Open Access Article
Open Access ArticleRetracted Article: Long non-coding RNA TUG1 alleviates high glucose induced podocyte inflammation, fibrosis and apoptosis in diabetic nephropathy via targeting the miR-27a-3p/E2F3 axis
Yang Lia,
Denggao Huangb,
Linlin Zhengb,
Hui Caob,
Yuanhui Gaob,
Yongmei Yanga and
Zhongcheng Fan *c
*c
aDepartment of Nephropathy and Rheumatology, Central South University Xiangya School of Medicine Affiliated Haikou Hospital, Haikou, Hainan, China
bDepartment of Central Lab, Central South University Xiangya School of Medicine Affiliated Haikou Hospital, Haikou, Hainan, China
cDepartment of Orthopedic, Central South University Xiangya School of Medicine Affiliated Haikou Hospital, Yipinshuiyuan 5-1102, Wudong Road, Haidian Island, Meilan District, Haikou, Hainan 570208, China. E-mail: zhao19727t@yeah.net; Tel: +86 898 66151128
First published on 19th November 2019
Abstract
Diabetic nephropathy (DN) is the most common cause of end-stage renal disease (ESRD) in developed countries. The long non-coding RNA (lncRNA) taurine upregulated gene 1 (TUG1) is associated with mitochondrial function in podocytes in DN. However, the detailed mechanism of TUG1 in DN has not yet been fully elucidated. In this study, we found that the expression levels of TUG1 and E2F transcription factor 3 (E2F3) were downregulated, and microRNA-27a-3p (miR-27a-3p) level was upregulated in DN tissues and HG-treated podocytes. Function analysis indicated that TUG1 overexpression inhibited inflammation, fibrosis and apoptosis in HG-induced podocytes. Rescue experiments demonstrated that miR-27a-3p reversed the suppression effects of TUG1 on apoptosis, fibrosis and inflammation in HG-stimulated podocytes, while E2F3 abrogated the promotion effects of miR-27a-3p on apoptosis, fibrosis and inflammation in HG-induced podocytes. Mechanical analysis revealed that TUG1 regulated E2F3 expression via sponging miR-27a-3p. In conclusion, our findings suggested that TUG1 alleviates high glucose induced inflammation, fibrosis and podocyte apoptosis in diabetic nephropathy via targeting the miR-27a-3p/E2F3 axis, providing a potential therapeutic target for treatment of DN.
Introduction
Diabetic nephropathy (DN), a microvascular complication in diabetes, is the most common cause of end-stage renal disease (ESRD) in developed countries.1 The morbidity of DN accounts for approximately 45% of newly diagnosed end-stage kidney failure, and the 5 year survival rate of end-stage kidney disease is less than 20%.2 It is noteworthy that renal fibrosis, the activation of myofibroblasts, podocyte dysfunction, thickening of the basement membrane and the accumulation of extracellular matrix (EMC) proteins are considered to be the typical characteristics of DN.3,4 Podocytes have been identified as a group of high differentiation glomerular epithelial cells and are closely related to the early pathological mechanism of DN pathogenesis.5 Besides, the function of renal is directly destroyed by the enhancement of inflammation.6 Despite the significant progress in treatment methods, it is still necessary to further probe the possible molecular mechanisms of DN to provide a novel therapeutic strategy for disease treatment.Long non-coding RNAs (lncRNAs), a family of non-protein-coding RNA molecules longer over than 200 nucleotides in size, was identified as important mediators in the progression of various diseases, including DN. For example, lncRNA MALAT1 overexpression could contribute to high glucose (HG)-induced EMT and fibrosis by regulating miR145/ZEB2 axis.7 Simultaneously, lncRNA Rpph1 was proved to be able to promote inflammation and cell proliferation by interacting with in DN.8 LncRNA taurine upregulated gene 1 (TUG1), an evolutionarily conserved long intergenic non-coding RNA, was firstly identified as a pivotal regulatory factor in the formation of photoreceptors and retinal development.9 Previous reports have demonstrated that TUG1 was downregulated in the DN rat model and associated with mitochondrial function in podocyte in DN.10,11 Moreover, a prior study displayed that TUG1 could alleviate podocyte apoptosis of DN rats by binding the TRAF5.12 However, the detailed mechanism of TUG1 in DN has not yet been fully elucidated.
MicroRNAs (miRNAs or miRs) are short non-coding RNAs with about 19–24 nucleotides which participating in the regulation of gene expression by competitively binding to the 3′-untranslated region (3′UTR) of target genes.13 More recently, abnormal expression of miRNAs has been highlighted to be implicated in DN, suggesting that miRNAs were vital in DN development.14,15 miR-27a-3p is a form of mature miR-27a which was upregulated and induced podocyte injury by PPAR γ-mediated β-catenin in the pathogenesis of DN.16 But the biological function of miR-27a-3p in DN is still unknown.
E2F transcription factor 3 (E2F3), a member of the E2F family, works as a vital regulator in numerous diseases and implicates in physiological processes containing the cell cycle, apoptosis and metastasis.17–20 In a recent publication, E2F3 was verified to be aberrantly decreased in podocyte under the treatment of HG, and E2F3 repressed podocyte injury by the upregulation of miR-503 in DN,21 implying that E2F3 exerted a significant function in DN progress.
