Open Access Article
Open Access ArticleA label-free RTP sensor based on aptamer/quantum dot nanocomposites for cytochrome c detection†
Dongxia
Li
 *,
Junping
Guo
,
Liang
Zhao
,
Guoxian
Zhang
and
Guiqin
Yan
*,
Junping
Guo
,
Liang
Zhao
,
Guoxian
Zhang
and
Guiqin
Yan
 *
*
Shanxi Normal University, Linfen 041004, P. R. China. E-mail: lidx_139@163.com; gqyan2013@163.com
First published on 8th October 2019
Abstract
Given the outstanding room-temperature phosphorescence (RTP) of Mn–ZnS quantum dots (QDs) and the specific recognition performance of the aptamer, we built phosphorescent composites from aptamers conjugated with polyethyleneimine quantum dots (PEI-QDs) and applied them to cytochrome c (Cyt c) detection. Specifically, QDs/CBA composites were generated from the electrostatic interaction between the positively-charged PEI-QDs and the negatively-charged Cyt c binding aptamer (CBA). With the presence of Cyt c, the Cyt c can specifically bind with the QDs/CBA composites, and quench the RTP of QDs through photoinduced electron-transfer (PIET). Thereby, an optical biosensor for Cyt c detection was built, which had a detection range of 0.166–9.96 μM and a detection limit of 0.084 μM. This aptamer-mediated phosphorescent sensor with high specificity and operational simplicity can effectively avoid the interference of scattering light from complex substrates. Our findings offer a new clue for building biosensors based on QDs and aptamers.
1. Introduction
Cytochrome c (Cyt c) is a hemachrome-containing metal micromolecular protein composed of 104 amino acids (∼12 kDa) and exists in the cytoplasm in between the inner membrane and outer membrane of mitochondria.1,2 Cyt c with high water solubility is an electron carrier that is a key part of the electron transport chains.3,4 Owing to its oxido-reduction ability, Cyt c is an important part of electron transport chains in living cells.5 After the permeability of mitochondrial membranes is disturbed, Cyt c acts as a precursor of apoptosis programmed cell death,6,7 so monitoring Cyt c may be a key factor for the discovery of the cell apoptosis process.8,9 Thus, detecting Cyt c as an appropriate marker can be considered in research on the participation of mitochondria in cell apoptosis. As reported, the serum Cyt c levels of patients can be adopted as a potential clinical marker for diagnosis of cell injury and death,7,10,11 and the release of Cyt c can also be used to screen anticancer drugs. So far, Cyt c analysis has attracted wide attention and been explored by various techniques, including fluorimetry,12–16 electrochemistry,17–21 enzyme-linked immunosorbent assay11,22,23 and flow cytometry.24 However, the detection process is restricted by operational complexity and low specificity, so it is urgent to develop convenient and sensitive Cyt c detection methods.Quantum dots (QDs) are a novel type of optical nanomaterials that possess high application prospects. Owing to their large Stokes shift, narrow emission spectrum and high luminescence efficiency, QDs have been commonly applied into DNA molecular hybridization25,26 and into detection of small molecules,27,28 proteins29,30 and enzymatic activity.31,32 Room-temperature phosphorescent (RTP) QDs possess longer lifetime and much rare phosphorescence and thus can avoid the interference from the background fluorescence of biological systems or from the autofluorescence and scattering light of biological samples.33–35 Moreover, the detection based on QDs and RTP does not need any deoxidizing agent or inductive agent and avoids any complex pretreatment. Thus, RTP QDs are more promising for the detection of biological samples or biomolecules.
To enhance the specificity and sensitivity between the QDs and the target and to broaden the application of QDs into biological detection, researchers have tried various small molecules, biomacromolecules, antibodies and macromolecular polymers to surface modify QDs, forming functional QDs.36 Also specific biomolecules that can be easily synthesized and stored are used to modify QDs, which is undoubtedly the most effective measure. Aptamer, a single-stranded oligonucleotide, which can specifically bind with and strictly identify the target molecules,37 can be used to replace antibodies and as a specific target identification biological sensor element.38–41 Aptamer, which can be easily synthesized and stably stored, has been highly valued in bioanalysis since it only requires simple modification before application.42 The aptamer-based electrochemistry has been used to successfully detect Cyt c.20,43,44 Despite the high sensitivity, this method requires multistep electrode pre-modification, but such covalent modification is usually time- and labor-intensive and is restricted by extreme conditions, which complicate the experiments.
