Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nitrogen donor ligand for capping ZnS quantum dots: a quantum chemical and toxicological insight
Vivek Pandey†
 *a,
Vinay Kumar Tripathi†b,
Keshav Kumar Singha,
Tejasvi Bhatiac,
Nitesh Kumar Upadhyayc,
Bela Goyald,
Gajanan Pandeye,
Inho Hwangb and
Poonam Tandon
*a,
Vinay Kumar Tripathi†b,
Keshav Kumar Singha,
Tejasvi Bhatiac,
Nitesh Kumar Upadhyayc,
Bela Goyald,
Gajanan Pandeye,
Inho Hwangb and
Poonam Tandon *a
*a
aDepartment of Physics, University of Lucknow, Lucknow-226007, India
bDepartment of Animal Science and Biotechnology, Chonbuk National University, Jeonju, 561-756, Republic of Korea
cVivacious – Centre for Nanoscience and Education Research, Lucknow-226020, India
dAll India Institute of Medical Sciences, Rishikesh-249201, India
eDepartment of Applied Chemistry, Babasaheb Bhimrao Ambedkar University, Lucknow-226025, India
First published on 11th September 2019
Abstract
Nanoparticles having strong optical and electronic properties are the most widely used materials in sensor development. Since the target analyte interacts directly with the surface of the material, the choice of ligand for functionalizing the surface of the material is the key for its further applications. The functionalized surface of the material makes it suitable for required applications as it controls the size of the particle during its growth from the solution phase. Biomolecule capped nanomaterials are favourable for various applications in bio-sensing. In the present work, an attempt has been made to explore the biologically active molecule imidazole as capping agent for ZnS semiconductor nanoparticles or quantum dots (QDs). This work explores the possibility of replacing conventional thiol-zinc bonding and hence paves new pathways for biomolecules having the possibility of being efficient capping agents. Computational chemistry has been used to study the mechanism of bonding between one of the nitrogen atoms of imidazole and the zinc ion of the ZnS QDs. The quantum chemical insight not only explores the most spontaneous interaction of zinc ion and imidazole molecule so as to act as an efficient capping agent but also explains the probable bonding site for nitrogen–zinc chemistry. The tailormade Mn doped ZnS QDs are one of the most promising materials for probe and sensor development. The ZnS core having non-toxicity and the emission in longer wavelength due to manganese makes this material highly useful biologically. The aqueous route of synthesis has been employed to obtain a highly homogeneous and pure material which was further characterized by UV (Ultra Violet spectroscopy), Spectrofluorometer, Transmission Electron Microscope and X-ray Diffraction. The toxicity at the cellular and genetic levels was also investigated to prove the potential of the imidazole capped Mn doped ZnS QD as a biocompatible material.
Introduction
The evolution of functionalized materials in smaller dimensions has revolutionized the nanotechnology world with their unique properties. Among various nanomaterials, quantum dots (QDs), due to their small size, ease of functionalization, stability and tunable optical properties, are key materials that have been extensively studied for imaging and sensing applications.1–3 The core material of QDs very commonly comprises of oxides or sulphides of zinc, cadmium and mercury belonging to group II of the transition metals.4 For the application of nanoparticles in biological fields they have to pass through the toxicity tests.5 Hence toxicity assessment is a prerequisite for the application of QDs in living cells. The toxicity of the QDs depends on various parameters like core material, capping agent and synthetic route. Cadmium based QDs have shown toxicity at all levels due to leakage of the metal ion in the matrix.6 Although the emergence of zinc sulphide/cadmium sulphide core–shell QDs solved many of the problems of core containing material but there is still a need for some more advancement in the development of biocompatible nanoparticles.7 Thus the use of biomolecules as surfactants will help in extending the research of obtaining non-toxic and highly applicable QDs from the zinc sulphide core.8 Further, in order to enhance the optical properties of the ZnS QDs, doping of the ZnS lattice with manganese has been routinely done which gives longer wavelength emission spectra in the orange region of the electromagnetic spectrum and the material is also non-toxic in nature.9 Thus ZnS core doped with manganese and capped with a suitable molecule QDs have paved the pathway to using biologically active materials in nanotechnology research.10Various routes have been investigated to synthesize small sized, homogeneous QDs but among all existing methods, aqueous synthesis of the particles involving low temperature and greener methodologies has various advantages over the conventional ones.10 The recent advances in the synthetic chemistry of ZnS QDs involving surfactants or capping agents for controlling the growth is expected to lead to the existence of newly generated advanced nanomaterials with highly improved quality.11