In this paper, we aimed to confirm the role of TUG1 in DN, and to clarify whether the effect of TUG1 on high glucose (HG)-treated podocyte injury was mediated by regulating miR-27a-3p/E2F3 axis.
Materials and methods
Clinical specimens and podocyte culture
Thirty-three tissues samples of DN and paired normal samples were obtained from 33 patients who undergone surgical operation at Central South University Xiangya School of Medicine Affiliated Haikou Hospital. All patients signed the written informed consents. This research was approval of Central South University Xiangya School of Medicine Affiliated Haikou Hospital committee.Human podocytes (podocytes) were purchased from Otwo Biotech (Shenzhen, China), and podocytes were maintained in McCoy's 5A medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Invitrogen) and 1% antibiotic (100 U mL−1 penicillin and 100 μg mL−1 streptomycin; Solarbio, Beijing, China) in an incubator containing 5% CO2 at 37 °C. Human embryonic kidney 293 cell line (293T) was acquired from Cell Resource Center (Shanghai, China) and cultured in a 5% CO2 incubator at 37 °C in Dulbecco's modified Eagle's medium (DMEM; HyClone, Beijing, China). Besides, for subsequent experiments, podocytes were exposed with 33 mM glucose for 24 h to trigger DN cell model.
Cell transfection
To over-express TUG1, the cDNA sequences of TUG1 were cloned in pcDNA3.1 empty vector (pcDNA; Invitrogen), termed as pcDNA3.1-TUG1 (TUG1). siRNA against TUG1(si-TUG1), siRNA against E2F3 (si-E2F3) and corresponding scrambled negative control (si-NC), miR-27a-3p mimic (miR-27a-3p), miR-27a-3p inhibitor (in-miR-27a-3p), and scrambled negative controls (miR-NC, in-miR-NC) were purchased from GenePharma (Shanghai, China). Transient transfection of the indicated oligonucleotides and plasmids into treated podocytes was implemented by Lipofectamine 2000 reagents (Invitrogen) based on the operation manual.RNA extraction and quantitative real-time PCR (RT-qPCR)
Total RNA was isolated form tissues and podocytes (after 48 h transfection) using TRIzol reagent (Invitrogen). Extracted RNA was used for synthesis of complementary DNA (cDNA) by M-MLV reverse transcriptase (Promega, Madison, Wisconsin, USA). The quantitative analysis of TUG1 and E2F3 was conducted using SYBR Premix ExTaq II Kit (Takara Bio, Inc., Otsu, Japan), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used to standardize expression levels of TUG1 and E2F3. miR-27a-3p level was measured with the TaqMan microRNA assay (Applied Biosystems, Carlsbad, CA, USA), and U6 snRNA was served as the endogenous control. The relative expression levels of TUG1, E2F3 and miR-27a-3p were calculated using the 2−ΔΔCt method.The following primer sequences were used in RT-qPCR:
TUG1 sense: 5′-TAGCAGTTCCCCAATCCTTG-3′, antisense: 5′-CACAAATTCCCATCATTCCC-3′;
E2F3 sense: 5′-CACTTCCACCACCTCCTGTT-3′, antisense: 5′-TGACCGCTTTCTCCTAGCTC-3′ (antisense);
GAPDH sense: 5′-CACCCACTCCTCCACCTTTG-3′, antisense: 5′-CCACCACCCTGTTGCTGTAG-3′;
miR-27a-3p sense: 5′-ACACTCCAGCTGGGTTCACAGTGGCTAAG-3′, antisense: 5′-TGGTGTCGTGGAGTCG-3′ (antisense);
U6 sense: 5′-CTCGCTTCGGCAGCACA-3′, antisense: 5′-AACGCTTCACGAATTTGCGT-3′.
