Open Access Article
Open Access ArticleQualitative and quantitative assessment of genotoxins using SRRz lysis reporter under the control of a newly designed SOS responsive promoter in Escherichia coli†
Pengfei Yuana,
Junqing Donga,
Weibin Zhaoa,
Min Zhuo*a,
Shuang Li *a,
Shaobin Huang
*a,
Shaobin Huang b and
Jianjun Li
b and
Jianjun Li c
c
aSchool of Biology and Biological Engineering, South China University of Technology, Higher Education Mega Center, Guangzhou 510006, China. E-mail: zhuomin@scut.edu.cn; shuangli@scut.edu.cn; Fax: +86 20 3938 0601; Tel: +86 20 3938 0601
bSchool of Environment and Energy, South China University of Technology, Higher Education Mega Center, Guangzhou 510006, China
cState Key Laboratory of Applied Microbiology Southern China, Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, Guangdong Institute of Microbiology, Guangzhou 510070, China
First published on 4th November 2019
Abstract
A new bacterial genotoxicity detection strain was constructed, in which the cell lysis gene of SRRz from a lambda phage was controlled by a new designed SOS responsive element, designated as Escherichia coli BL21/pUC-PST. The biosensor responded only after 0.5 h contact with mutagens and the changes in cell culture turbidity could be easily differentiated with the naked eyes from the control sample. This SOS/SRRz system presented a dose-dependent manner to five model DNA-damaging agents with an improved detection sensitivity. The limits of detection (LODs) were 0.026 μM for mitomycin C, 320.4 μM for azinphos-methyl, 34.4 μM for methyl methanesulfonate, 4.6 μM for dithianone and 6.0 μM for dichlofluanid, which were much lower than previously reported. By performing binary and ternary mixture experiments, the toxic equivalency concept was validated in the E. coli SOS/SRRz system by comparison with bioanalytical equivalent concentrations (BEQ) and overall toxic equivalent concentration (TEQmixture) using Cr(VI) as the reference compound. Pearson analysis indicated that a strong correlation existed between the TEQmixture and BEQ values. Thus the TEQmixture could be presented as the Cr(VI) equivalent concentration from its dose–effect lysis profiles for the environmental sample. The proposed genotoxicity reporter strain allows for easier qualitative characterization and quantitative interpretation of the TEQmixture values using Cr(VI) as the reference for environmental water samples.
1. Introduction
The advancement of industrialization and agricultural activities produces various chemicals and has posed high health and environmental risks for their uncontrolled discharge to the air, soil and water. Many thousands of chemicals, including pharmaceutical products, pesticides and petroleum products, are mutagenic or cancerogenic because of their ability of inducing inheritable somatic mutations.1 These chemicals are often presented in trace amounts in the environment. There is a widespread desire to evaluate the genotoxicity and carcinogenicity of these chemicals. Although many modern chemistry analysis methods, such as mass spectrometry, have been developed, it remains impracticable to monitor every chemical presented in samples because of limitations in analytical capacity, complexity, cost and time.2,3 Moreover, these chemical monitoring methods are generally targeted to specific individual or groups of compounds, and do not provide information about health impacts or cumulative effects from complex mixture interactions.4 These push forward the development of sensitive, cost-effective and relatively simple bacterial bioassays which can indicate the presence of unknown genotoxic chemicals, also estimate the contribution of unknown chemicals and chemicals below detection limits to the overall mixture effect.Bacteria have been proved to be more attractive to be used in mutagenicity assays than methods based on tissue culture and animal experiments, i.e. micronucleus test and comet assay. In vitro bacterial genotoxin assays are useful methods to evaluate the genotoxic properties of compounds due to their sensitivity, simplicity and flexibility. Bacterial genotoxicity tests are generally classified into two classes, gene mutation assays (Salmonella and E. coli), including reverse and forward mutation assays5,6 and DNA-damage assays, including the SOS test systems7,8 and umu-test.9 The SOS Chromotest, developed at Pasteur Institute (France), was activated by DNA damage and respond to genotoxic chemicals.10 The method was standardized according to the International Standardization Organization (ISO) (ISO/CD 13829) (ISO, 2000) and DIN (DIN 38415-3). Now, it has been extensively employed to investigate and monitor genotoxicity in different environmental samples including tap water,11 reclaimed wastewater12 industrial wastewater13 and airborne particles.14 The SOS Chromotest assays were adapted to a 96-well microplate for high throughput screening of samples in the early of 1990s.15
In the common SOS Chromotest assays, a reporter gene lacZ under the SOS responsive promoter of umuDC16 or sfiA7 is induced and expressed when the test strain was exposed to chemical carcinogen. Then it is possible to check the inductivity of reporter gene expression by measuring the β-galactosidase activity using UV/vis spectrometry or fluorometry.17 However, the SOS test system is unable to direct evaluate samples containing a high concentration of chromophoric or colored dissolved organic matters because it depends on the spectrophotometric detection.18 In addition, enzyme extraction is another key-step in the SOS/umu bioassay to measure the β-galactosidase activity. Conventional protein extraction detergents, such as sodium dodecyl sulfate (SDS) and the combination of SDS with Z-buffer, have been widely used in umu test. Due to the report that SDS treatment may result in the denaturation of β-galactosidase,19 a more effective and commercially available protein extraction detergent, BugBuster Master Mix containing nuclease and lysozyme, was applied to the SOS Chromotest assay.20
Mitomycin C (MMC) and methyl methanesulfonate (MMS) are direct-acting alkylating agents that may induce SOS response in E. coli through forming cross-links to adjacent guanines in DNA.21,22 MMC may strongly induces the SOS response, while MMS showed lower mutagenic activities in the SOS/umu-test.15,21,22 Azinphos-methyl is classified by the World Health Organization (WHO 2010) as groups 1 (acute toxicity) and 3 (environmental toxicity). Results indicated that the azinphos-methyl might induce mutation through frameshift mutation and pair base substitution.23 While in some studies the response is negative,24 and in another it is weak positive.25 Dithianon and dichlofluanid have been used as multi-site contact fungicides since 1965. The previous study showed that dithianon and dichlofluanid were genotoxic.15,26 However, the Joint FOA/WHO meeting concluded the contradictory data that dithianon is unlikely to be genotoxic in vivo.27
In this study, a new SOS response element was designed to control the expression of phage-derived SRRz lysis gene. The applicability of developed SOS/SRRz system was evaluated by comparing its performance with the conventional SOS/umu bioassay through examining the genetic toxicity of some pure chemicals and their combinations. The results suggest that the SOS/SRRz system may be useful for evaluating genotoxicity of chemical carcinogens.