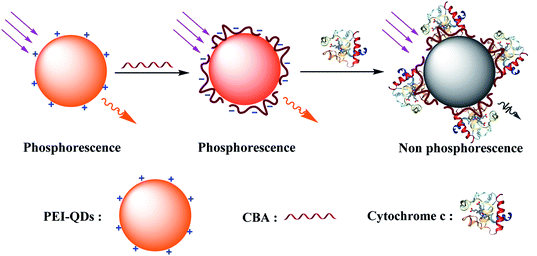
In this study, the nanocomposites from polyethyleneimine-capped Mn-doped ZnS QDs (PEI-QDs) and Cyt c binding aptamer (CBA) were prepared and used as Cyt c RTP sensors. The construction process is illustrated in Fig. 1. Specifically, QDs/CBA nanocomposites were formed on basis of the electrostatic interaction between the positively-charged PEI-QDs and the negatively-charged CBA, and this process did not need the fixation of CBA molecular covalence onto surface of QDs, but the electrostatic coupling further stabilized the QDs/CBA composites. After Cyt c was introduced into the system, the Cyt c binded with QDs/CBA to form a ternary compound, and the electron transfer between QDs and Cyt c quenched the RTP of QDs. Thus, the specific Cyt c detection can be realized by monitoring the phosphorescent signals of QDs. This sensor did not need any complex biological crosslinking and can be operated conveniently. Moreover, based on the RTP, the intervention from the self-fluorescence and scattering light of the biological systems can be significantly weakened, which modestly improved the sensitivity and practicability of this sensor.
2. Experimental
2.1 Materials and reagents
Polyethyleneimine (PEI, Aladdin Bio-Chem Technology Co. Ltd., China), Zn(Ac)2·2H2O, Mn(Ac)2·4H2O and Na2S·9H2O (Tianjin Kemiou Chemical Reagent Co. Ltd., China) were used to prepare PEI-capped Mn-doped ZnS QDs (PEI-QDs). Cyt c and IFN-γ were purchased from Shanghai Sangon Biotechnology Co. Ltd. (China). Human serum albumin (HSA) and bovine serum albumin (BSA) were purchased from Aladdin Bio-Chem Technology Co. Ltd. (China). Alkaline phosphatase (ALP) were bought from Sigma-Aldrich (USA). The 76-mer Cyt c binding aptamer (CBA) with the sequence of 5′-ATC GAT AAG CTT CCA GAG CCG TGT CTG GGG CCG ACC GGC GCA TTG GGT ACG TTG TTG CCG TAG AAT TCC TGC AGC C-3′ was synthesized by Shanghai Sangon Biotechnology Co. Ltd. (China). The human serum samples was purchased from Chongqing Manuik Technology Co. Ltd. (China). All other chemicals were of analytical grade, and the resistivity of water used in this study was higher than 18 MU cm.2.2 Instruments
The morphology and microstructures of QDs were characterized by a transmission electron microscope (JEM-2100FTEM, Japan). Phosphorescence spectra were measured by a Cary Eclipse fluorescence spectrophotometer (Varian Inc., CA, USA) equipped with a plotter unit and a quartz cell (1 × 1 cm2) in the phosphorescence mode. The crystalline structures of QDs were studied by a D/Max-2500 powder X-ray diffractometer (XRD, Rigaku, Japan) Ultraviolet/visible (UV-Vis) absorption spectra were collected using a UV-29100 UV-Vis spectrophotometer (Shimadzu, Japan). In addition, The zeta potential was measured by a ZS90 Zetasizer Nanoscale device and the dynamic light scattering (DLS) measurements were conducted on a Malvern Zetasizer Nano ZS90 DLS system (Malvern Instruments Ltd., Worcestershire, UK).2.3 Synthesis of PEI-capped Mn-doped ZnS QDs