Cell apoptosis assay
The flow cytometry assay was employed to evaluate the influence of HG, TUG1, miR-27a-3p and E2F3 on the apoptosis rate of podocytes. Briefly, podocytes were harvested after 48 h transfection, and then treated podocytes were resuspended in 200 μL binding buffer and 5 μL Annexin V-FITC/1 μL PI (Life Technologies, New York, USA), following by incubation for 15 min in the dark. The apoptotic cells were treated with the FACSan flow cytometry (BD Bioscience) and Cell Quest software.Western blot assay
Podocytes were lysed using pre-cold RIPA buffer (Thermo Fisher Scientific, Rockford, IL, USA) with protease inhibitors (Sigma, St Louis, MO, USA). And then, 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate these podocytes lysates, whereafter, these isolated proteins were transferred onto a polyvinylidene fluoride (PVDF) membrane (Merck KGaA, Darmstadt, Germany). The membranes were sealed with 5% nonfat milk for 1 h at room temperature, and incubated with primary antibodies against E2F3 (ab152126, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 dilution, Abcam, Cambridge, UK), B-cell lymphoma-2 (Bcl-2, ab32124, 1
2000 dilution, Abcam, Cambridge, UK), B-cell lymphoma-2 (Bcl-2, ab32124, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution, Abcam), FN (ab18265, 1
1000 dilution, Abcam), FN (ab18265, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution, Abcam), Bcl-2 related X protein (Bax, ab69643, 1
1000 dilution, Abcam), Bcl-2 related X protein (Bax, ab69643, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution, Abcam), α-SMA (Cat. 19245, 1
1000 dilution, Abcam), α-SMA (Cat. 19245, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution, Cell Signaling Technology, Danvers, MA, USA), Col I (Cat. 84336, 1
1000 dilution, Cell Signaling Technology, Danvers, MA, USA), Col I (Cat. 84336, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution, Cell Signaling Technology) or β-actin (Cat. 3700, 1
1000 dilution, Cell Signaling Technology) or β-actin (Cat. 3700, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 dilution, Cell Signaling Technology) overnight at 4 °C, following by incubation with horseradish peroxidase (HRP)-linked secondary antibody (ab205718, 1
2000 dilution, Cell Signaling Technology) overnight at 4 °C, following by incubation with horseradish peroxidase (HRP)-linked secondary antibody (ab205718, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 dilution, Abcam) were incubated at 37 °C for 1 h. Protein bands were detected using an ECL reagent (Pierce Biotechnology, Rockford, IL, MA, USA).
000 dilution, Abcam) were incubated at 37 °C for 1 h. Protein bands were detected using an ECL reagent (Pierce Biotechnology, Rockford, IL, MA, USA).
Enzyme-linked immunosorbent assay (ELISA)
Podocyte were cultured in 24-well plates and carried out with different treatments for 24 h, and centrifuged and collected the supernatant of podocytes. And then, the levels of IL-1β, TNF-α and IL-6 in podocytes supernatant were detected by ELISA assay kits (Boster, Wuhan, China) referring to the operation manual. The fluorescence signals of IL-1β, TNF-α and IL-6 at 450 nm were assessed using a microplate reader.Dual luciferase reporter assay
Partial TUG1 sequence and E2F3 3′UTR harboring putative miR-27a-3p targeting site were cloned into psiCHECK-2 vector (Promega), namely TUG1 WT/MUT, E2F3 3′UTR WT/MUT reporter plasmids. Afterwards, 293T cells were co-transfected with TUG1 WT/MUT or E2F3 3′UTR WT/MUT and miR-NC, miR-27a-3p, in-miR-NC or in-miR-27a-3p and incubated for 48 h. At last, podocytes were harvested, and luciferase activities in cell lysates were analyzed using a dual luciferase reporter assay kit (Promega).Statistical analysis