2. Experimental section
2.1 Chemicals and reagents
Mitomycin C (MMC, CAS 50-07-7), methyl methanesulfonate (MMS, 66-27-3), azinphos-methyl (CAS 86-50-0), dithianone (CAS 3347-22-6), dichlofluanid (CAS 1085-98-9), 3,4-benzo[a]pyrene (BaP, CAS 50-32-8) and 2-aminoanthracene (2-AA, CAS 613-13-8) and dimethyl sulfoxide (DMSO, CAS 67-68-5) were all purchased from Sigma-Aldrich (St. Louis, MI, USA). All other chemical reagents used were analytical grade and purchased from standard companies. For lower water solubility, the tested chemicals were dissolved in DMSO followed by dilution with PBS to working concentration. The final DMSO working concentration was kept at 8% using serial dilutions with DMSO–PBS mixture.For molecular cloning manipulation, DNA polymerase of PrimeSTAR was purchased from TaKaRa (Dalian, China). FastDigest restriction enzymes were obtained from Fermentas of Thermo Scientific (Waltham, MA, USA). SanPrep Column Plasmid Mini-preps Kit and SanPrep Column PCR Product Purification Kit (Sangon, Shanghai, China) were used for the preparation of plasmids and PCR products.
2.2 Construction of genotoxin-inducible vector
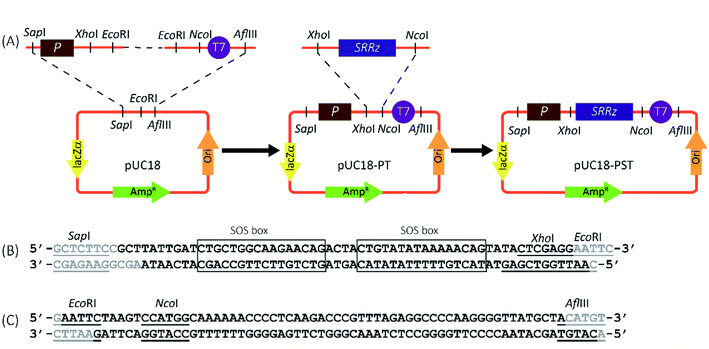
The genotoxin responsive vector, pUC18-PST, was constructed as shown in Fig. 1A. For the construction of genotoxin-inducible promoter (Psos-box, Fig. 1B), two complementary oligonucleotides (5′-GCTTATTGACATGCTGGCAAGAACAGACTACTGTATATAAAAACAGTATACTCGAGG-3′ and 5′-AATTGGTCGAGTATACTGTTTTTATATACAGTAGTCTGTTCTTGCCAGCATGTCAATA-3′) were chemically synthesized and mixed in equal molar amounts. The mixture was heated at 95 °C for 5 min and gradually cooled to 25 °C within an hour. The common terminator (T7) was obtained using the similar procedures (5′-AATTCTAAGTCCATGGCAAAAAACCCCTCAAGACCCGTTTAGAGGCCCCAAGGGGTTATGCTA-3′, 5′-CATGTAGCATAACCCCTTGGGGCCTCTAAACGGGTCTTGAGGGGTTTTTTGCCATGGACTTAG-3′, Fig. 1C). The resulting double-stranded DNA fragments containing the SapI-EcoRI and EcoRI-AflIII overhangs for Psos-box and T7, respectively. The annealed parts were phosphorylated by Shrimp Alkaline Phosphatase (rSAP, New England Biolabs, Ipswich, MA, USA), and ligated through the EcoRI sticky ends. Then the ligation product was inserted between the SapI and AflIII sites of pUC18 (TaKaRa, Dalian, China), generating the plasmid pUC18-PT. The SRRz cluster (1540 nt) from Enterobacteria phage λ was synthesized by Sangon Biotech (Shanghai, China) and inserted into pUC18-PT to yield pUC18-PST (between the XhoI and NcoI sites). The construction vector was chemically transformed into Escherichia coli (E. coli) BL21 (Novagen, Madison, WI, USA), generating BL21/pUC18-PST as genotoxin tester strain.2.3 Cell viability determination
The recombinant E. coli BL21 cells harboring pUC18-PST were cultured overnight at 37 °C in Luria–Bertani (LB) medium (1% tryptone, 0.5% yeast extract, 1% NaCl) supplemented with 100 μg mL−1 ampicillin. Saturated overnight cultures (in the range of OD600 2.0–3.0) were diluted 100-fold into test tube containing fresh cells and grown at 37 °C, 250 rpm for about 1.5 h to reach an OD600 of 0.3–0.4. 198 μL aliquots of cell culture were transferred to 96-well microplates (round-bottom, Corning, NY). 2 μL of different chemicals or mixture were then added to each well. 0.1 μM MMC, 600 μM azinphos-methyl, 150 μM MMS, 18 μM dithianone and 10 μM dichlofluanid were used, respectively in cell viability determination.The contents of the wells were mixed and covered with a lid. After incubation at 37 °C at 250 rpm in a shaker equipped with accessories specially for shaking 96-well plates (Zhichu, Shanghai China), cell density was measured as turbidity at 600 nm at indicated interval time using a SpectroMAX® M5 Microplate reader (Molecular Devices, Sunnyvale, CA, USA). The cell viability for each chemical was calculated by the absorbance at 600 nm of the sample divided by the absorbance of the controls (DMSO added into the cell culture) at the same time (eqn (1)).
