12 By treatment of alkynes 1, Togni reagent, and sodium benzenesulfinates 2 in DMSO under metal-free conditions at room temperature, a series of (E)-β-trifluoromethyl vinylsulfones 3 were obtained in moderate to good yields. On the basis of this strategy, the library of (E)-β-trifluoromethyl vinylsulfones was constructed easily with high efficiency.Biological activity
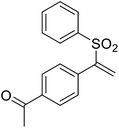
We chose (E)-(3,3,3-trifluoro-1-(phenylsulfonyl)prop-1-en-1-yl)benzene as our structural template and modified the aryl groups. Subsequently, 15 derivatives were synthesized, which included various functional groups on both aryl groups (Ar1 and Ar2) (Table 1, compounds 3-1 to 3-15). To our delight, these compounds had an excellent inhibitory activity against different tumor cells and especially against ES-2, HO-8910 and K562 with less than 10 μM of IC50 values. As outlined in Table 1, it was apparent that compounds 3-1 to 3-4 offered reasonable potency profiles when the electron-withdrawing groups were introduced to Ar1, while the electron-donating groups on Ar1 (compounds 3-5 and 6) would result in decreased activities. But the much stronger electron-withdrawing groups of cyano, fluoro, and trifluoromethyl didn't give better activity improvements (compounds 3-4, 3-7 to 3-9, and 3-11). A same electronic effect was also found on Ar2 (compounds 3-16 to 3-20), but the activity changes were much minor than that on Ar1.To further validate the activities of electron-withdrawing groups, more substituents were introduced to investigate the structure–activity relations (Table 2, compounds 3-16 to 3-30). With the same results, the changes of Ar2 didn't make significant activity changes (compounds 3-16 to 3-20). Compound 3-27, with an acetyl group on p-position of Ar1, all of the IC50 values on ES-2, HO-8910, A2780, and K562 decreased to nM level. However, the compound became more inactive when the acetyl group was connected on the m-position (compound 3-28). When the acetyl group was attached on Ar2, the activity was affected very little (compounds 3-20, 21, 23, and 25). Interestingly, when a strong electron-withdrawing carboxyl group was introduced to Ar1 (compounds 3-25 and 3-26), the activity was dramatically decreased. Therefore, it was reasonable to conclude that an acetyl group on the p-position of Ar1 was essential for the inhibitory activity, which might be due to a better noncovalent binding to the biological target by a moderate electronic withdrawing effect of the acetyl group.
Moreover, we synthesized compounds without double bond or trifluoromethyl group to identify the key structure of this skeleton (compounds 3-29 and 30). The results showed that the proliferation inhibition activities of these two compounds were both obviously inhibited compared with 3-27, about 40-, 11-, and 43-fold decrease for 3-29, and 60-, 16-, and 86-fold decrease for 3-30, on ES-2, HO-8190, and K562, respectively. Therefore, it showed that β-trifluoromethyl vinylsulfone along with an acetyl group on the p-position of Ar1 was essential for the antitumor activity. Subsequently, varies different electron-withdrawing groups were introduced to Ar2 with the structure of compound 3-27 as the structural template (Table 3, compounds 31-41). Unfortunately, no compound showed better profiles than that of compound 3-27.
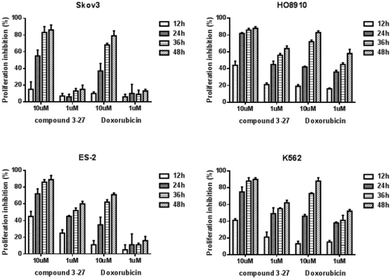
The further time courses of the proliferation inhibitions on Skov3, HO-8910, ES-2, and K562 cells of compound 3-27 were conducted with different concentrations (10 and 1 μM) for different time periods (12, 24, 36, and 48 h). As shown in Scheme 3, the cellular growth was obviously inhibited in a dose- and time-dependent manner. All of tested cell lines were more sensitive to compound 3-27 treatment than doxorubicin.