The PEI-capped Mn–ZnS QDs were synthesized as reported.45,46 Firstly, to a three-necked bottle, 0.75 g of PEI and 20 mL of water were added; then 1 mmol of Zn(Ac)2 and 0.04 mmol of Mn(Ac)2 were added. The mixture was adjusted into pH 8 by adding 1 M HCl, magnetically stirred under argon gas protection and at room temperature, then heated to 80 °C for 30 min of reaction. After that, 5 mL of 0.2 M Na2S was added, followed by reaction at 80 °C for 1 h. The solution was cooled and adjusted by adding 1 M HCl to pH 8. The resulting QDs solution was aged at 50 °C and in air for 2 h, then QDs were precipitated in an equal volume of anhydrous ethanol, and centrifuged at high speed. After that, the precipitates were washed by ethanol three times. Then powder QDs were obtained after vacuum-drying at room temperature 24 h.2.4 Preparation of QDs/CBA nanocomposites
The QDs/CBA nanocomposites were prepared according to a reported method47 with some modification. Firstly, CBA (100 μM) was dissolved in a buffer solution (pH 7.4, 20 mM Tris–HCl, 0.1 M NaCl, 5 mM MgCl2), forming a 100 μM CBA solution, which was heated at 70 °C for 4 min and then cooled to room temperature. Then the PEI–Mn–ZnS QDs were dissolved in a Tris–HCl buffer solution (pH 7.4, 20 mM) and added with the CBA solution under stirring for 15 min. After ultrafiltration and several times of purification, the resulting solution was dissolved in a Tris–HCl buffer solution, forming a 1.0 mg mL−1 QDs/CBA solution (calculated as per the QDs concentration).2.5 Sensing detection of Cyt c
For RTP sensing of Cyt c with the QDs/CBA probe as-prepared, the QDs/CBA phosphorescent probe was used to detect Cyt c. Into a series of 10 mL colorimetric tubes, 500 μL of the Tris–HCl buffer solution (pH 7.4, 20 mM), 100 μL of the QDs/CBA solution, and different amounts of Cyt c were added. All tubes were diluted to 5 mL, uniformly shaken and incubated at room temperature for 30 min, followed by phosphorescence intensity detection (at excitation wavelength = 295 nm).2.6 Sample detection
To confirm the feasibility of the RTP sensor in reality, we selected human serum samples as the testing environment. To 50 μL of serum samples, different concentrations of Cyt c were added to form spiked samples, which were detected following the above procedures. The final Cyt c spiked concentrations were 0.83, 3.32 and 6.64 μM. Each experiment was conducted in triplicate. The samples were only diluted 100 times, without any other pretreatment.3. Results and discussion
3.1. Characterizations of PEI-capped Mn-doped ZnS QDs and QDs/CBA
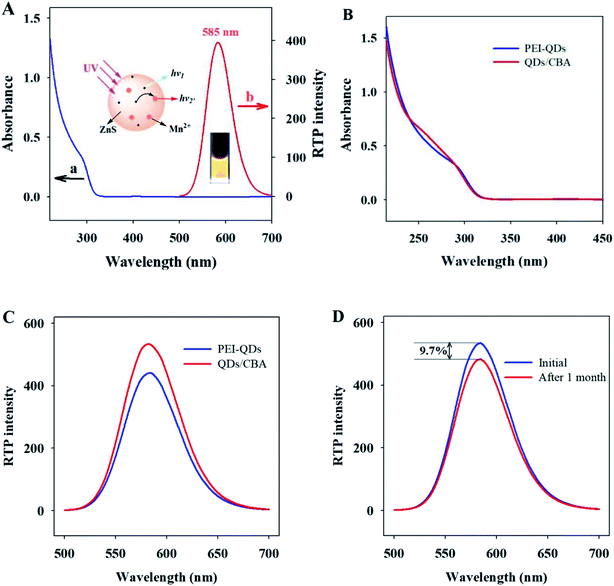
The shape and size of the PEI-capped Mn–ZnS QDs (PEI-QDs) were characterized by TEM. The PEI-QDs looked like uniform near-spherical particles (Fig. S1A†). XRD shows the QDs have three evident diffraction peaks, corresponding to the three crystal faces (111), (220) and (311) of cubic blende (Fig. S1B†), indicating the PEI-capped Mn–ZnS QDs as-synthesized exist as typical cubic crystals.48 The above results suggest the PEI-capped Mn–ZnS QDs were successfully synthesized.Further characterizations of optical properties of PEI-capped Mn–ZnS QDs are shown in Fig. 2A. The ultra-violet spectra (curve a) show these QDs have characteristic absorption peak at 295 nm and the maximum phosphorescence emission peak at 585 nm (curve b). This RTP of QDs is induced by the transition of electrons from the emission triplet state 4T1 to the 6A1 ground state. First, ZnS is excited, so the excited state electrons are captured by the cavity Mn2+, then electrons and holes form a compound on Mn2+, exciting Mn2+ and finally releasing energy in the form of RTP.