 | (1) |
2.4 β-Galactosidase activity assays
The β-galactosidase activity was assayed as previous method9 with minor modifications. The reaction system consisted of 0.2 mL cell lysates and 0.4 mL 4 g L−1 o-nitrophenyl-β-D-galactopyranoside (ONPG; Sigma, St. Louis, MO, USA) dissolved in 0.1 M phosphate buffer, pH 7.0 and 1.8 mL of buffer B (16.1 g Na2HPO4, 5.5 g NaH2PO4·H2O, 0.75 g KCl, 0.25 g MgSO4·7H2O, 1 g sodium dodecylsulfate, 2.7 mL β-mercaptoethanol in 1 L H2O, pH = 7.0). After incubated at 28 °C for 20 min, the reaction was terminated by the adding 1.6 mL of 1 M Na2CO3. The absorbance at 420 nm and 550 nm were recorded and the β-galactosidase activity is calculated according to the ref. 15:
 | (2) |
2.5 Dose–lysis profile measurement
The overnight culture of E. coli BL21/pUC18-PST (in the range of OD600 2.0–3.0) was diluted with fresh LB medium containing 50 μg mL−1 ampicillin and grown at 37 °C to the early exponential phase, followed by the addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM to induce the indigenous β-galactosidase expression. After 1 h induction, specific concentrations of test chemicals (MMC, MMS, azinphos-methyl, dithianone, dichlofluanid or K2Cr2O7) were added to the cell cultures and incubated at 37 °C for another 2 h. The supernatant was harvested by centrifugation for extracellular β-galactosidase activity assay. The cell pellets were recovered and resuspended in equal volume of 0.1 M Z buffer (pH 7.5) to release intracellular β-galactosidase. The lysis efficiency (LE) was derived from comparing the extracellular enzyme activity to the sum of the extracellular and intracellular activity (eqn (3)).
 | (3) |
From the lysis efficiency, a log-logistic concentration–effect curve (eqn (4)) can be fitted using Prism 8.0 software (GraphPad, San Diego, CA, USA). The minimum values must be greater than 0.
 | (4) |
After running this fitting program, the related parameters, the maximum and minimum values, EC50 and slope are all output automatically through Prism 8.0 software.
The genotoxic potency was also expressed as the induction ratio (IR). The IR was defined as the ratio of the sample lysis efficiency divided by the average lysis efficiency of the control (DMSO).
 | (5) |
The semi-logarithm concentration–induction ratio curves were also plotted through the log-logistic concentration–effect curve in Prism 8.0 software. The ECIR1.5 values were deduced from this nonlinear regression curve when IR is equal to 1.5 (eqn (6))
 | (6) |
Limit of detection (LOD) is the lowest analyst concentration likely to be reliably distinguished from the solvent and at which detection is feasible. LOD is determined by utilizing both the mean value and standard deviation (SD) when replicates of a DMSO sample containing no analyte are tested (eqn (7)).28
| LOD = meanDMSO + 3 × SDDMSO | (7) |
2.6 Mixture experiments
For hexavalent chromium [Cr(VI)] is a known respiratory carcinogen that is capable of interacting with DNA to yield genotoxic and mutagenic effects,29 it served as the reference chemical in this study.The toxic equivalency concept is a widely applied method to express the toxicity of complex mixtures of compounds.30 The toxic equivalent concentration (TEQ) is the sum of the analytically determined concentration of chemicals sample i multiplied by each chemical's relative effect potency (ECIR1.5 ref/ECIR1.5i) in relation to a specific potent reference compound [Cr(VI)] (eqn (8)).30
 | (8) |
The bioanalytical equivalent concentration (BEQ) for each mixture sample is the sum of the potency-scaled concentrations of unknown chemicals that have toxic action as the reference compound and act concentration-additively. It is often calculated from the concentration–responsive curve of the reference compound and translated into concentrations of the reference compound.
Chemical mixture experiments were performed at a fixed concentration of the chosen five model genotoxicants, and cell lysis efficiencies induced by these chemicals were determined.
The correlation between the pair of TEQ-Cr(VI) and BEQmixture was directedly computed by Correlation Matrix Analysis in GraphPad Software, and the Pearson's correlation coefficient (PCC) r and one-tailed p values were derived at the confidence interval 95%. The PCC values were automatically output, which was displayed as Spearman r in the GraphPad software.