Every independent experiment was repeated three times. All data were presented as the mean ± standard deviation (S.D.). Pearson correlation analysis was used to analyze the expression interaction. Data were detected with SPSS 18.0 (IBM, Chicago, IL, USA). Statistical differences were analyzed using Student's t test or one way of variance (ANOVA). P < 0.05 meant that the difference was statistically significant.Results
TUG1 and E2F3 was downregulated, and miR-27a-3p was upregulated in DN tissues and podocytes treated with HG
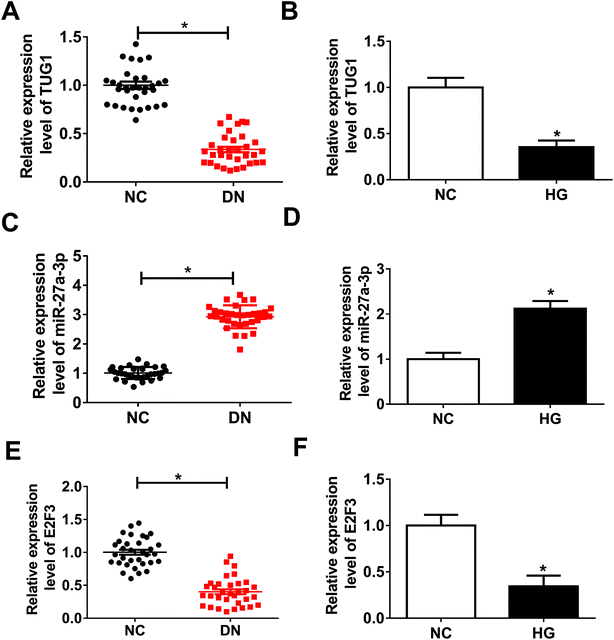
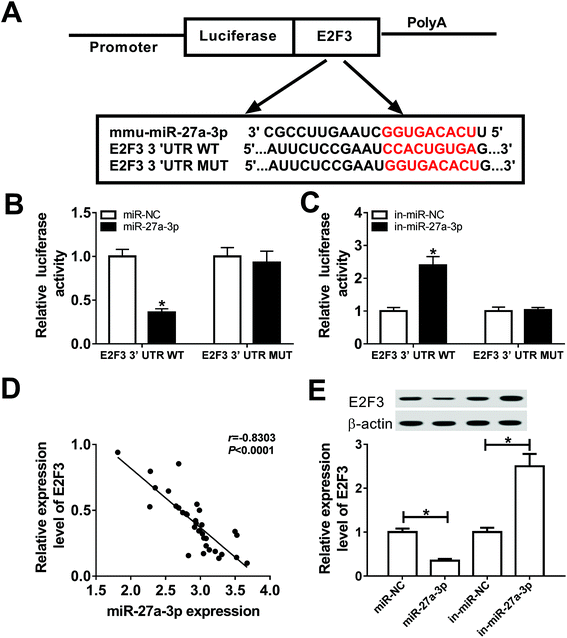
Firstly, to investigate the role of TUG1, miR-27a-3p and E2F3 in DN, their expression patterns were detected by RT-qPCR assay. Compared with NC group, TUG1 (Fig. 1A) and E2F3 (Fig. 1E) expressions were obviously decreased and miR-27a-3p (Fig. 1C) was evidently increased in renal tissues from DN patients. Then, we further validated that TUG1 (Fig. 1B) and E2F3 (Fig. 1F) were low expressed, and miR-27a-3p (Fig. 1D) was highly expressed in podocytes treated with HG with respect to podocytes without treatment. These data indicated that the involvement of TUG1, miR-27a-3p and E2F3 in DN progression.TUG1 mitigated HG-induced apoptosis, fibrosis and inflammation
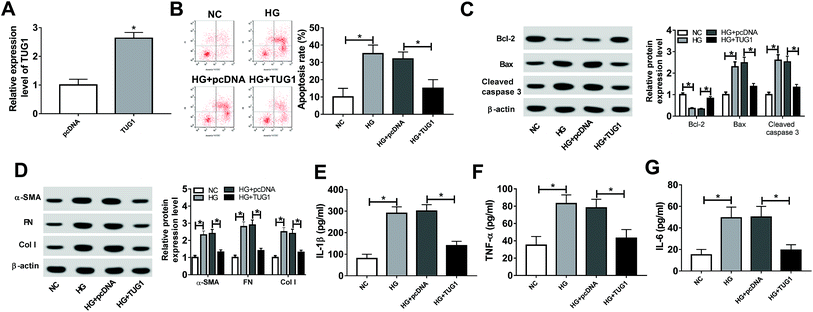
Next, to explore the function of TUG1 in HG-induced podocytes, the overexpression plasmid of TUG1 was constructed. According to the result shown in Fig. 2A, the expression level of TUG1 was remarkably upregulated in podocytes transfected with pcDNA3.1-TUG1 in comparison with cells transfected with empty vector. Thus, we employed the overexpression systems to further probe the role of TUG1 in apoptosis, fibrosis and inflammation of podocytes treated with HG. The results of flow cytometry assays exhibited that the high expression of TUG1 effectively reversed HG-induced increase in apoptotic rate in podocytes (Fig. 2B). Similar to flow cytometry results, TUG1 overexpression markedly abolished HG-triggered downregulation of the Bcl-2, and upregulation of Bax and cleaved caspase 3 in podocytes (Fig. 2C). Then, we further investigated that the effect of TUG1 on fibrosis of podocytes treated with HG. The data showed that HG improved the expression levels of fibrogenesis-associated proteins, α-SMA, FN and Col I, while recover of TUG1 expression greatly weakened these effects (Fig. 2D). In addition, the inflammatory cytokines IL-1β, TNF-α and IL-6 were detected in HG-treated podocytes. The ELISA analysis pointed out that HG accelerated IL-1β IL-6 and TNF-α expression levels, which was apparently attenuated by regaining of TUG1 (Fig. 2E–G). All these data indicated that TUG1 mitigated HG-induced apoptosis, fibrosis and inflammation of podocytes.miR-27a-3p served as a target of TUG1
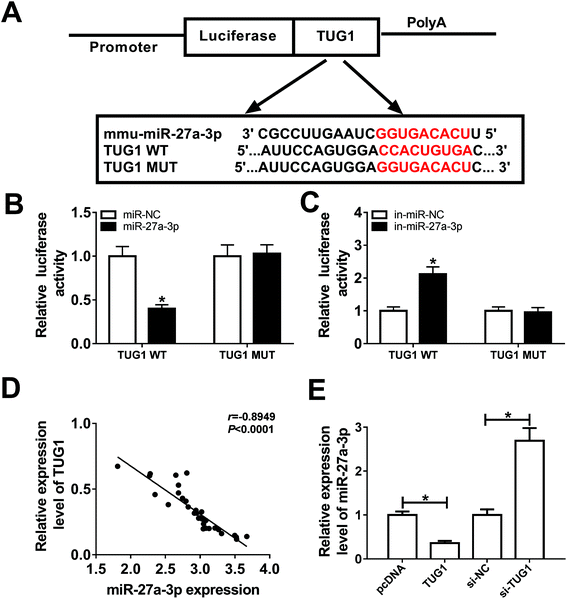
It is widely accepted that lncRNAs could exert the function by interacting with miRNAs. Therefore, online prediction website DIANA-LncBase v2 was used to search the potential target miRNAs of TUG1. As a result, miR-27a-3p contained some complementary bases pairing with TUG1 (Fig. 3A). To confirm the potential interaction between miR-27a-3p and TUG1, TUG1 WT or TUG1 MUT reporter plasmid with miR-NC, miR-27a-3p, in-miR-NC or in-miR-27a-3p was co-transfected into 293T cells. After 48 h transfection, luciferase reporter assay suggested that ectopic expression of miR-27a-3p reduced the luciferase activity of TUG1 WT reporter systems, but miR-27a-3p deficiency elevated the luciferase activity of TUG1 WT reporter systems (Fig. 3B and C). Nevertheless, upregulation or downregulation of miR-27a-3p had no marked effect on the luciferase activity of TUG1 MUT reporter systems (Fig. 3B and C). Furthermore, miR-27a-3p expression level was strikingly inversely associated with TUG1 level in DN tissues and podocytes (Fig. 3D and E). Collectively, these results indicated that TUG1 interacted with miR-27a-3p to hinder its expression.miR-27a-3p reversed the suppression effects of TUG1 on apoptosis, fibrosis and inflammation in HG-stimulated podocytes