Furthermore, special attention was paid to potential cell toxicities caused by the tested compounds. The cell toxicities of several active compounds were evaluated on human bone marrow mesenchymal stem cells (hBMSCs), compared with doxorubicin (Table 4). Except compound 3-31, all of these tested compounds had much lower toxicities on hBMSCs. A representative example, compound 3-27 had much higher inhibition activities on tumor cells with only 1/5 toxicity on hBMSCs than doxorubicin, which indicates a potential development of such a new kind of skeleton as an alternative chemo drug.
| Compounds | 3-27 | 3-32 | 3-36 | 3-37 | 3-39 | 3-40 | Doxorubicin |
| IC50 (μM) | 5.4 ± 0.61 | 0.65 ± 0.45 | 15.3 ± 0.72 | 6.5 ± 0.56 | 6.8 ± 1.57 | 8.4 ± 0.91 | 0.96 ± 0.24 |
Conclusions
In summary, we have identified several hits with a structural β-trans-trifluoromethyl vinylsulfone, which shows promising biological effects on antitumor with low cell toxicities, even the biological target is still unknown. The moderate electron-withdrawing groups, such as chloro, bromo, or acetyl group, on the p-position of Ar1 and with Ar2 unsubstituted both benefit the activity. More studies of structure–activity relationship, biological mechanism, and in vivo activity are likely to be subsequently conducted on such a lead compound.Experimental
Cell culture
Human ovarian cancer cells (ES-2, HO-8910, Skov3, and A2780), human alveolar basal epithelial cells (A549), human hepatocellular carcinoma cells (Bel-7402), human myelogenous leukemia cells (K562) and human bone marrow mesenchymal stem cell (hBMSCs) were obtained from the Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Cancer cells were maintained in RPMI-1640 supplemented with antibiotics (100 units per mL penicillin A and 100 μg mL−1 streptomycin) and 10% FBS in an atmosphere of 5% CO2 at 37 °C. hBMSCs was maintained in high glucose DMEM with 10% FBS. The medium was changed every three days. Exponentially growing cells were plated at a final concentration of 1 × 104 cells per well in 96-well plates for cell proliferation assay.Cell viability
The cell growth inhibitory effect of tested compounds determined using the MTT assay. After incubation for 24 h in 96-well plates, cells were treated with various concentrations (25, 12.5, 6.25, 3.12, 1.56, 0.78, 0.39, and 0.20 μM) of tested compounds for 48 hours, and the MTT solution (0.5 mg mL−1) was added. After 4 h of incubation at 37 °C for MTT-formazan formation, the supernatant was removed and 100 μL of dimethyl sulfoxide (DMSO) was added into each well. Absorbance at 490 nm was determined spectrophotometrically by using a microplate reader (Epoch, BioTek Instruments, Inc., VT, USA). Each concentration was performed in triplicate. Antitumor activity was evaluated using IC50 determined by non-linear regression analysis.General experimental procedure for the synthesis of (E)-β-trifluoromethyl vinylsulfones from alkyne, Togni reagent, and sodium benzenesulfinate
Under nitrogen atmosphere, a mixture of alkyne 1 (0.2 mmol) and Togni reagent (0.22 mmol) in DMSO (1.0 mL) was stirred for several minutes. Then sodium benzenesulfinate 2 (0.4 mmol) in DMSO (2.0 mL) was added to the solution. The mixture was stirred overnight at room temperature. After completion of reaction as indicated by TLC, water (10 mL) was added and the mixture was extracted by EtOAc (3 × 10 mL). The solvent was evaporated and the residue was purified by flash column chromatography (EtOAc/n-hexane, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8) to give the desired product 3. Data of typical examples are shown below.