Each experiment (individual compounds and mixtures) was carried out in three independent replicates on different plates.
3. Results and discussion
3.1 Construction of SOS responsive cell lysis strain
The genotoxin responsive expression cassette was constructed first (Fig. 1A). The SOS responsive promoter (Psos-box, 47 bp) containing two consensus SOS-box sequences was obtained through annealing of two chemically synthesized complementary sequences (Fig. 1B).31 The sticky-end ligation was carried out through the overhanging base pairs on the Psos-box promoter and T7 terminator (Fig. 1C) to yield pUC18-PT. The SRRz lysis gene cluster containing the XhoI-NcoI cohesive-ends was inserted into pUC18-PT to generate the genotoxin detection vector of pUC18-PST.The promoter sequence from any of these genes could in theory be coupled to a suitable reporter gene to create a biosensor, but obviously the specific choices of the promoter and reporter will greatly affect the performance of the system. As many people know, SOS response is controlled by the LexA repressor. Expression of the SOS genes depends on the properties of LexA-binding sequence (SOS box) in the promoter, which shares high homology but is distinct from gene to gene. The preferred SOS response promoters in the early SOS-based genotoxicity assays are from the recA, umuDC, sulA and colD, and most of the length of these promoter sequences are longer than 300 bp.31,32 There are many redundant sequence for the SOS transcription initiation. In this study, only 47 bp promoter sequence containing two reported SOS boxes (deduced from umuDC′31) was employed (Fig. 1B), which functioned normally and perfectly with low leaky expression and good induced target expression. To our best knowledge, this is the shortest SOS-inducible promoter used in the genotoxin detection.
The high copy number of vector pUC18 was chosen as the backbone for the expression of the lysis gene cluster SRRz from bacteriophage Lambda in E. coli.33 Phage lysis is a ubiquitous biological process. The lysis cluster coding for holin (S),34 endolysin (R) and spanins (Rz and Rz1) could lead the E. coli to death.35
3.2 Characterization of genotoxicity-inducible bacterial lysis system
To test the efficiency of the new constructed genotoxicant assay strain, different strengths of genotoxic substances based on previous research,24 i.e. MMS, MMC, azinphos-methyl, dithanone, and dichlofluanid were employed. SOS induction was performed by measuring the cell density (OD600) over time in E. coli BL21 and BL21/pUC18-PST treated with 0.1 μM MMC, 600 μM azinphos-methyl, 150 μM MMS, 18 μM dithianone and 10 μM dichlofluanid, respectively. Results showed that the cell viability in the wild-type and recombinant E. coli strains was significant distinct (Fig. 2). The cell viabilities dramatically decreased to about 0.6 when BL21/pUC18-PST cells after 0.5 h incubation with the different five mutagens, indicating that the expression of SRRz was initiated because of the contact with genotoxicants. As time went on, the cell viabilities of BL21/pUC18-PST treated with MMC, azinphos-methyl and dichlofluanid still decreased to various degrees (0.2–0.4) but later started to increase again (Fig. 2A, B and E). For MMS and dithianone treatments, cell viabilities did not rise but remained at 0.4–0.6. The culture turbidity changes were the results of SRRz expression induced by mutagens. However, the cell viabilities for the wild-type E. coli BL21 without SOS-response vector always remained above 0.9 during the entire test period (4 h). The results implied that the five chemicals at the tested concentrations were not cytotoxic to E. coli cells. It is worth mentioning that the cell turbidities of the recombinant BL21/pUC18-PST cultured with genotoxins appeared to be much clearer compared with that cells incubated with DMSO (Fig. 2F). The changes in cell turbidity could be easily observed with the naked eyes, indicating that the SOS/SRRz for genotoxicity detection could be easily achieved without cumbersome steps and advanced facilities. Cell growth curves of different strains were determined and showed in Fig. S1.† Results indicated that the leaky expression of SRRz could be ignored for no apparent difference in growth patterns between E. coli BL21/pUC-18 and the cells containing pUC18-PST was observed.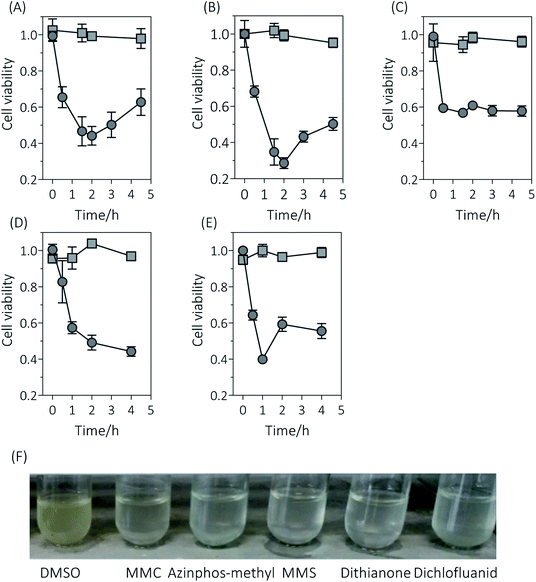 | ||
| Fig. 2 Response of E. coli strains to genotoxicants. Cell viabilities of E. coli BL21/pUC18 (square) and BL21 containing the SOS-responsive lysis vector pUC18-PST (circle) incubated with MMC (A), azinphos-methyl (B), MMS (C), dithianone (D) and dichlofluanid (E). (F) Cell culture turbidities of E. coli BL21/pUC18-PST incubated with the tested chemicals for 1 h. “DMSO” indicated that cells were cultured with 8% DMSO. Cell viabilities were calculated as eqn (1). Experiments were done in triplicates. | ||
In this study, no S9 mix (S9 microsomal fraction for metabolic activation) was added as previous umu-test,36 which is an important factor for efficient activation of indirect mutagens. It was reported36 that only 1 μg mL−1 BaP and 0.3 μg mL−1 2-AA could result the induction of the umu operon in the presence of S9 mix. However, 31.5 μg mL−1 BaP and 19.3 μg mL−1 2-AA could not initiate the SOS responsive SRRz expression in E. coli BL21/pUC-PST (Fig. S2†). This result implied that the constructed system was not suitable for detecting the promutagens.
