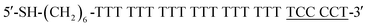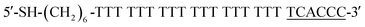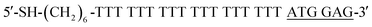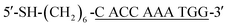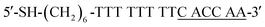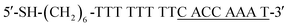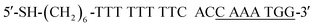Open Access Article
Open Access ArticleAn aptasensor based on the microscopic enumeration of encoding gold nanoparticles for the detection of C-reactive protein†
Yuanfang Zhaoab,
Jingru Zhaoa,
Tian Jinc,
Shuqing Sun *ad,
Wenlan Liue and
Ying Tan
*ad,
Wenlan Liue and
Ying Tan *c
*c
aInstitute of Optical Imaging and Sensing, Shenzhen Key Laboratory for Minimal Invasive Medical Technologies, Graduate School at Shenzhen, Tsinghua University, Shenzhen, 518055, P. R. China. E-mail: sun.shuqing@sz.tsinghua.edu.cn
bOpen FIESTA, Graduate School at Shenzhen, Tsinghua University, Shenzhen, 518055, P. R. China
cState Key Laboratory of Chemical Oncogenomics, Key Laboratory of Chemical Biology, Graduate School at Shenzhen, Tsinghua University, Shenzhen, 518055, P. R. China
dDepartment of Physics, Tsinghua University, Beijing 100084, P. R. China
eThe Central Laboratory, The First Affiliated Hospital of Shenzhen University, Shenzhen Second People's Hospital, Shenzhen 518035, China
First published on 24th October 2019
Abstract
C-reactive protein (CRP) is a crucial clinical biomarker for inflammatory and cardiovascular diseases. Therefore, the sensitive, selective and convenient detection of CRP is of great significance. Using gold nanoparticles (AuNPs) and combining the specific interaction between an aptamer and CRP, we developed a simple and convenient assay for CRP detection. The aptamer-based probe was fabricated through the hybridization of CRP-aptamer immobilized on magnetic beads (MBs) to a short complementary DNA (cDNA) chain attached to AuNPs to form a MB–Aptamer–AuNP sandwich structure. Upon the addition of CRP, aptamer–cDNA dehybridization occurred due to the strong interaction between CRP and the aptamer, resulting in the release of AuNPs, which were subjected to DFM imaging and subsequently counted using the MATLAB program. The number of AuNPs was therefore positively correlated to the concentration of CRP and a detection limit as low as 2.71 nM was achieved. The current approach could also exclude the disturbance of other proteins, including thrombin, IgG, Lys and BSA. In addition, the concentration of CRP detected was in good agreement with the amount cast in bovine and mouse serum, indicating that the proposed probe is robust and accurate, and it is very promising for practical applications where CRP detection is necessary. The current strategy is also promising for the detection of other proteins where a suitable aptamer is selected.
Introduction
C-reactive protein (CRP) is a major acute-phase reactant protein produced in the liver that rises sharply in the plasma when the human body is infected or damaged.1 The serum CRP level is a sensitive and objective indicator of bacterial infection and an important predictor of early joint destruction; it is also associated with atherosclerosis and cerebral infection.2–4 The current techniques for the clinical detection of CRP are mainly antibody-based techniques associated with low sensitivity, expensive reagents and instruments.5–9 Sensors based on aptamer-CRP interactions have been developed recently to avoid the use of expensive antibodies.Aptamers are single-stranded oligonucleotides with dozens of nucleotides (20–80 nucleotides) selected from specific oligonucleotide libraries by the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) technology.10,11 They can form secondary or tertiary structures by self-folding, such as a stem, a convex ring, a hairpin, a G-quadruplex, a pseudo-junction and a tetragonal ring, which can specifically bind to the targets through spatial structure matching.12 Compared to antibodies, aptamers are easily obtained and modified for a wide range of target molecules with excellent stability.13,14 In recent years, some aptamer-based assays have emerged and shown excellent performances over others.15–17 Vance et al. reported the detection of CRP using an aptamer-functionalized surface plasmon resonance (SPR) chip,18 and Centi et al. developed a disposable electrochemical assay using an aptamer as the capture probe.19 On the other hand, due to the strong plasmonic resonance effect, gold nanoparticles (AuNPs) scatter light very strongly and each AuNP can be individually identified using dark-field microscopy (DFM). A digital sensor system based on the enumeration of encoding AuNPs can thus be designed, in which the number of AuNPs are correlated with the concentration of target molecules. This approach is cost-effective and sensitive for biomolecule detection with simplicity.20–24
Herein, we developed a simple and sensitive assay for the detection of CRP based on the digital counting of encoding AuNPs using DFM. First, the complementary DNA (cDNA)-modified AuNPs were coupled to the aptamer-functionalized magnetic beads (MBs) to form an aptamer-based probe with a sandwich structure through hybridization between the aptamer and cDNA. Followed by magnetic separation, the probes were isolated and added to a sample solution, and AuNPs were displaced from the magnetic beads by the target molecules. Second, after removing the magnetic microspheres, the free AuNPs in the supernatant were subjected to DFM imaging and counted by the MATLAB program. The concentration of CRP correlated to the number of AuNPs was then obtained. Fig. 1 shows a schematic diagram of the fabrication process of the probes and the mechanism for CRP detection through the competitive displacement of AuNPs on MBs.