8) to give the desired product 3. Data of typical examples are shown below.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial supports from the Natural Science Foundation of Science and Technology Commission of Shanghai Municipality (No. 19ZR1407100) and Double First-Class Construct Project of Fudan University (No. IDH1615098) are gratefully acknowledged.Notes and references
- (a) X. Dneg, H. Cao, C. Chen, H. Zhou and L. Yu, Sci. Bull., 2019, 64, 1280 CrossRef; (b) M. Liu, Y. Li, L. Yu, Q. Xu and X. Jiang, Sci. China: Chem., 2018, 61, 294 CrossRef CAS; (c) T. Prochnow, A. Maroneze, D. F. Back and G. Zeni, J. Org. Chem., 2019, 84, 2891 CrossRef CAS; (d) L.-H. Lu, S.-J. Zhou, W.-B. He, W. Xia, P. Chen, X. Yu, X. Xu and W.-M. He, Org. Biomol. Chem., 2018, 16, 9064 RSC; (e) S. Kodama, T. Saeki, K. Mihara, S. Higashimae, S.-i. Kawaguchi, M. Sonoda, A. Nomoto and A. Ogawa, J. Org. Chem., 2017, 82, 12477 CrossRef CAS; (f) C. Ma, J.-Y. Zhou, Y.-Z. Zhang, Y. Jiao, G.-J. Mei and F. Shi, Chem.–Asian J., 2018, 13, 2549 CrossRef CAS; (g) C. Ma, F. Jiang, F.-T. Sheng, Y. Jiao, G.-J. Mei and F. Shi, Angew. Chem., Int. Ed., 2019, 58, 3014 CrossRef CAS PubMed; (h) X. Gong, J. Chen, X. Li, W. Xie and J. Wu, Chem.–Asian J., 2018, 13, 2543 CrossRef CAS PubMed; (i) D. Ren, B. Liu, X. Li, S. Koniarz, M. Pawlicki and P. J. Chmielewski, Org. Chem. Front., 2019, 6, 908 RSC; (j) Z. Wang, L. Yang, H.-L. Liu, W.-H. Bao, Y.-Z. Tan, M. Wang, Z. Tang and W.-M. He, Youji Huaxue, 2018, 38, 2639 CrossRef CAS; (k) Y. Zheng, M. Liu, G. Qiu, W. Xie and J. Wu, Tetrahedron, 2019, 75, 1663 CrossRef CAS.
- S. Woo, J. Kim, M. Moon, S. Han, S. Yeon, J. Choi, B. Jang, H. Song, Y. Kang, J. Kim, J. Lee, D. Kim, O. Hwang and K. Park, J. Med. Chem., 2014, 57, 1473 CrossRef CAS.
- J. A. Lee, J. H. Kim, S. Y. Woo, H. J. Son, S. H. Han, B. K. Jang, J. W. Choi, D. J. Kim, K. D. Park and O. Hwang, Br. J. Pharmacol., 2015, 172, 1087 CrossRef CAS.
- (a) L. Ni, S. Zheng, P. K. Somers, L. K. Hoong, R. R. Hill, E. M. Marino, K.-L. Suen, U. Saxena and C. Q. Meng, Bioorg. Med. Chem. Lett., 2003, 13, 745 CrossRef CAS; (b) W. Xie, Y. Wu, J. Zhang, Q. Mei, Y. Zhang, N. Zhu, R. Liu and H. Zhang, Eur. J. Med. Chem., 2018, 145, 35 CrossRef CAS; (c) W. Xie, S. Xie, Y. Zhou, X. Tang, J. Liu, W. Yang and M. Qiu, Eur. J. Med. Chem., 2014, 81, 22 CrossRef CAS; (d) W. Xie, H. Zhang, J. He, J. Zhang, Q. Yu, C. Luo and S. Li, Bioorg. Med. Chem. Lett., 2017, 27, 530 CrossRef CAS; (e) T. Guo, Y. Liu, Y.-H. Zhao, P.-K. Zhang, S.-L. Han and H.-M. Liu, Tetrahedron Lett., 2016, 57, 3920 CrossRef CAS.
- (a) B. A. Frankel, M. Bentley, R. G. Kruger and D. G. Mccafferty, J. Am. Chem. Soc., 2004, 126, 3404 CrossRef CAS; (b) D. Chen, Y. Shan, J. Li, J. You, X. Sun and G. Qiu, Org. Lett., 2019, 21, 4044 CrossRef CAS; (c) Y. Zheng, M. Liu, G. Qiu, W. Xie and J. Wu, Tetrahedron, 2019, 75, 1663 CrossRef CAS; (d) R. Liu, W. Xie, H. Zhou, Y. Zhang and G. Qiu, J. Org. Chem., 2019, 84(18), 11763–11773 CrossRef CAS; (e) Y.-H. Wang, B. Ouyang, G. Qiu, H. Zhou and J.-B. Liu, Org. Biomol. Chem., 2019, 17, 4335 RSC; (f) G. Qiu, Z. Chen, W. Xie and H. Zhou, Eur. J. Org. Chem., 2019, 4327 CrossRef CAS; (g) Y.-C. Wang, R.-X. Wang, G. Qiu, H. Zhou, W. Xie and J.-B. Liu, Org. Chem. Front., 2019, 6, 2471 RSC; (h) Y.-H. Wang, G. Qiu, H. Zhou, W. Xie and J.-B. Liu, Tetrahedron, 2019, 75, 3850 CrossRef CAS; (i) R.-X. Wang, Z. Fang, G. Qiu, W. Xie and J.-B. Liu, Synthesis, 2019 DOI:10.1055/s-0039-1690155.