Based on the SOS regulation mechanism, Quillardet7 first constructed the bacterial genotoxicity assays by means of a sfiA::lacZ operon fusion in E. coli. The tester was rapid (a few hours) and did not require survival of the tester strain as previous Ames test.37 Later on, methods based on the activation of the bacterial SOS response for the genotoxin detection have been developed. The reporter elements could be lacZ (galactosidase),36 luxCDABE (luciferase),38 phoA (alkaline phosphatase)39 and gfp (green fluorescence protein),40 which can be measured colorimetrically, luminometrically, electrochemically or by emission of fluorescence light, respectively. All these test systems seem to have some practical advantages compared Salmonella/microsome assay.18 However, the results obtained need a few hours and some optical or electronic instruments. In this study, lysis cluster SRRz were employed downstream of the SOS-responsive promoter. When the recombinant E. coli cells incubated with genotoxins, an obvious change occurring in cell culture turbidity in 0.5 h could be easily seen with the naked eyes (Table 1). This new constructed E. coli BL21/pUC18-PST have a great future in genotoxin qualitative detection because of its low cost, short time, good applicable without special equipment, etc.
| Test method | Cell | Detection type | Detection timea | Note | Reference |
|---|---|---|---|---|---|
| a Cell cultivation time was not included. | |||||
| Micronucleus test | Mammalian or plant cells | Aneugenic and chromosome-breaking agents | Several days depending on the experiment cells | Need to cultivate animals/plants or mammalian/plant cells | 46–48 |
| Comet assay | Mammalian cell | DNA damage | 3–4 h | Need to cultivate animals or mammalian cells | 49 |
| Ames test | Salmonella typhimurium TA100, TA98, TA1535, and TA1537 | Gene mutations | >2 d | 6 and 50 | |
| SOS/umu-C′′::lacZ | E. coli or Salmonella typhimurium | DNA damage | 2 h | 18 | |
| SOS/recA′::luxCDABE | E. coli | DNA damage | 3–4 h | Need a microplate luminometer for detecting chemiluminescence | 38 |
| SOS/sulA::phoA | E. coli | DNA damage | 3 h | Need electrochemical equipments | 39 |
| SOS/umuC′::gfp | E. coli | DNA damage | 5 h | Need fluorescence spectrophotometer | 40 |
| SOS/SRRz | E. coli | DNA damage | 0.5–1 h (turbidity) | Naked eyes | This study |
| 2 h (enzyme assay) | β-Galactosidase activity assay | This study | |||
3.3 Dose–effect of genotoxicity on cell lysis
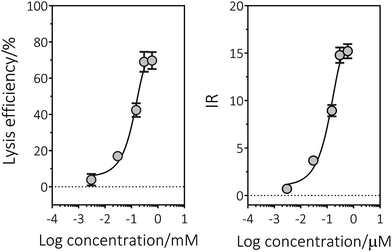
In order to estimate the dose–effect of chemicals more accurately, the β-galactosidase basically expressed in E. coli BL21 was determined as the extracellular and intracellular activity. Because β-galactosidase is endocellular enzyme, the detected extracellular enzyme activity for E. coli BL21/pUC18-PST was the result of cell lysis induced by mutagens.As shown in the semi-log plots (Fig. 3), the test strain BL21/pUC18-PST displayed an apparent concentration-dependent cell lysis response to the five genotoxicants, which are previously reported as “positive” with at least one of the Ames tester strains.15 The cell lysis efficiencies reached the largest values at the highest concentration used, which ranged from 67% (dithianone) to 81% (MMS). For the tested chemicals are weakly polar and nonpolar compounds, they were dissolved in DMSO. The analytical noise resulted from DMSO in the absence of tested chemicals were determined as 8.26 ± 2.31%. LOD is estimated by utilizing the eqn (7)28 and presented in Table 2. The lowest LOD was observed as 0.026 μM for MMC using β-galactosidase activities as the response parameters, which was roughly comparable with the classical strain TA1535/pSK1002 possessing the plasmid with fused gene umuC′-′lacZ. To be noted that the LOD values derived from the β-galactosidase activities were all much lower than Reifferscheid et al. reported for azinphos-methyl, MMS, dithianone and dichlofluanid.15 However, the LOD values calculated from the cell turbidities (OD600) were not satisfying, which might be due to the low sensitivity of OD determination. Results indicated that the constructed SOS/SRRz strain has potential applications in genotoxin detection.