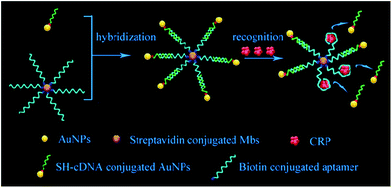 | ||
| Fig. 1 Schematic diagram of the fabrication process of probes and mechanism for CRP detection through competitive displacement of AuNPs on MBs. | ||
Materials and methods
Reagents and apparatus
Streptavidin-modified magnetic beads were purchased from Thermo Fisher Co., Ltd. AuNPs with a diameter of 60 nm were bought from Nanoseedz Co., Ltd. Recombinant Human CRP was obtained from Novoprotein Co., Ltd. (γ-Aminopropyl)triethoxysilane (APTES) was supplied by Chen Gong Co., Ltd (Nanjing, China). Bovine serum albumin (BSA), immunoglobulin G (IgG), lysozyme (Lys), thrombin (Thr) and Tris(2-carboxyethyl)phosphine (TCEP) were purchased from Sigma-Aldrich Co., Ltd (USA). Fetal bovine serum (Cat. No. S0415) was purchased from Millipore Biochrom Co., Ltd. Mouse serum was collected from 2 week-old male NIH mice, which were purchased from Guangdong Medical Animal Center (Guangzhou, China). All animal experiments followed the animal protocols approved by the Animal Care and Use Committee of Tsinghua University (No. 17-lf1#, Beijing, China). Animals received care following the NSFC regulations concerning the use of experimental animals. University protocols and experiments were approved by the Animal Ethics Committee of NSFC. A 40-mer DNA aptamer25 that specifically binds CRP was custom synthesized and HPLC-purified by Sangon Biotech Co., Ltd (Shanghai, China) after modifying with biotin at the 5′-terminal. The sequence of the modified DNA aptamer is 5′-biotin–(CH2)6–CGA AGG GGA TTC GAG GGG TGA TTG CGT GCT CCA TTT GGT G-3′. The complementary DNA strand (cDNA) modified with 5′-terminal thiol groups was synthesized and HPLC-purified by Sangon Biotech Co., Ltd (Shanghai, China). Their sequences are shown in Table 1. All other reagents are of analytical grade. A solution of 0.2 mM phosphate buffered saline (PBS) (pH 7.4) was used as the incubating and washing buffer. All solutions were prepared with ultrapure water produced by a Milli-Q system (≥18.2 MΩ, Millipore, USA).DFM images were obtained using an optical microscope (BX53, Olympus) specially equipped with Colorful CCD (DP73, Olympus, Japan) and analyzed by Olympus cell senstry software. Scanning electron microscopy (SEM) images were obtained by a ZEISS SUPRA®55 field emission scanning electron microscope (Carl Zeiss, Germany). Energy dispersive spectra (EDS) were recorded by an Oxford X-Max 20 spectrometer. Ultraviolet-visible (UV-vis) absorption spectra were recorded using an UV-2450 Spectrophotometer (Shimadzu Corporation, Japan).