CBA, the aptamer of Cyt c, contains a 76-base oligonucleotide chain and due to the presence of phosphate groups, possesses abundant negative charge on surface, which can bind via electrostatic interaction to the surface of QDs, forming the QDs/CBA as-required. Ultraviolet analysis showed after QDs and CBA binded, the evident characteristic peak of DNA appeared at 260 nm, while the ultraviolet peak of QDs at 295 nm did not change significantly (Fig. 2B), indicating the binding between QDs and CBA did not affect the phosphorescent emission. The RTP emission peak of QDs/CBA at room temperature appeared at 585 nm (Fig. 2C), suggesting the relative position of emission spectrum did not change after the modification, but the RTP intensity of QDs/CBA was slightly enhanced, which may be because the modification with DNA decreased the surface defect status of QDs and thereby intensified the RTP. Moreover, after the new probe was stored in a refrigerator at 4 °C for 1 month, its phosphorescent intensity was only reduced by 9.7%, indicating this RTP probe has high optical stability (Fig. 2D).
3.2 Mechanism on the interactions between QDs/CBA and Cyt c
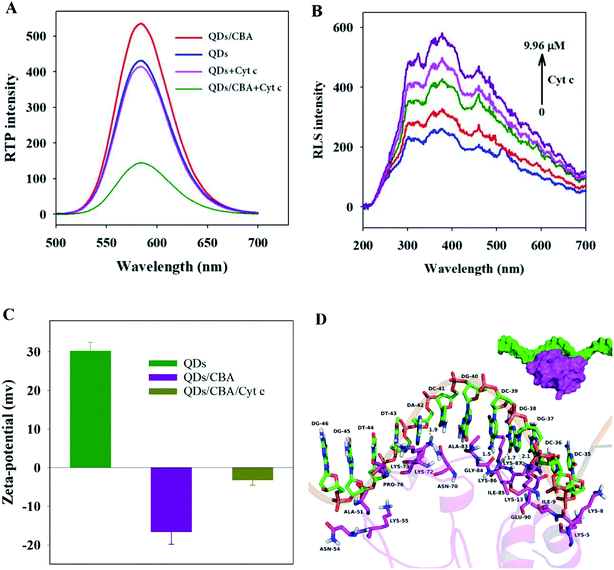
To validate the feasibility of the QDs/CBA sensor into Cyt c detection, we studied the phosphorescent emission spectra of QDs and QDs/CBA. Clearly, the addition of Cyt c did not significantly weaken the RTP intensity of QDs, but with the presence of CBA, the Cyt c can considerably quench the RTP (Fig. 3A). The CBA as a capture element in the system induced the specific binding of the QDs/CBA sensor with Cyt c, which led to photoinduced electron transfer. When the QDs were excited from the ground state, the Cyt c (III) captured electrons and was reduced into Cyt c (II), which blocked the radiative recombination between the excited electrons and the holes of QDs, thereby quenched the RTP. The corresponding photoinduced electron transfer process is as below.| Mn–ZnS + hν → Mn–ZnS* | (1) |
| Mn–ZnS* + Cyt c (III) → Mn–ZnS + Cyt c (II) | (2) |
Fig. 3B shows the RLS images of QDs/CBA and Cyt c. Clearly, within 200–700 nm, after the addition of Cyt c, the RLS intensity was enhanced with the rise of Cyt c concentration, indicating Cyt c bound to the surface of QDs/CBA to form larger scattering particles. To probe into the interaction between QDs/CBA and Cyt c, we detected the zeta potentials of PEI-QDs, QDs/CBA, and QDs/CBA/Cyt c. The zeta potentials of PEI-QDs and QDs/CBA at pH 7.4 were +30.2 and −16.6 respectively, and the zeta potential significantly decreased, which indicate the efficient static binding between PEI-QDs and CBA as well as the variation of apparent surface charge (Fig. 3C). The zeta potential rose from −16.6 to −5.2 after the addition of Cyt c, which was because the Cyt c molecules contained abundant lysine residues, which existed as cation and bound with QDs/CBA, leading to the rise of zeta potential in the system. DLS spectra (Fig. S2†) showed the particle sizes changed significantly, but after PEI-QDs bound with CBA, the particle sizes were enlarged from 10.1 ± 1.7 to 37.8 ± 9.6 nm, and after the addition of Cyt c, the particle sizes considerably rose to 91.3 nm. The above results suggest that QDs, CBA and Cyt c can form the CBA-bridged ternary composites.