 | ||
Fig. 3 The dose–lysis curves illustrated E. coli BL21/pUC18-PST genotoxicity of five chemicals and fitted with dose–response model: Y = Min. + (Max. − Min.)/(1 + 10(log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EC50 − log EC50 − log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) X) × slope) under the constraint that the bottom parameter must be greater than 0 (eqn (4)). The statistical analysis of these data is shown in Table 2. (A) MMC, (B) azinphos-methyl, (C) MMS, (D) dithianone, (E) dichlofluanid. All the experiments were carried out in triplicate. X) × slope) under the constraint that the bottom parameter must be greater than 0 (eqn (4)). The statistical analysis of these data is shown in Table 2. (A) MMC, (B) azinphos-methyl, (C) MMS, (D) dithianone, (E) dichlofluanid. All the experiments were carried out in triplicate. | ||
| Chemicals | CAS number | Highest concentration detected (mM) | LOD (μM) | Max. IR | Induction of genotoxic response | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| β-Galactosidasea | OD600b | Ref. 15 | In this study | Ref. 15 | EC50 (mM) | ECIR1.5 (μM) | P value | |||
| a The LODs were calculated from the dose–response curves using the β-galactosidase activities as the response parameters.b The LODs were calculated from the dose–response curves using the cell turbidities (OD600) as the response parameters. | ||||||||||
| MMC | 50-07-7 | 0.001 | 0.026 | 0.1221 | 0.01 | 15.9 | 14.0 | 0.0018 | 0.1221 | 0.0012 |
| Azinphos-methyl | 86-50-0 | 60 | 320.4 | 2057.0 | 600 | 15.4 | 2.6 | 21.31 | 2057.0 | 0.0001 |
| MMS | 66-27-3 | 15 | 34.4 | 66.0 | 150 | 16.6 | 12.0 | 1.187 | 66.0 | 0.0016 |
| Dithianone | 3347-22-6 | 1.8 | 4.6 | 14.4 | 18 | 13.9 | 2.0 | 1.02 | 14.4 | 0.0003 |
| Dichlofluanid | 1085-98-9 | 1 | 6.0 | 32.5 | 10 | 15.0 | 3.5 | 1.049 | 32.5 | <0.0001 |
| K2Cr2O7 | 0.0006 | 0.0271 | 0.1747 | 15.6 | 0.0013 | 0.1747 | <0.0001 | |||
The induction ratio (IR) of 1.5 is defined by the EN ISO guideline (International Standard Organization, 2000) as the threshold of genotoxic effect, indicating the sample was not cytotoxic. The effect concentration that induced IR = 1.5 (or ECIR1.5) was interpolated from the sigmoidal concentration–effect curve (Fig. S3†) and also presented in Table 2. For the cytotoxicity assay, the highest detected concentration in dose–effect experiment, 1 μM MMC, 60 mM azinphos-methyl, 15 mM MMS, 1.8 mM Dithianone and 1 mM dichlofluanid were added into E. coli BL21 culture medium separately. No obvious decrease in cell viability compared with control sets was noticed (Fig. S4,† p < 0.001). And the derived values of ECIR1.5 are approximately 20 times lower than the cytotoxicity assay concentrations, indicating that there is a wide window between induced cell lysis and cytotoxicity.
In this study, the five tested chemicals showed strong or weak genotoxic (Table 2). However, in different studies using different detection procedure, even the same substance might present various levels of genotoxicity as introduction section described. This is ascribed to the limitations of current genotoxicity testing procedures which affect the reliability of the methods. A single test is not sufficient for the evaluation all relevant genotoxic substances. To improve the accuracy, the predictions from various models could be combined.41
Since Oda et al.36 developed the SOS/umu-test strain for the SOS-inducing carcinogens detection, this method has received great attention and acceptance.9,18,42 Considering the cell lysis requirements for the intracellular expressed β-galactosidase activity measurement, we introduced cell lysis cluster (SRRz) under the control of genotoxicity inducible promoter. Research demonstrated that cell lysis efficiency was dependent on the genotoxic effects (Fig. 3). The released β-galactosidase increased as the genotoxin concentration increased, thus the IR values of the tested five chemicals displayed a dose-dependent manner in the detection concentration range. The five maximum IR values were in the range of 13.8 to 16.0, which are all higher than those literature reported (Table 1). Especially, the truncated SOS responsive promoter (Psos-box, 47 bp) showed a much more intense response to azinphos-methyl, dithianone and dichlofluanid than the traditional umu operon.36
3.4 Mixture effects of the known components
Hexavalent chromium [Cr(IV)] is a well-known mutagen and carcinogen, and therefore served as the reference chemical to calculate the bioanalytical equivalent concentrations (BEQ) of mixed genotoxicity. The sigmoidal concentration–effect curve was plotted as Fig. 4, and the LOD and ECIR1.5 values were derived as 0.6 μM and 0.17 μM, respectively (Table 2).To establish whether the genotoxicity of a combination of chemicals will deviate from an effect expected for additivity, the mixtures of five model genotoxicants at specific concentrations (Table S1†) were prepared. All the mixtures could induce apparent cell lysis through OD600 detection. The bioanalytical equivalent concentrations (BEQ) and the toxic equivalent concentration (TEQ) using Cr(VI) as reference were determined. The scatterplot shows a strong relation between TEQ-Cr(VI) and BEQ-Cr(VI) (Fig. 5). The low TEQ-Cr(VI) values typically accompanied with low BEQ-Cr(VI) (calculated according to the dose–effect fitting curves), and vice versa. Furthermore, this relation is roughly linear. Through Pearson correlation evaluation, the PCC value between TEQ-Cr(VI) and BEQ-Cr(VI) were determined as 0.8182 and 0.8424, respectively. Results indicated that the correlation between TEQ-Cr(VI) and BEQ-Cr(VI) is very strong. This results clearly indicated that the toxic equivalent concentration of Cr(VI) in aqueous phase sample could be derived from the observed lysis efficiency induced by mixtures through the dose–response curve of Cr(VI).