Glass slide cleaning and modification
Glass slides were first soaked in piranha solution (H2SO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O2 = 7
H2O2 = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) for over 6 h to remove stains. Following ultrasonic cleaning three times with ultrapure water, the glass slides were soaked in a 10% (v/v) (γ-aminopropyl)triethoxysilane (APTES) ethanol solution for 6 h. Finally, the slides were ultrasonically cleaned three times again with pure water and dried in a vacuum oven for 3 h.
3) for over 6 h to remove stains. Following ultrasonic cleaning three times with ultrapure water, the glass slides were soaked in a 10% (v/v) (γ-aminopropyl)triethoxysilane (APTES) ethanol solution for 6 h. Finally, the slides were ultrasonically cleaned three times again with pure water and dried in a vacuum oven for 3 h.
Preparation of the aptamer-based probe
The scattering characteristics of AuNPs change with their shape and size. AuNPs of 30–60 nm in diameter scatter light around 530 nm and appear green under DFM.20,26 In order to clearly see the signal of AuNPs in DFM and to obtain a stable aptamer-based probe, AuNPs of 60 nm in diameter were used. The preparation of the aptamer-based probe includes three parts: the modification of AuNPs with SH-cDNA and the modification of MBs with an aptamer, followed by the assembly of the probe by mixing MBs and AuNPs to form MBs-aptamer/cDNA-AuNPs. The experimental details are described below.First, we will describe the modification of AuNPs with cDNA. Ten μL of 10 mM TCEP solution was added into 100 μL 10 μM cDNA in TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH = 8.0) to activate the thiol group of cDNA and the mixture was shaken at room temperature (RT) for 1 h. Then, 400 μL of 40 pM AuNPs was added into the as-activated cDNA solution and incubated at RT for 16 h. During this process, 10 μL of 2 M NaCl (0.1 M PBS, 0.1% SDS, pH = 7.5) solution was used to neutralize the negative charges of cDNA and promote the attachment of cDNA onto AuNPs. The salinization process was repeated 6 times with an interval of 20 min. Finally, the mixture was incubated on a shaking bed at RT for 16 h.
Second, we will describe the modification of MBs with the aptamer. Ten μL 10 mg mL−1 MBs modified with streptavidin was washed 3 times using 60 μL B&W Buffer (10 mM Tris–HCl, 1 mM EDTA, 2 M NaCl, pH = 7.5) by the magnetic separation technique, and 5 mg mL−1 MBs was obtained by adding 20 μL hybridization solution (10 mM PBS, 0.01 wt% SDS, pH = 7.4). Then, 100 μL 1 μM solution of biotin-conjugated CRP-aptamer diluted from 1 μL 100 μM was added to the as-prepared dispersion of MBs and incubated at RT for 30 min, resulting in the attachment of the aptamer to MBs through biotin-avidin interactions. Free aptamer in solution was removed by magnetic separation and the aptamer-modified MBs were washed at least 3 times before use.
Third, we will describe the assembly of the probe. cDNA-functionalized AuNPs were centrifuged at 5000 rpm for 7 minutes followed by the addition of 100 μL hybridization solution. This process was repeated 3 times to remove free cDNA in the supernatant. Similarly, MBs were washed with hybridization solution 3 times to remove the unconjugated CRP-aptamer by the magnetic separation technique, followed by the addition of 100 μL hybridization solution. The as-obtained solutions of AuNPs and MBs were mixed and allowed to incubate at RT for 1 h. The supernatant of the mixture was extracted by the magnetic separation technique 3 times to remove the AuNPs that did not attach to MBs.