Furthermore, molecular docking studies (refer to ESI† for the analysis method) illustrate the binding mode between the Cyt c and CBA from the molecular level. Fig. 3D have shown that, a hydrophobic interaction was observed between the residues Ala-51 and Pro-76 of the Cyt c and the nucleotides DT-44, DG-45 and DG-46 of the CBA. The residues Lys-73 and Lys-87 of the Cyt c formed cation–π interactions with the nucleotides DA-42 and DC-36 of the CBA, respectively, whereas the residue Lys-13 of the Cyt c formed cation–π interactions with the nucleotides DG-37 and DG-38 of the CBA. In addition, anion–π interactions were observed between the residue Glu-90 of the Cyt c and the nucleotides DC-36 and DG-37 of the CBA. Importantly, four hydrogen bond interactions were shown between the residue Lys-73 of the Cyt c and the DA-42 of the CBA (bond length: 1.9 Å), the Gly-84 of the Cyt c and the DC-39 of the CBA (bond length: 1.5 Å), the Lys-86 and Lys-13 of the Cyt c and the DG-38 of the CBA (bond length: 1.7 and 2.1 Å). All of these interactions induced Cyt c to form a stable compound with CBA.
3.3. Optimization of detection strategy
To further stabilize the system, we investigated the effects of pH, time and salinity on the interaction between QDs/CBA and Cyt c during the reactions. The interaction between CBA and proteins was associated with the incubation time. Firstly, the RTP intensity variation of the QDs/CBA or Cyt c at different incubation time was studied. Clearly, the RTP intensity (P0/P) abruptly rose within the first 30 min of incubation and then basically stabilized (Fig. S3A†). Thus, the subsequent tests of QDs/CBA and Cyt c were conducted after 30 min of incubation. Then the effects of pH on the QDs/CBA and Cyt c interaction were studied. The RTP intensity (P0/P) maximized at pH 7.4 (Fig. S3B†), so the pH of buffer solutions was set at 7.4 in the subsequent tests. As showed in Fig. S3C,† the ionic strength also affects the interaction between QDs/CBA and Cyt c. With the presence of 0–10 mM NaCl, the RTP intensity stabilized, but was weakened when NaCl concentration was above 10 mM, which was because the binding between PEI-QDs and CBA was mainly dependent on electrostatic interaction, so the high concentration of NaCl largely affected the system stability.3.4 Phosphorescence sensing of QDs/CBA for Cyt c
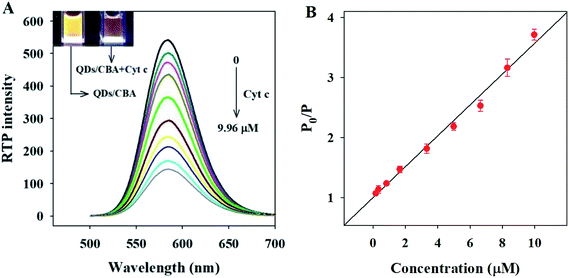
Fig. 4A shows the RTP intensity of the QDs/CBA nanocomposites changing with the Cyt c concentration. Clearly, as the dosage of Cyt c gradually rose, the RTP intensity of the QDs/CBA nanocomposites was regularly quenched. Due to the specificity between Cyt c and the aptamer, the addition of Cyt c led to the formation of an aptamer-bridged QDs/CBA/Cyt c ternary composite. After the QDs were excited, the electrons and holes were separated. Cyt c as an excellent electron acceptor can block the electron–hole recombination and thereby induce the electron transfer between QDs and Cyt c, leading to the quenching of RTP. The inset of Fig. 4A shows the digital images under ultraviolet light. Clearly, after the addition of Cyt c, the bright orange phosphorescence was quenched, indicating this system can be used as an RTP sensor for Cyt c detection.Based on the above results, we established a Cyt c detection sensor that utilized the RTP intensity of QDs as signals. Fig. 4B shows the RTP quenched value (P0/P) was linearly correlated with Cyt c concentration within a certain range, under the optimal conditions. This sensor had a linear detection range of 0.166–9.96 μM Cyt c, with a linear equation P0/P = 0.2572CCyt c+ 1.0035 (R = 0.995) and a detection limit of 0.084 μM. The analytical performances of this method were compared with other studies on Cyt c detection summarized in Table S1.† Clearly, compared with other label-free fluorescence methods,12–14,49 our new method is superior with higher sensitivity and the avoidance of any complicated molecular imprinting processing. Noticeably, though the linear detection range and detection limit of our method are similar to those of the dye molecular-labeled fluorescence method,50,51 our method did not need any label or signal magnification and avoided interference from the background biological fluorescence, and thus is more feasible for Cyt c detection in biological samples.