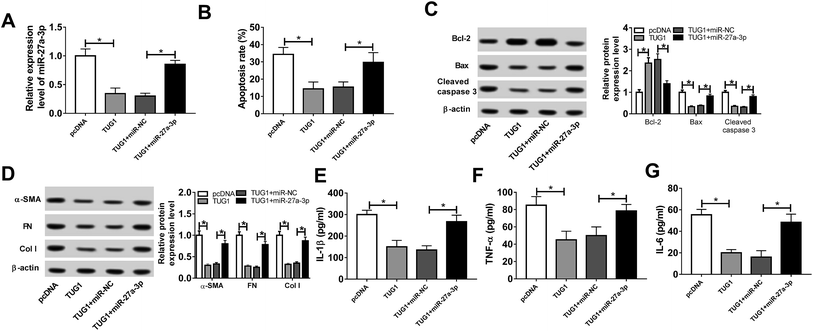
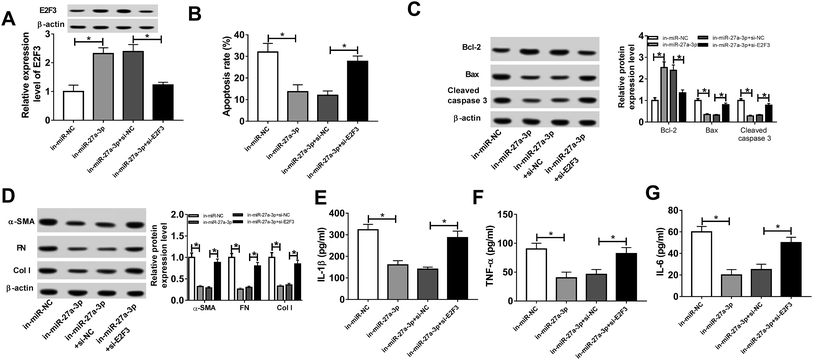
Then, to probe the mechanism of TUG1 involved in apoptosis, fibrosis and inflammation of HG-triggered podocytes, we executed remedial experiments by transfecting pcDNA3.1, pcDNA3.1-TUG1, pcDNA3.1-TUG1 + miR-NC or pcDNA3.1-TUG1 + miR-27a-3p into podocytes prior to HG treatment. As presented in Fig. 4A, overexpression of TUG1 conspicuously inhibited miR-27a-3p expression level, while rescue of miR-27a-3p distinctly abated the effect in HG-induced podocytes. Subsequently, cells apoptosis, fibrosis and inflammation were detected. Flow cytometry results suggested that TUG1 upregulation prominently retarded apoptotic rate in HG-treated podocytes, however, recover of miR-27a-3p expression obviously counteracted this effect (Fig. 4B). Similarly, TUG1 overexpression led to an overt increase in the expression of Bcl-2, and an evident decrease in the expression of Bax and cleaved caspase 3, while the changes of apoptosis-associated proteins were eliminated after the co-transfected with miR-27a-3p (Fig. 4C). Besides, TUG1-elicited inhibitory effects on fibrosis and inflammation were substantially abrogated after reintroduction with miR-27a-3p, presenting that the highly expression of TUG1 repressed the expression level of fibrogenesis-associated proteins (α-SMA, FN and Col I) and inflammatory cytokines (IL-1β, TNF-α and IL-6) and introduction of miR-27a-3p effectively relieved these effects (Fig. 4D–G). All of these suggested that miR-27a-3p upregulation reversed TUG1-elicited apoptosis, fibrosis and inflammation in HG-treated podocytes.E2F3 was a target of miR-27a-3p
DIANA microT-CDS was used to search for the potential targets of miR-27a-3p. As showed in Fig. 5A, E2F3 was found to contain some binding sequences with miR-27a-3p. Next, luciferase reporter assay was implemented to verify the bioinformatics prediction. In this assay, luciferase reporter plasmids, including wild-type or mutated E2F3 3′UTR with miR-27a-3p or in-miR-27a-3p, were co-transfected into 293T cells and then the luciferase activities were detected after 48 h transfection. The data indicated that miR-27a-3p upregulation remarkably reduced the luciferase activity of E2F3 WT reporter, while miR-27a-3p downregulation notably enhanced the luciferase activity of E2F3 WT reporter in 293T cells (Fig. 5B and C). However, high or low expression of miR-27a-3p exhibited little effects on the luciferase activity of E2F3 MUT reporter (Fig. 5B and C). What's more, we analyzed the effect of miR-27a-3p on E2F3 expression in HG treatment of the DN tissues and podocytes. As expected, miR-27a-3p expression level was negatively correlated with E2F3 expression level in DN tissues treated with HG (Fig. 5D). Western blot results also displayed that the overexpression of miR-27a-3p curbed E2F3 protein level, and the depletion of miR-27a-3p accelerated E2F3 protein level in HG-induced podocytes (Fig. 5E). Taken together, these results testified that miR-27a-3p directly targeted E2F3 and regulated its expression.miR-27a-3p-triggered apoptosis, fibrosis and inflammation were abolished by E2F3
Subsequently, we further identified whether miR-27a-3p could exert its function by targeting E2F3 in treated podocytes. Podocytes were transfected with in-miR-NC, in-miR-27a-3p, in-miR-27a-3p + si-NC or in-miR-27a-3p + si-E2F3, following by exposed to HG for 24 h. As presented in Fig. 6A, miR-27a-3p knockdown obviously facilitated E2F3 expression level, which was effectively weakened by suppression of E2F3. Moreover, downregulation of miR-27a-3p resulted in an apparent decline in apoptotic rate, while silence of E2F3 notably abolished this effect (Fig. 6B). In parallel, the lower expression of miR-27a-3p triggered an overt increase of Bcl-2 expression, and a striking decrease of Bax and cleaved caspase 3 expression in treated podocytes, while these effect were undermined after downregulating E2F3 (Fig. 6C). Additionally, the introduction of si-E2F3 prominently upregulation in-miR-27a-3p-mediated suppressive effect on α-SMA, FN, Col I, IL-1β, TNF-α and IL-6 protein expression levels in treated podocytes (Fig. 6D–G). All these data revealed that E2F3 could reverse miR-27a-3p-induced apoptosis, fibrosis and inflammation in HG-treated podocytes.TUG1 worked as a molecular sponge of miR-27a-3p to influence E2F3 expression in HG-treated podocytes