The detection of CRP
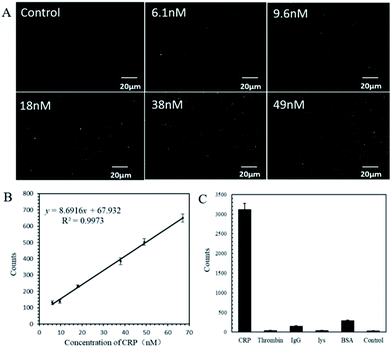
For the detection of CRP using the fabricated aptamer-based probe, CRP with a certain concentration was added to the solution of the probe and incubated for 30 min. Due to the specific interaction between CRP and the aptamer, the aptamer in the probe folded adaptively to form a three-dimensional structure, which bound closely to the target CRP and destroyed the paired bases between the aptamer and cDNA on MBs and AuNPs, respectively, resulting in the release of AuNPs from MBs. Upon magnetic separation, the AuNPs in the supernatant were extracted and subjected to DFM imaging. The numbers of AuNPs displaced by CRP were counted using the MATLAB software. Typically, 10 μL CRP with a gradient concentration in each group was tested. Three μL of the supernatant containing displaced free AuNPs was pipetted onto a clean slide for DFM imaging. The images recorded using DFM were imported into the MATLAB program to quantify the number of AuNPs. The MATLAB program converted the RGB image into HSV format to extract the H value and obtain the number of free AuNPs in the DFM image. A linear relationship between the CRP concentrations and the counts of AuNPs was obtained.Selection of aptamer and cDNA
This paper deals with the competitive replacement of AuNPs on aptamer-based probes; thus, it is necessary to screen the complementary DNA (cDNA) of aptamer that is better for this purpose. There are two key points for the successful detection of CRP. One is to prepare stable MB-aptamer/cDNA-AuNP sandwich-structured probes, and the other is that AuNPs on MBs can be successfully displaced by CRP. To prepare a stable probe, MBs and AuNPs need to be linked together by strong base complementary pairing. Competitive replacement of AuNPs on MBs by CRP requires that the specific interactions between the CRP and aptamer should be greater than that between the aptamer and cDNA.In order to obtain the most suitable complementary paring between CRP-aptamer and cDNA, two rounds of screening were performed. The first round of screening aimed to find the most suitable complementary position between CRP-aptamer and its cDNA. We designed three cDNAs, namely, cDNA-1, cDNA-2 and cDNA-3 (shown in Table 1) paired with the aptamer in different complementary positions. In this step, cDNA-1, cDNA-2 and cDNA-3 were attached to AuNPs and the AuNPs modified with cDNA-1, cDNA-2 and cDNA-3 were conjugated to MBs modified with the aptamer.
The complementary base number of cDNA in the whole detection system is an important factor. In the second screening, we designed four cDNAs, namely, cDNA-4, cDNA-5, cDNA-6 and cDNA-7 (shown in Table 1) with different lengths. In this step, cDNA-4, cDNA-5, cDNA-6 and cDNA-7 were attached to AuNPs, and the AuNPs modified with cDNA-4, cDNA-5, cDNA-6 and cDNA-7 were conjugated to MBs modified with the aptamer. Then, the same concentration of CRP was added into the MB-aptamer/cDNA-AuNP sandwich-structured probes.
Result and discussion
The selection results of aptamer and cDNA
As shown in Fig. S1A,† the color of the supernatant from AuNPs modified with different cDNAs is still pink and indicates the successful attachment of cDNAs on AuNPs. Fig. S1B† shows a photograph of the supernatant after the addition of MBs to the solution of AuNPs, in which changes in the color of some solutions can be observed. When AuNPs were conjugated onto MBs, the number of free AuNPs was reduced and the color of the supernatant changed. As more AuNPs attached to MBs, less free AuNPs lest in the supernatant, the color of the supernatant became lighter. After incubation, as shown in Fig. S1C,† the change in the color of the supernatant with cDNA-1 and cDNA-2 was negligible, while the one with cDNA-3 became lighter in color, which indicated that the formation of the sandwich-structured probe using cDNA-3 was more efficient than that using cDNA-1 and cDNA-2. To further check this point, SEM analysis was performed. As shown in Fig. 2, no AuNP can be found on MBs using cDNA-1 and cDNA-2 (Fig. 2A and B), while it is clear that a number of bright dots are present on MBs using cDNA-3 (Fig. 2C), indicating the successful conjugation of AuNPs to MBs using cDNA-3. This is observed because the 3′-terminal of the aptamer is located outside the surface of MBs, which has smaller steric hindrance between the aptamer, cDNA, AuNPs, and magnetic nanoparticles. We thus chose the 3′-terminal of the aptamer as the complementary position.