3.5 Specificity analysis
To study the identification selectivity of the QDs/CBA composites over Cyt c, we investigated how bovine serum albumin (BSA), human serum albumin, alkaline phosphatase, and interferon will affect the RTP of this sensor under the same conditions. BSA, human albumin, alpase alkaline phosphatase or interferon did not significantly affect the RTP of the QDs CBA composites (Fig. 5). In comparison, the RTP was significantly decreased with the presence of Cyt c. These results indicate the new sensor is highly selective for Cyt c detection.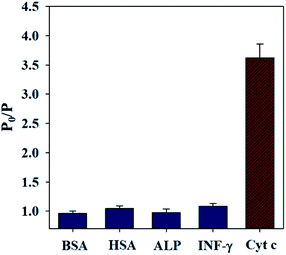 | ||
| Fig. 5 Effects of 1 × 10−6 mol L−1 BSA, 1 × 10−6 mol L−1 HAS, 50 U L−1 ALP, 1 × 10−6 mol L−1 IFN-γ and 9.96 μM Cyt c on the RTP intensity of QDs/CBA composites. | ||
3.6 Sample analysis
To confirm the practical detection ability of this new RTP sensor, we conducted spiked recovery experiments with human serum samples. The human serum samples, after being diluted by 100 times, were directly sent into Cyt c analysis. Each experiment was conducted in triplicate and did not require any complicated pretreatment. The spiked experiments (Table 1) show the average recovery rate is 93.1–102.9% under the optimal conditions, indicating this RTP sensor can be used to detect Cyt c in biological fluids.| Type of samples | Spiked (μM) | Found (μM) | RSD (%) | Recovery (%) |
|---|---|---|---|---|
| Human serum | 0.00 | Not detected | — | — |
| 0.83 | 0.85 | 5.1 | 102.9 | |
| 3.32 | 3.09 | 2.8 | 93.1 | |
| 6.64 | 6.39 | 4.3 | 96.2 |
4. Conclusions
The composites of QDs/CBA were prepared from PEI-QDs and Cyt c binding aptamer, and used as a Cyt c RTP sensor. While in the presence of Cyt c, the QDs/CBA composites could bind specifically with Cyt c to form the ternary composites of QDs/CBA/Cyt c, and quenching RTP of the QDs was observed. This sensor had a detection range of 0.166–9.96 μM and a detection limit of 0.084 μM. It was simple, easy to operate, and the QDs-CBA composites had highly selective response to Cyt c over other proteins. In addition, the method was also successfully applied to Cyt c detection in biological samples of serum with good recovery. The dependence on the RTP of QDs effectively avoided the interference from the autofluorescence and scattering light of biological fluids, so contributed to the Cyt c detection in complex biological systems.Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grants 31700862 and 31700876), the Fund for Construction Program of Chemical Advantage and Key discipline of Shanxi Province of China (Grant 912019).References
- A. M. Davies, J. G. Guillemette, M. Smith, C. Greenwood, A. G. P. Thurgood, A. G. Mauk and G. R. Moore, Biochemistry, 1993, 32, 5431–5435 CrossRef CAS.