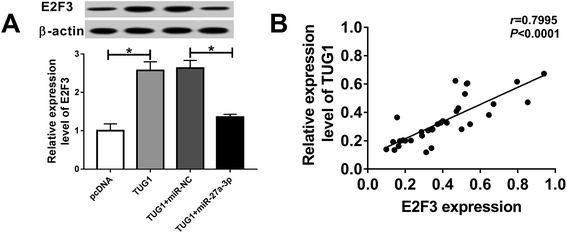
Based on the above findings, we inferred that TUG1 could exert its suppressive role partly by the miR-27a-3p/E2F3 axis. To confirm the assumption, we explored whether TUG1 affecting the expression of E2F3 by miR-27a-3p. As showed in Fig. 7A, high expression of TUG1 elevated the expression of E2F3, and introduction of miR-27a-3p abolished TUG1-induced increase in E2F3 expression level in HG-treated podocytes. Beside, TUG1 expression level was positively associated with E2F3 expression level in stimulated podocytes (Fig. 7B). In a word, these data suggested that TUG1 served as ceRNA to sequester miR-27a-3p from E2F3.Discussion
Recent years, the biological role of lncRNA in multiple diseases has been a hotspot in the research field. Accumulative evidences have indicated that lncRNAs act as diagnostic biomarkers and possible therapeutic targets in DN.22 LncRNA TUG1, located at chromosome 22q12, had been documented to be downregulated in HG-treated MPC5 cells, and TUG1 was tightly correlated with podocytes apoptosis of DN rats.12 Previous documents confirmed that TUG1 worked as an inhibiting factor by modulating proliferation, fibrosis and apoptosis in DN.23,24 Nevertheless, the precise biological function and mechanism of TUG1 are still unclear in DN. Furthermore, miR-27a-3p, a form of mature miR-27a, exerted the positive effect on podocytes injury through binding to FOXO1 in DN.25 Meanwhile, E2F3, a DP-binding protein, was reported to serve as the tumor suppressor role in pancreatic cancer and gastric cancer.26,27 However, it is unknown whether miR-27a-3p and E2F3 are involved in the TUG1-mediated DN.Thus, our study firstly investigated TUG1, miR-27a-3p and E2F3 expression levels in DN tissues and HG-stimulated podocytes. Data exhibited that TUG1 and E2F3 expression were apparently high expressed and miR-27a-3p was markedly lower expressed in DN tissues and HG-stimulated podocytes versus their corresponding counterparts. Then, the effects of TUG1 on apoptosis, fibrosis and inflammation in HG-treated podocytes were detected. The results suggested that TUG1 blocked apoptosis, fibrosis and inflammation in HG-induced podocytes. That is to say, TUG1 served as an inhibition factor in DN progress. Next, we explored the mechanisms basis of TUG1 in DN development. It has been acknowledged that lncRNAs can exert its function by interacting with miRNAs.28–30 Results of bioinformatics analysis and dual-luciferase reporter assay testified that miR-27a-3p was a potential target of TUG1. miR-27a-3p has been pointed out to be closely associated with HG-caused cell viability and apoptosis.29 Furthermore, our study displayed that miR-27a-3p expression was inversely correlated with TUG1 expression in DN tissues and podocytes treated with HG. Hence, we further probed whether the effects of TUG1 on apoptosis, fibrosis and inflammation of HG-triggered podocytes were mediated via modulating miR-27a-3p. Function analysis disclosed that TUG1 inhibited the apoptosis, fibrosis and inflammation and restoration of miR-27a-3p expression partially abolished suppressive effects of TUG1 on HG-stimulated podocytes. In this paper, we firstly verified that TUG1 worked as an inhibited factor in HG-treated podocytes by hindering miR-27a-3p expression.