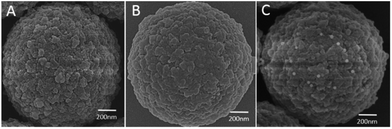 | ||
| Fig. 2 SEM image: (A) cDNA-1-modified AuNPs are coupled to MB, (B) cDNA-2-modified AuNPs are coupled to MB, and (C) cDNA-3-modified AuNPs are coupled to MB. | ||
The second screening was based on the first screening, where we chose suitable length complementary bases at the 3′-terminal of the aptamer. The color of the 60 nm AuNP solution was pink. When AuNPs were agglomerated, the color of the solution turned blue and purple or it even became colorless. Generally, AuNPs agglomerate in a solution of 0.1–0.2 M NaCl. Upon the successful attachment of cDNA, AuNPs do not agglomerate and the solution remains pink. As shown in Fig. S2A,† the supernatant color of AuNPs modified with different cDNAs is still pink and indicates the successful modification of AuNPs with cDNA-5, cDNA-6 and cDNA-7; in contrast, the cDNA-4-modified AuNP solution became almost colorless, indicating that cDNA-4 failed to attach to AuNPs and thus, a subsequent discussion of cDNA-4 will not be conducted. Fig. S2B† shows the supernatant photographs right after the addition of MBs to the solution of AuNPs, and changes in the color of some solutions can be observed. After incubation, Fig. S2C† shows that the supernatants using cDNA-5, cDNA-6 and cDNA-7 become colorless, which indicates the formation of the sandwich-structured probes. To further check the successful formation of the fabricated probes, SEM analysis was performed to determine whether AuNPs were attached to MBs through aptamer and cDNA hybridization. As shown in Fig. 3, it is clear that a number of bright dots are attached onto MBs using cDNA-5, cDNA-6 and cDNA-7 (Fig. 3A–C), indicating the successful conjugation of AuNPs to MBs. As the number of complementary bases increased, the base pairing force became stronger between cDNA and aptamer, and AuNPs attached more easily to MBs.
 | ||
| Fig. 3 SEM image: (A) cDNA-5-modified AuNPs are coupled to MB, (B) cDNA-6-modified AuNPs are coupled to MB, and (C) cDNA-7-modified AuNPs are coupled to MB. | ||
Therefore, cDNA-5, cDNA-6 and cDNA-7 were selected as the cDNAs to fabricate stable probes.
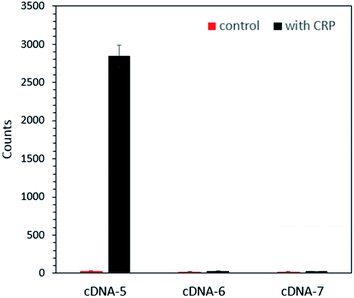
To verify whether AuNPs on the aptamer-based probe could be successfully displaced by CRP to free particles, the same amount of CRP was added into the same amount of the as-prepared different probes reported above. After incubation for 5 minutes, the supernatant was subjected to DFM imaging. Fig. S3† shows a typical DFM image of the AuNPs displaced by CRP and the control group. As can be seen in the DFM image, few AuNPs are observed for the control group, indicating that AuNPs barely dropped from the probe without the addition of CRP and illustrating the good stability of the probe. For the samples in which CRP was added, it is clear that the probe fabricated using cDNA-5 released the most AuNPs, which can be seen from the bar chart of the number of AuNPs shown in Fig. 4. This result indicated that the use of cDNA-5 will result in the most efficient probe. From these results, it became clear that when the number of complementary bases was more than 8 bps (base pairs), such as that observed for cDNA-6 and cDNA-7, it is difficult to replace AuNPs by CRP. In addition, according to the results, cDNA-5 was selected as the cDNA of CRP-aptamer for probe fabrication and CRP detection in this assay system.