- H. Hayashi and R. A. Capaldi, Biochim. Biophys. Acta, Biomembr., 1972, 282, 166–173 CrossRef CAS.
- S. Amani and A. Naeem, Amino Acids, 2014, 46, 1839–1851 CrossRef CAS.
- Y. Kushnareva, A. N. Murphy and A. Andreyev, Biochem. J., 2002, 368, 545–553 CrossRef CAS.
- Y. Hatefi, Annu. Rev. Biochem., 1985, 54, 1015–1069 CrossRef CAS.
- G. C. Brown and V. Borutaite, Biochim. Biophys. Acta, Bioenerg., 2008, 1777, 877–881 CrossRef CAS.
- J. C. Goldstein, C. Muñoz-Pinedo, J. E. Ricci, S. R. Adams, A. Kelekar, M. Schuler, R. Y. Tsien and D. R. Green, Cell Death Differ., 2005, 12, 453 CrossRef CAS.
- J.-C. Martinou, S. Desagher and B. Antonsson, Nat. Cell Biol., 2000, 2, E41 CrossRef CAS.
- C. B. L. Campos, B. A. Paim, R. G. Cosso, R. F. Castilho, H. Rottenberg and A. E. Vercesi, Cytometry, Part A, 2006, 69, 515–523 CrossRef.
- K. Barczyk, M. Kreuter, J. Pryjma, E. P. Booy, S. Maddika, S. Ghavami, W. E. Berdel, J. Roth and M. Los, Int. J. Cancer, 2005, 116, 167–173 CrossRef CAS.
- C. Y. Kadam and S. A. Abhang, Clin. Chim. Acta, 2015, 438, 98–102 CrossRef CAS.
- Y.-J. Yan, X.-W. He, W.-Y. Li and Y.-K. Zhang, Biosens. Bioelectron., 2017, 91, 253–261 CrossRef CAS.
- T. Guo, Q. Deng, G. Fang, C. Liu, X. Huang and S. Wang, Biosens. Bioelectron., 2015, 74, 498–503 CrossRef CAS.
- W. Zhang, X.-W. He, Y. Chen, W.-Y. Li and Y.-K. Zhang, Biosens. Bioelectron., 2011, 26, 2553–2558 CrossRef CAS PubMed.
- M. Cao, C. Cao, M. Liu, P. Wang and C. Zhu, Microchim. Acta, 2009, 165, 341–346 CrossRef CAS.
- M. Shamsipur, F. Molaabasi, S. Hosseinkhani and F. Rahmati, Anal. Chem., 2016, 88, 2188–2197 CrossRef CAS.
- G.-X. Wang, Y. Qian, X.-X. Cao and X.-H. Xia, Electrochem. Commun., 2012, 20, 1–3 CrossRef CAS.
- T. Wang, S. Zhang, C. Mao, J. Song, H. Niu, B. Jin and Y. Tian, Biosens. Bioelectron., 2012, 31, 369–375 CrossRef CAS.
- X.-W. Hu, C.-J. Mao, J.-M. Song, H.-L. Niu, S.-Y. Zhang and H.-p. Huang, Biosens. Bioelectron., 2013, 41, 372–378 CrossRef CAS.
- X. L. Zhang, Z. R. Tang, Y. P. Dong and C. M. Wang, Talanta, 2018, 179, 139–144 CrossRef CAS.
- M. R. Karimi Pur, M. Hosseini, F. Faridbod, M. R. Ganjali and S. Hosseinkhani, Sens. Actuators, B, 2018, 257, 87–95 CrossRef CAS.
- A. Langs-Barlow, S. Selvaraj, O. Ogbuagu, V. Shabanova, E. D. Shapiro and E. Paintsil, Mitochondrion, 2015, 20, 71–74 CrossRef CAS.
- J. Javid, R. Mir, P. K. Julka, P. C. Ray and A. Saxena, Tumor Biol., 2015, 36, 4253–4260 CrossRef CAS.
- M. A. King, A. Eddaoudi and D. C. Davies, Cytometry, Part A, 2007, 71, 668–674 CrossRef.
- Y. Miao, J. Lv and G. Yan, Biosens. Bioelectron., 2017, 94, 263–270 CrossRef CAS PubMed.