So far, the ceRNA hypothesis point out lncRNAs can act as a molecular sponge of miRNAs to seclude miRNAs away from target mRNAs.31,32 Accordingly, we searched for the potential targets of miR-27a-3p, the bioinformatics prediction results manifested that miR-27a-3p possessed some binding sequences with E2F3, which was further confirmed by the luciferase reporter assay. E2F3 has been indicated to link with podocytes injury.21 In this study, we also found that E2F3 expression level was negatively correlated with miR-27a-3p expression level in the treatment of DN tissues and podocytes. Further rescue experiments in HG-triggered podocytes testified that miR-27a-3p-induced apoptosis, fibrosis and inflammation were partly abrogated following the E2F3 reintroduction, indicating that miR-27a-3p contributed to HG-induced apoptosis, fibrosis and inflammation by targeting E2F3.
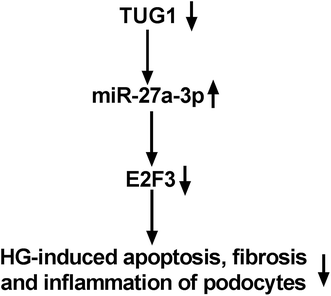
Finally, to further proved that TUG1 could work as a ceRNA of miR-27a-3p to impact the expression level of E2F3, the effect of TUG1 on E2F3 expression was detected. In this paper, we firstly verified that TUG1 expression level was positively related to E2F3 expression in HG-treated DN tissues. Then, western blot results demonstrated that exogenous expression of TUG1 improved E2F3 level in HG-provoked podocytes, while this effect was attenuated after co-transfection with miR-27a-3p (Fig. 8).
Conclusions
Taken together, this study reveals that TUG1 performed as the ceRNA of miR-27a-3p to upregulate E2F3 expression, thereby suppressing HG-induced inflammation, fibrosis and podocyte apoptosis in DN. Our findings elucidate a potential molecular basis of TUG1 in DN, promising an underlying therapeutic target for treatment of DN.Conflicts of interest
The authors declare that they have no financial conflicts of interest.Abbreviations
| DN | Diabetic nephropathy |
| lncRNAs | Long non-coding RNAs |
| miRNAs | MicroRNAs |
| ESRD | End-stage renal disease |
| TUG1 | Taurine upregulated gene 1 |
| lncRNAs | Long non-coding RNAs |
| E2F3 | E2F transcription factor 3 |
| miR-27a-3p | MicroRNA-27a-3p |
| EMC | Extracellular matrix |
| HG | High glucose |
| 3′UTR | 3′-Untranslated region |
| FBS | Fetal bovine serum |
| 293T | Human embryonic kidney 293 cell line |
| DMEM | Dulbecco's modified Eagle's medium |
| pcDNA | pcDNA3.1 empty vector |
| PVDF | Polyvinylidene fluoride |
| HRP | Horseradish peroxidase |
| ELISA | Enzyme-linked immunosorbent assay |
Acknowledgements
This work is supported by Hainan Provincial Natural Science Foundation of China, No. 819QN384.References
- K. Umanath and J. B. Lewis, Am. J. Kidney Dis., 2018, 71, 884–895 CrossRef PubMed.
- F. Conserva, P. Pontrelli, M. Accetturo and L. Gesualdo, J. Nephrol., 2013, 26, 811–820 CrossRef PubMed.
- B. Najafian, C. E. Alpers and A. B. Fogo, Contrib. Nephrol., 2011, 170, 36–47 CrossRef PubMed.
- W. H. Yiu, M. Lin and S. C. Tang, Kidney Int. Suppl., 2014, 4, 20–25 CrossRef CAS PubMed.
- K. Susztak, A. C. Raff, M. Schiffer and E. P. Bottinger, Diabetes, 2006, 55, 225–233 CrossRef CAS PubMed.
- R. Pichler, M. Afkarian, B. P. Dieter and K. R. Tuttle, Am. J. Physiol. Renal. Physiol., 2017, 312, F716–F731 CrossRef CAS PubMed.