- I. D. Kerr, J. H. Lee, C. J. Farady, R. Marion, M. Rickert, M. Sajid, K. C. Pandey, C. R. Caffrey, J. Legac, E. Hansell, J. McKerrow, C. S. Craik, P. J. Rosenthal and L. S. Brinen, J. Biol. Chem., 2009, 284, 25697 CrossRef CAS.
- D. C. Meadows and J. Gervay-Hague, Med. Res. Rev., 2006, 26, 793 CrossRef CAS.
- (a) S. Purser, P. R. Moore, S. Swallow and V. Gouverneur, Chem. Soc. Rev., 2008, 37, 320 RSC; (b) D. O'Hagan, Chem. Soc. Rev., 2008, 37, 308 RSC; (c) W. K. Hagmann, J. Med. Chem., 2008, 51, 4359 CrossRef CAS; (d) J. Wang, M. SanchezRosello, J. L. Aceña, C. del Pozo, A. E. Sorochinsky, S. V. Fustero, A. Soloshonok and H. Liu, Chem. Rev., 2014, 114, 2432 CrossRef CAS; (e) Y. Zhou, J. Wang, Z. Gu, S. Wang, W. Zhu, J. L. Aceña, V. A. Soloshonok, K. Izawa and H. Liu, Chem. Rev., 2016, 116, 422 CrossRef CAS.
- (a) X. Fu, Y. Meng, X. Li, M. Stepień and P. J. Chmielewski, Chem. Commun., 2018, 54, 2510 RSC; (b) X. Li, Y. Meng, P. Yi, M. Stepień and P. J. Chmielewski, Angew. Chem., Int. Ed., 2017, 56, 10810 CrossRef CAS; (c) B. Liu, T. Yoshida, X. Li, M. Stepień, H. Shinokubo and P. J. Chmielewski, Angew. Chem., Int. Ed., 2016, 55, 13142 CrossRef CAS; (d) Z. Deng, X. Li, M. Stepień and P. J. Chmielewski, Chem. –Eur. J., 2016, 22, 4231 CrossRef CAS; (e) K. Deng, X. Li and H. Huang, Electrochim. Acta, 2016, 204, 84 CrossRef CAS; (f) B. Liu, X. Li, M. Stepień and P. J. Chmielewski, Chem. –Eur. J., 2015, 21, 7790 CrossRef CAS PubMed; (g) B. Liu, H. Fang, X. Li, W. Cai, L. Bao, M. Rudolf, F. Plass, L. Fan, X. Lu and D. M. Guldi, Chem. –Eur. J., 2015, 21, 746 CrossRef CAS; (h) B. Liu, X. Li, J. Maciołek, M. Stepień and P. J. Chmielewski, J. Org. Chem., 2014, 79, 3129 CrossRef CAS; (i) B. Liu, X. Li, X. Xu, M. Stepień and P. J. Chmielewski, J. Org. Chem., 2013, 78, 1354 CrossRef CAS; (j) X. Li, B. Liu, P. J. Chmielewski and X. Xu, J. Org. Chem., 2012, 77, 8206 CrossRef CAS; (k) X. Li, B. Liu, X. Yu, P. Yi, R. Yi and P. J. Chmielewski, J. Org. Chem., 2012, 77, 2431 CrossRef CAS PubMed; (l) X. Li, B. Liu, P. Yi, R. Yi, X. Yu and P. J. Chmielewski, J. Org. Chem., 2011, 76, 2345 CrossRef CAS.