- J. Lv, Y. Miao, J. Yang, J. Qin, D. Li and G. Yan, Biosens. Bioelectron., 2017, 91, 560–565 CrossRef CAS.
- J. Yang, Z.-Z. Lin, H.-P. Zhong, X.-M. Chen and Z.-Y. Huang, Sens. Actuators, B, 2017, 252, 561–567 CrossRef CAS.
- S. Luo, Y. Miao, J. Guo, X. Sun and G. Yan, Microchim. Acta, 2019, 186, 249 CrossRef.
- F. f. Liu, Y. Yu, B. x. Lin, X. g. Hu, Y. j. Cao and J. z. Wu, Spectrochim. Acta, Part A, 2014, 131, 9–16 CrossRef CAS.
- P. Yan, X. Li, Y. Dong, B. Li and Y. Wu, Analyst, 2019, 144, 2891–2897 RSC.
- Z. Hu, J. Chen, Y. Li, Y. Wang, Q. Zhang, E. Hussain, M. Yang, S. A. Shahzad, D. Yu and C. Yu, Talanta, 2017, 169, 64–69 CrossRef CAS.
- D. Li, J. Qin and G. Yan, Sens. Actuators, B, 2018, 255, 529–535 CrossRef CAS.
- Y. Gong and Z. Fan, J. Lumin., 2015, 160, 299–304 CrossRef CAS.
- J. Chen, Y. Zhu and Y. Zhang, Spectrochim. Acta, Part A, 2016, 164, 98–102 CrossRef CAS.
- J. Qin, X. Sun, D. Li and G. Yan, RSC Adv., 2019, 9, 12747–12754 RSC.
- J. Zhou, Y. Liu, J. Tang and W. Tang, Mater. Today, 2017, 20, 360–376 CrossRef CAS.
- Y. Lu, X. Li, L. Zhang, P. Yu, L. Su and L. Mao, Anal. Chem., 2008, 80, 1883–1890 CrossRef CAS.
- S. Zhang, J. Xia and X. Li, Anal. Chem., 2008, 80, 8382–8388 CrossRef CAS.
- J. Yang, Z. Zhang and G. Yan, Sens. Actuators, B, 2018, 255, 2339–2346 CrossRef CAS.
- H. Jin, R. Gui, J. Gong and W. Huang, Biosens. Bioelectron., 2017, 92, 378–384 CrossRef CAS.
- P. Jolly, P. Damborsky, N. Madaboosi, R. R. G. Soares, V. Chu, J. P. Conde, J. Katrlik and P. Estrela, Biosens. Bioelectron., 2016, 79, 313–319 CrossRef CAS.
- F. Ding, Y. Gao and X. He, Bioorg. Med. Chem. Lett., 2017, 27, 4256–4269 CrossRef CAS.
- N. Bin, W. Li, X. Yin, X. Huang and Q. Cai, Talanta, 2016, 160, 570–576 CrossRef CAS.
- Y.-C. Liu, S.-Q. Cui and Z.-S. Yang, Anal. Sci., 2006, 22, 1071–1074 CrossRef CAS.
- H. Yan and H.-F. Wang, Anal. Chem., 2011, 83, 8589–8595 CrossRef CAS PubMed.
- M. Shao and H.-F. Wang, Analyst, 2013, 138, 4618–4623 RSC.
- S. Li, Z. Gao and N. Shao, Talanta, 2014, 129, 86–92 CrossRef CAS.
- J. Zhuang, X. Zhang, G. Wang, D. Li, W. Yang and T. Li, J. Mater. Chem., 2003, 13, 1853–1857 RSC.
- R. M. Amin, S. A. Elfeky, T. Verwanger and B. Krammer, Biosens. Bioelectron., 2017, 98, 415–420 CrossRef CAS.
- T.-T. Chen, X. Tian, C.-L. Liu, J. Ge, X. Chu and Y. Li, J. Am. Chem. Soc., 2015, 137, 982–989 CrossRef CAS.
- J. Tang, C. Huang, J. Shu, J. Zheng, D. Ma, J. Li and R. Yang, Anal. Chem., 2018, 90, 5865–5872 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra05761g |
| This journal is © The Royal Society of Chemistry 2019 |