- B. Liu, L. Qiang, G. D. Wang, Q. Duan and J. Liu, Eur. Rev. Med. Pharmacol. Sci., 2019, 23, 3478–3486 CAS.
- P. Zhang, Y. Sun, R. Peng, W. Chen, X. Fu, L. Zhang, H. Peng and Z. Zhang, Cell Death Dis., 2019, 10, 526 CrossRef PubMed.
- T. L. Young, T. Matsuda and C. L. Cepko, Curr. Biol., 2005, 15, 501–512 CrossRef CAS PubMed.
- J. Long, S. S. Badal, Z. Ye, Y. Wang, B. A. Ayanga, D. L. Galvan, N. H. Green, B. H. Chang, P. A. Overbeek and F. R. Danesh, J. Clin. Invest., 2016, 126, 4205–4218 CrossRef PubMed.
- L. J. Duan, M. Ding, L. J. Hou, Y. T. Cui, C. J. Li and D. M. Yu, Biochem. Biophys. Res. Commun., 2017, 484, 598–604 CrossRef CAS PubMed.
- X. Lei, L. Zhang, Z. Li and J. Ren, Drug Des., Dev. Ther., 2018, 12, 2785–2793 CrossRef CAS PubMed.
- D. P. Bartel, Cell, 2018, 173, 20–51 CrossRef CAS PubMed.
- Y. Zhang, S. Zhao, D. Wu, X. Liu, M. Shi, Y. Wang, F. Zhang, J. Ding and Y. Xiao, J. Diabetes Res., 2018, 2018, 4728645 Search PubMed.
- S. Z. Zhang, X. J. Qiu, S. S. Dong, L. N. Zhou, Y. Zhu, M. D. Wang and L. W. Jin, Eur. Rev. Med. Pharmacol. Sci., 2019, 23, 1248–1256 Search PubMed.
- Z. Zhou, J. Wan, X. Hou, J. Geng, X. Li and X. Bai, Cell Death Dis., 2017, 8, e2658 CrossRef CAS PubMed.
- B. De Felice, F. Manfellotto, C. Garbi, M. Santoriello and M. Nacca, Mol. Med. Rep., 2018, 17, 7081–7088 CAS.
- P. Chen, R. Wang, Q. Yue and M. Hao, Biochem. Biophys. Res. Commun., 2018, 503, 2956–2962 CrossRef CAS PubMed.
- N. Sharma, C. Timmers, P. Trikha, H. I. Saavedra, A. Obery and G. Leone, J. Biol. Chem., 2006, 281, 36124–36131 CrossRef CAS PubMed.
- M. Gan, J. Du, L. Shen, D. Yang, A. Jiang, Q. Li, Y. Jiang, G. Tang, M. Li, J. Wang, X. Li, S. Zhang and L. Zhu, In Vitro Cell. Dev. Biol.: Anim., 2018, 54, 304–310 CrossRef CAS PubMed.
- F. Zha, L. Bai, B. Tang, J. Li, Y. Wang, P. Zheng, T. Ji and S. Bai, J. Cell. Biochem., 2019, 120, 12574–12581 CrossRef CAS PubMed.
- J. Long and F. R. Danesh, Diabetes, 2018, 67, 552–553 CrossRef CAS PubMed.
- X. J. Zang, L. Li, X. Du, B. Yang and C. L. Mei, Eur. Rev. Med. Pharmacol. Sci., 2019, 23, 7519–7525 Search PubMed.
- H. Shen, Y. Ming, C. Xu, Y. Xu, S. Zhao and Q. Zhang, J. Cell. Physiol., 2019 DOI:10.1002/jcp.28153.
- X. Bai, J. Geng, X. Li, J. Liu, Z. Zhou and X. Liu, Antioxid. Redox Signaling, 2018, 29, 355–376 CrossRef CAS PubMed.
- J. Yang, H. F. Zhang and C. F. Qin, Eur. Rev. Med. Pharmacol. Sci., 2017, 21, 4050–4057 CAS.
- X. Li, H. Li, R. Zhang, J. Liu and J. Liu, Cell. Physiol. Biochem., 2015, 35, 2033–2042 CrossRef CAS PubMed.
- S. F. Sun, P. M. K. Tang, M. Feng, J. Xiao, X. R. Huang, P. Li, R. C. W. Ma and H. Y. Lan, Diabetes, 2018, 67, 731–744 CrossRef CAS PubMed.
- F. Zha, X. Qu, B. Tang, J. Li, Y. Wang, P. Zheng, T. Ji, C. Zhu and S. Bai, Aging, 2019, 11, 3716–3730 Search PubMed.
- J. Hu, H. Wu, D. Wang, Z. Yang and J. Dong, Biochimie, 2019, 157, 102–110 CrossRef CAS PubMed.
- J. H. Yoon, K. Abdelmohsen and M. Gorospe, Semin. Cell Dev. Biol., 2014, 34, 9–14 CrossRef CAS PubMed.
- H. S. Chiu, M. R. Martinez, M. Bansal, A. Subramanian, T. R. Golub, X. Yang, P. Sumazin and A. Califano, BMC Genomics, 2017, 18, 418 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2019 |