 | ||
| Fig. 5 Pearson analysis of TEQmixture with BEQ-Cr(VI) at the confidence interval 95% with one-tailed p values. (A) Two components were mixed. (B) Three components were mixed. All the chemical concentration in each sample is presented in Table S1.† | ||
For the real environmental aqueous samples are a “cocktail” of chemicals, containing a lot of contaminants that have potential to induce gene mutation. Despite the rapid development of chemical analysis methods, it was still difficult or infeasible to track every genotoxicants in water. Especially, there are many chemicals are not readily identified because of their unknown structure, lack of pure sample preparation and detection method.4,43,44 The rapid, less expensive and high-throughput technology was pursued, that could screen thousands of chemicals for potential genotoxicity in a relatively simple way. The first use of a toxicity equivalence-like method for risk assessment purposes was described by Eadon et al. (1986).45 In this study, we selected five representative chemicals including MMC, azinphos-methyl, MMS, dithianone as well as dichlofluanid to address the risk assessments. The good correlation between TEQ-Cr(VI) and BEQ-Cr(VI) preliminarily demonstrated that the SOS/SRRz system could be used in risk assessment for environment samples, by converting the concentration of unknown genotoxic compounds into equivalent concentrations of Cr(VI).
4. Conclusions
In this study, we demonstrated that the activity of the new-constructed strain of E. coli BL21/pUC-PST can be used in genotoxin detection. For qualitative detection, it could be done simply through observing the turbidity of the bacteria with naked eyes after 0.5 h contact with the genotoxic substance. While, for quantitative testing, determination of the extracellular and intracellular β-galactosidase activity also can help achieve. While, as also demonstrated here, the truncated SOS responsive promoter may display improved detection sensitivity. The SOS/SRRz cell lysis system provides a potential approach on water samples for estimating the SOS-mediated genotoxic risks.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key R & D Program of China (2018YFA0901504), the Natural Science Foundation of China (21878104 and U1701243), the Project on the Integration of Industry, Education, and Research of Guangzhou, China (201704020183 and 201903010086) and the Science and Technology Planning Project of Guangdong Province, China (2017A010105019).References
- S. Wu, S. Powers, W. Zhu and Y. A. Hannun, Nature, 2015, 529, 43–47 CrossRef.
- F. Dardenne, R. Smolders, W. De Coen and R. Blust, Environ. Sci. Technol., 2007, 41, 1790–1796 CrossRef CAS.
- C. L. Lemieux, A. S. Long, I. B. Lambert, S. Lundstedt, M. Tysklind and P. A. White, Environ. Sci. Technol., 2015, 49, 1797–1805 CrossRef CAS.
- A. Jia, B. I. Escher, F. D. L. Leusch, J. Y. M. Tang, E. Prochazka, B. Dong, E. M. Snyder and S. A. Snyder, Water Res., 2015, 80, 1–11 CrossRef CAS.
- T. R. Skopek, H. L. Liber, J. J. Krolewski and W. G. Thilly, Proc. Natl. Acad. Sci. U. S. A., 1978, 75, 410–414 CrossRef CAS.
- D. M. Maron and B. N. Ames, Mutat. Res., 1983, 113, 173–215 CAS.
- P. Quillardet, O. Huisman, R. D'Ari and M. Hofnung, Proc. Natl. Acad. Sci. U. S. A., 1982, 79, 5971–5975 CrossRef CAS.
- J. L. Fuentes, A. G. Forero, N. Q. Ruiz, C. A. P. Medina, N. R. Castellanos, D. A. F. Nino, D. A. C. Garcia, Y. C. Campo and E. E. Stashenko, Photochem. Photobiol. Sci., 2017, 16, 1424–1434 RSC.
- G. Reifferscheid and J. Heil, Mutat. Res., 1996, 369, 129–145 CAS.
- C. Janion, Int. J. Biol. Sci., 2008, 4, 338–344 CrossRef CAS.
- L. Shen, J. Y. Wu, G. F. Lin, J. H. Shen, J. Westendorf and H. Huehnerfuss, Chemosphere, 2003, 52, 1641–1646 CrossRef CAS.
- Q. Chai, A. Hu, Y. Qian, X. Ao, W. Liu, H. Yang and Y. F. Xie, Chemosphere, 2018, 191, 335–341 CrossRef CAS.
- H. Dizer, E. Wittekindt, B. Fischer and P. D. Hansen, Chemosphere, 2002, 46, 225–233 CrossRef CAS.
- Y. Oda, K. Funasaka, M. Kitano, A. Nakama and T. Yoshikura, Environ. Mol. Mutagen., 2004, 43, 10–19 CrossRef CAS.
- G. Reifferscheid, J. Heil, Y. Oda and R. K. Zahn, Mutat. Res., 1991, 253, 215–222 CAS.
- Y. Oda, H. Yamazaki, M. Watanabe, T. Nohmi and T. Shimada, Mutat. Res., 1995, 334, 145–156 CAS.
- J. Courcelle, J. R. Donaldson, K. H. Chow and C. T. Courcelle, Science, 2003, 299, 1064–1067 CrossRef CAS.
- Y. Oda, Genes Environ., 2016, 38, 24 CrossRef.
- M. P. D. Méo, V. Miribel, A. Botta, M. Laget and G. Duménil, Mutagenesis, 1988, 3, 277–283 CrossRef.