The characterization of the probe
The cDNA-modified AuNPs and aptamer-modified MBs established a stable probe through the interaction of the complementary base pairing between the aptamer and cDNA. To further verify the successful attachment of AuNPs onto MBs, SEM analysis of the probe was performed. Fig. S4† shows the SEM image of MBs before and after conjugation with AuNPs. It shows that there is no obvious bright particle on the surface of MBs on the left control image, while bright particles appear on the surface of MBs on the right image. EDS analysis, shown in Fig. S5,† confirmed that the bright particles on MBs were indeed AuNPs.Sensitivity and specificity of the CRP detection assay
To test the sensitivity of the as-prepared probes, the detection of 0–2.17 μM CRP was carried out (Fig. S6 and S7†). Among them, Fig. 5A shows the typical DFM images obtained for each concentration (0–49 nM) of CRP. From the DFM images, it can be seen that the higher the concentration of CRP, the more AuNPs can be displaced. Fig. 5B shows the linear relationship between the number of AuNPs and the concentration of CRP in the range of 0–70 nM. The simulated functional relation is y = 9.0608 x + 50.0233 (y represents the number of AuNPs; x represents the concentration of CRP) with a correlation coefficient of R2 = 0.9973. The limit of detection (LOD) of CRP was calculated to be 2.71 nM, which was much lower than that in human serum (9 nM).27 When the concentration of CRP added to the probe solution was high enough, the color of the supernatant changed from colorless to pink, as shown in Fig. S6,† indicating that more AuNPs were displaced into the solution. In this case, the number of AuNPs was hard to recognize and count by the MATLAB program and as shown in Fig. S7,† a platform of the nonlinear curve appeared between the AuNP counts and CRP concentrations.In addition to sensitivity, sensors for biomolecule detection also need to be highly selective. Other proteins were used to check the selectivity of the as-prepared probe towards CRP. Specifically, 10 μL of 0.43 μM CRP was added to 10 μL probe solutions and solutions of BSA, thrombin, IgG, and Lys with concentrations 10 times higher than that of CRP were added to the other 4 probe solutions for comparison. Ten μL buffer was added to the last probe solution as a control experiment. Fig. S8† shows the typical DFM images obtained from each experiment described above. One can clearly see that numerous AuNPs were released from the CRP group and only a few AuNPs were released from the control group and the rest of the protein groups. This indicates that the specificity of the probe to CRP was far better than that to other proteins. Repetition experiments were carried out and the bar chart is shown in Fig. 5C, which confirms that the as-prepared probes are specific to CRP over other proteins.
The detection of CRP in serum
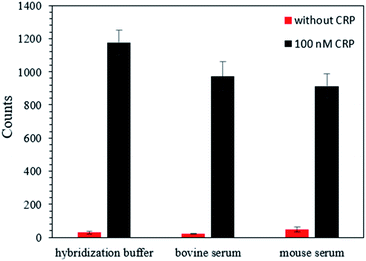
To assess the potential practical applications of this strategy, we performed CRP detection in bovine serum and mouse serum, which are complex media containing various ingredients. The results, shown in Fig. 6, indicated that the AuNP counts increased upon the addition of 100 nM CRP, and the values for the bovine serum and mouse serum were similar to those from the hybridization buffer. These findings suggest that this strategy can be used for the detection of CRP in complex biological samples.Conclusions
In summary, a stable aptamer-based probe with a strategy combining AuNP counting with DFM to detect CRP was proposed. The aptamer-based probes were highly sensitive and selective towards CRP. An LOD value of 2.71 nM was achieved, which was much lower than that in human serum (9 nM). Furthermore, the as-prepared aptamer-based probe could be used to test CRP in complex biological media including bovine serum and mouse serum containing various ingredients. This strategy offers straightforward fabrication of probes and a facile and convenient detection process, which paves a way for an accurate, convenient and low-cost approach for CRP detection. The approach reported here is also promising for the detection of other proteins with suitable aptamers.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Grants 21573124 and 21874082) and the Innovation Foundation for Science and Technology of Shenzhen (Grants JCYJ20170413104646428 and JCYJ20170817172150505).References
- M. B. Pepys and M. L. Baltz, Adv. Immunol., 1983, 34, 141–212 CrossRef CAS.
- D. Thompson, M. B. Pepys and S. P. Wood, Structure, 1999, 7, 169–177 CrossRef CAS.