- For recent examples, see: (a) X. Gong, M. Wang, S. Ye and J. Wu, Org. Lett., 2019, 21, 1156 CrossRef CAS; (b) X. Wang, M. Yang, W. Xie, X. Fan and J. Wu, Chem. Commun., 2019, 55, 6010 RSC; (c) S. Ye, D. Zheng, J. Wu and G. Qiu, Chem. Commun., 2019, 55, 2214 RSC; (d) S. Ye, Y. Li, J. Wu and Z. Li, Chem. Commun., 2019, 55, 2489 RSC; (e) X. Gong, X. Li, W. Xie, J. Wu and S. Ye, Org. Chem. Front., 2019, 6, 1863 RSC; (f) J. Zhang, W. Xie, S. Ye and J. Wu, Org. Chem. Front., 2019, 6, 2254 RSC; (g) S. Ye, T. Xiang, X. Li and J. Wu, Org. Chem. Front., 2019, 6, 2183 RSC; (h) S. Ye, X. Li, W. Xie and J. Wu, Asian J. Org. Chem., 2019, 8, 893 CrossRef CAS; (i) S. Ye, X. Li, W. Xie and J. Wu, Eur. J. Org. Chem., 2019 DOI:10.1002/ejoc.201900396; (j) J. Zhang, X. Li, W. Xie, S. Ye and J. Wu, Org. Lett., 2019, 21, 4950 CrossRef CAS; (k) Y. Zong, Y. Lang, M. Yang, X. Li, X. Fan and J. Wu, Org. Lett., 2019, 21, 1935 CrossRef CAS; (l) F.-S. He, Y. Wu, X. Li, H. Xia and J. Wu, Org. Chem. Front., 2019, 6, 1873 RSC.
- (a) C. Wu, L.-H. Lu, A.-Z. Peng, G.-K. Jia, C. Peng, Z. Cao, Z. Tang, W.-M. He and X. Xu, Green Chem., 2018, 20, 3683 RSC; (b) L.-H. Lu, S.-J. Zhou, M. Sun, J.-L. Chen, W. Xia, X. Yu, X. Xu and W.-M. He, ACS Sustainable Chem. Eng., 2019, 7, 1574 CrossRef CAS; (c) K.-J. Liu, S. Jiang, L.-H. Lu, L.-L. Tang, S.-S. Tang, H.-S. Tang, Z. Tang, W.-M. He and X. Xu, Green Chem., 2018, 20, 3038 RSC; (d) L.-Y. Xie, S. Peng, J.-X. Tan, R.-X. Sun, X. Yu, N.-N. Dai, Z.-L. Tang, X. Xu and W.-M. He, ACS Sustainable Chem. Eng., 2018, 6, 16976 CrossRef CAS; (e) L.-Y. Xie, S. Peng, F. Liu, J.-Y. Yi, M. Wang, Z. Tang, X. Xu and W.-M. He, Adv. Synth. Catal., 2018, 360, 4259 CrossRef CAS; (f) L.-Y. Xie, S. Peng, L.-H. Lu, J. Hu, W.-H. Bao, F. Zeng, Z. Tang, X. Xu and W.-M. He, ACS Sustainable Chem. Eng., 2018, 6, 7989 CrossRef CAS; (g) C. Wu, H.-J. Xiao, S.-W. Wang, M.-S. Tang, Z.-L. Tang, W. Xia, W.-F. Li, C. Zhong and W.-M. He, ACS Sustainable Chem. Eng., 2019, 7, 2169 CrossRef CAS; (h) L.-Y. Xie, S. Peng, L.-L. Jiang, X. Peng, W. Xia, X. Yu, X.-X. Wang, Z. Cao and W.-M. He, Org. Chem. Front., 2019, 6, 167 RSC; (i) L.-Y. Xie, S. Peng, F. Liu, G.-R. Chen, W. Xia, X. Yu, W.-F. Li, Z. Cao and W.-M. He, Org. Chem. Front., 2018, 5, 2604 RSC; (j) C. Wu, Z. Wang, Z. Hu, F. Zeng, X.-Y. Zhang, Z. Cao, Z. Tang, W.-M. He and X. Xu, Org. Biomol. Chem., 2018, 16, 3177 RSC; (k) C. Wu, J. Wang, X.-Y. Zhang, G.-K. Jia, Z. Cao, Z. Tang, X. Yu, X. Xu and W.-M. He, Org. Biomol. Chem., 2018, 16, 5050 RSC.
- Y. Xiang, Y. Li, Y. Kuang and J. Wu, Adv. Synth. Catal., 2017, 359, 2605 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06368d |
| This journal is © The Royal Society of Chemistry 2019 |