- Z. Tian, Y. Oda, Y. Zhang, M. Yang and H. Li, Bull. Environ. Contam. Toxicol., 2015, 94, 370–375 CrossRef CAS.
- T. Dapa, S. Fleurier, M.-F. Bredeche and I. Matic, Genetics, 2017, 206, 1349–1360 CrossRef CAS.
- E. Eder, W. Kütt and C. Deininger, Chem.-Biol. Interact., 2001, 137, 89–99 CrossRef CAS.
- S. Gómez-Arroyo, L. Sánchez-Estrada, S. Andrade-Morales, J. Cortés-Eslava and R. Villalobos-Pietrini, Rev. Int. Contam. Ambiental, 2015, 31, 227–236 Search PubMed.
- N. E. Garrett, H. F. Stack and M. D. Waters, Mutat. Res., 1986, 168, 301–325 CAS.
- E. Zeiger, B. Anderson, S. Haworth, T. Lawlor, K. Mortelmans and W. Speck, Environ. Mol. Mutagen., 1987, 9, 1–109 CrossRef CAS.
- J. E. Gallagher, D. Hellmich, M. Hergenröder and R. K. Zahn, Environ. Mol. Mutagen., 1991, 17, 20–26 CrossRef.
- EFSA, EFSA J., 2015, 13, 4278 Search PubMed.
- D. A. Armbruster and T. Pry, Clin. Biochem. Rev., 2008, 29, S49–S52 Search PubMed.
- K. P. Nickens, S. R. Patierno and S. Ceryak, Chem.-Biol. Interact., 2010, 188, 276–288 CrossRef CAS.
- B. I. Escher, N. Bramaz, J. F. Mueller, P. Quayle, S. Rutishauser and E. L. Vermeirssen, J. Environ. Monit., 2008, 10, 612–621 RSC.
- A. Norman, L. Hestbjerg Hansen and S. J. Sørensen, Appl. Environ. Microbiol., 2005, 71, 2338–2346 CrossRef CAS PubMed.
- H. Shinagawa, T. Kato, T. Ise, K. Makino and A. Nakata, Gene, 1983, 23, 167–174 CrossRef CAS.
- R. Young, Microbiol. Rev., 1992, 56, 430–481 CAS.
- R. White, S. Chiba, T. Pang, J. S. Dewey, C. G. Savva, A. Holzenburg, K. Pogliano and R. Young, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 798–803 CrossRef CAS.
- J. Berry, M. Rajaure, T. Pang and R. Young, J. Bacteriol., 2012, 194, 5667–5674 CrossRef CAS.
- Y. Oda, S. Nakamura, I. Oki, T. Kato and H. Shinagawa, Mutat. Res., 1985, 147, 219–229 CAS.
- P. Gee, C. H. Sommers, A. S. Melick, X. M. Gidrol, M. D. Todd, R. B. Burris, M. E. Nelson, R. C. Klemm and E. Zeiger, Mutat. Res., 1998, 412, 115–130 CAS.
- A. C. Vollmer, S. Belkin, D. R. Smulski, T. K. Van Dyk and R. A. LaRossa, Appl. Environ. Microbiol., 1997, 63, 2566–2571 CAS.
- A. Biran, H. Ben Yoav, S. Yagur-Kroll, R. Pedahzur, S. Buchinger, Y. Shacham-Diamand, G. Reifferscheid and S. Belkin, Anal. Bioanal. Chem., 2011, 400, 3013–3024 CrossRef CAS.
- S. M. Thomas and T. Justus, Mutagenesis, 1999, 14, 351–356 CrossRef.
- H. Turkez, M. E. Arslan and O. Ozdemir, Expert Opin. Drug Metab. Toxicol., 2017, 13, 1089–1098 CrossRef CAS.
- H. Hori, D. Hirata, W. Fujii and Y. Oda, Environ. Mol. Mutagen., 2017, 58, 209–216 CrossRef CAS.
- B. I. Escher, C. van Daele, M. Dutt, J. Y. M. Tang and R. Altenburger, Environ. Sci. Technol., 2013, 47, 7002–7011 CrossRef CAS.
- N. Klüver, C. Vogs, R. Altenburger, B. I. Escher and S. Scholz, Chemosphere, 2016, 164, 164–173 CrossRef PubMed.
- G. Eadon, L. Kaminsky, J. Silkworth, K. Aldous, D. Hilker, P. O'Keefe, R. Smith, J. Gierthy, J. Hawley and N. Kim, Environ. Health Perspect., 1986, 70, 221–227 CrossRef CAS.
- OECD, Test No. 487: In vitro mammalian cell micronucleus test, OECD Publishing, Paris, 2010 Search PubMed.
- OECD, Test No. 474: Mammalian erythrocyte micronucleus test, OECD Publishing, Paris, 2014 Search PubMed.
- S. Hamada, M. Shigano, S. Kawakami, M. Ueda, H. Sui, K. Yamada, S. Hagio, A. Momonami, A. Maeda, Y. Terashima, W. Ohyama, T. Morita and M. Hayashi, Genes Environ., 2019, 41, 13 CrossRef.
- OECD, Test No. 489: In vivo mammalian alkaline comet assay, OECD Publishing, Paris, 2016 Search PubMed.
- OECD, Test No. 471: Bacterial reverse mutation test, OECD Publishing, Paris, 1997 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06202e |
| This journal is © The Royal Society of Chemistry 2019 |