- A. K. Shrive, D. Holden, D. A. A. Myles and T. J. Greenhough, Acta Crystallogr., Sect. D: Biol. Crystallogr., 1996, 52, 1049–1057 CrossRef CAS PubMed.
- N. S. Akkececi, G. Y. Cetin, H. Gogebakan and C. Acipayam, Med. Sci. Monit., 2019, 25, 1401–1409 CrossRef.
- M. E. H. Ahmed, G. Hamed, S. Fawzy and K. M. Taema, Medicina intensiva, 2019 Search PubMed.
- B. Babamiri, R. Hallaj and A. Salimi, Biosens. Bioelectron., 2018, 99, 353–360 CrossRef CAS.
- X. Ge, P. Li and Z. Wu, J Coll Physicians Surg Pak, 2019, 29, 189–190 CrossRef.
- M. H. Buch, Y. Seto, S. J. Bingham, V. Bejarano, D. Bryer, J. White and P. Emery, Arthritis Rheum., 2005, 52, 42–48 CrossRef CAS PubMed.
- Z. Cai, W. He, F.-J. Zhuang and Y. Chen, Int. J. Neurosci., 2019, 129, 423–429 CrossRef CAS.
- D. H. J. Bunka and P. G. Stockley, Nat. Rev. Microbiol., 2006, 4, 588–596 CrossRef CAS.
- K.-M. Song, S. Lee and C. Ban, Sensors, 2012, 12, 612–631 CrossRef.
- T. Mairal, V. C. Oezalp, P. L. Sanchez, M. Mir, I. Katakis and C. K. O'Sullivan, Anal. Bioanal. Chem., 2008, 390, 989–1007 CrossRef CAS.
- A. Chen and S. Yang, Biosens. Bioelectron., 2015, 71, 230–242 CrossRef CAS.
- Y. S. Kim, N. H. A. Raston and M. B. Gu, Biosens. Bioelectron., 2016, 76, 2–19 CrossRef.
- E. D. Bernard, K. C. Nguyen, M. C. DeRosa, A. F. Tayabali and R. Aranda-Rodriguez, Anal. Biochem., 2015, 472, 67–74 CrossRef CAS.
- J. Chen, Z. Fang, J. Liu and L. Zeng, Food Control, 2012, 25, 555–560 CrossRef CAS.
- J. A. Cruz-Aguado and G. Penner, J. Agric. Food Chem., 2008, 56, 10456–10461 CrossRef CAS.
- S. A. Vance and M. G. Sandros, Sci. Rep., 2014, 4, 5129 CrossRef CAS.
- S. Centi, L. B. Sanmartin, S. Tombelli, I. Palchetti and M. Mascini, Electroanalysis, 2009, 21, 1309–1315 CrossRef CAS.
- Z. Wu, R. Yang, D. Zu and S. Sun, Sci. Rep., 2017, 7, 14472 CrossRef.
- Y. Lai, S. Sun, T. He, S. Schlucker and Y. Wang, RSC Adv., 2015, 5, 13762–13767 RSC.
- S. Sun, D. Thompson, U. Schmidt, D. Graham and G. J. Leggett, Chem. Commun., 2010, 46, 5292–5294 RSC.
- Z. Mu, X. Zhao, Z. Xie, Y. Zhao, Q. Zhong, L. Bo and Z. Gu, J. Mater. Chem. B, 2013, 1, 1607–1613 RSC.
- G. Li, L. Zhu, Z. Wu, Y. He, H. Tan and S. Sun, Anal. Chem., 2016, 88, 10994–11000 CrossRef CAS.
- B. Wu, R. Jiang, Q. Wang, J. Huang, X. Yang, K. Wang, W. Li, N. Chen and Q. Li, Chem. Commun., 2016, 52, 3568–3571 RSC.
- T. Li, X. Xu, G. Zhang, R. Lin, Y. Chen, C. Li, F. Liu and N. Li, Anal. Chem., 2016, 88, 4188–4191 CrossRef CAS.
- A. Rota, C. Milani, B. Contiero, E. Artusi, B. S. Holst and S. Romagnoli, Anim. Reprod. Sci., 2019, 204, 111–116 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06203c |
| This journal is © The Royal Society of Chemistry 2019 |