Developing new capping agents that can reduce the toxic effects, improve the optical properties and also control the growth of quantum dots is a highly challenging task.12 Capping agents are very significant as they are the only part of the nanomaterial that interacts with the surface to the target molecule and helps in sensing the analyte with high selectivity and sensitivity.13 The metal ion of the nanomaterial acts like a Lewis acid and hence the target ion acting like Lewis base interacts with the metal ion forming a strong covalent bond. The functional group of the capping agent gives the whole species an ability to get functionalized for water solubility and biocompatibility.14 Conventionally, thiol containing capping agents are used to attach with the metal ion, but this leaves us with limited molecules to be used for controlling the Ostwald growth.15 Also sensing of metal ions becomes difficult if thiol group is blocked upon attaching the capping agent to the core metal of the host crystal lattice. There are various capping agents that have been explored, like mercaptophenylboronic,16 thioglycerol, L-cysteine,10 MPA (mercaptopropionic acid), MPTS (mercapto propyl trimethoxysilane),13 N-acetyl L-cysteine, glutathione,17 thioglycolic acid, cysteamine, etc. QDs based bio-conjugates are also developed for detection at high sensitivity.18 The physical properties of QDs make it a very strong material in electroluminescence sensing and other sensing mechanisms that involve optoelectronic properties.19–23 Recent years have also explored core–shell QDs capped with polymers or shells for sensing macrobiomolecules.24 Mostly the conventional capping agents have mercapto as donor site for attachment to the metal. Hence it is necessary to exploit other ligands, like nitrogen, to develop capping agents having nitrogen as donor atom.25 Imidazole molecule having two nitrogen atoms in the ring can serve as a ligand to donate its electron pair or charge to the zinc atom.26
Herein we have reported for the first time the imidazole capped QDs along with its toxicity profiling. The imidazole containing molecules are a major source of interest for chemists as well as for biologists since many compounds with imidazole or imidazole derivatives are used for pharmaceutical applications.27 The imidazole molecule is a planar 5-membered ring with 3C and 2N atom in conjugation. It is classified as a diazole heterocyclic aromatic molecule that is soluble in water and other polar solvents. Many natural products, especially alkaloids and other biological molecules (histidine) are having the imidazole ring. Imidazole group of drugs are commonly used as antifungal drugs. Nitrogen atom of the imidazole ring binds to the fungal cytochrome P450 enzyme and inhibits the conversion of lanosterol to ergosterol by inhibiting lanosterol 14-α-demethylase, thereby disrupting fungal cell membrane. Imidazoles are broad spectrum antifungals especially effective against Candida species. Topical antifungals like clotrimazole, miconazole etc. are effective against oral and vulvovaginal candidiasis. Ketoconazole is particularly used against systemic fungal infections. A recent study has demonstrated the utility of controlled release nanoparticle linked clotrimazole for vaginal candidiasis.28 Synthetic imidazole have many potential pharmacological uses.29 Imidazoles due to diverse synthetic properties with varying physical and chemical properties are very exciting surfactant that can be used with living cells.30
Zinc has a tendency to form bond with nitrogen atom of the imidazole ligand due to the basic character of imidazole and acid character of zinc ion enabling complex formation.31 The interaction of nitrogen of histidine and zinc was well established and have been used in sensing dopamine by Ravi Arunan et al.38 This feature of zinc was employed to synthesize and explore Mn:ZnS QDs capped with imidazole.
In the present work, for the first time an attempt was made to obtain highly efficient optical probe with low toxicity. The preference of the capping agent is based on the adaptability of imidazole, its water solubility and strong connectivity with biological world. As of our acquaintance, it is the foremost report of the synthesis and toxicological scrutiny of imidazole Mn:ZnS capped quantum dot. We have also performed computational studies to understand the mechanism of bonding between nitrogen atom of imidazole and zinc ion of the ZnS QDs.32–37 The synthesized QDs capped with imidazole ring containing nitrogen as heteroatom will have potential to be used in bio-mimicking where the enzymes like carbonic anhydrase can be mimicked by using zinc as the metal ion and imidazole as ligand.
Materials and methods
Computational details
The quantum chemical calculations to study the interaction of imidazole with ZnS have been done via the density functional theory (DFT). Structure of imidazole and Zn2+ (generate from ZnS in aqueous medium) and products obtained after reactions have been fully optimized to minimum energy at B3LYP/6-311++G (d, p) theoretical model. The functional B3LYP consist of Becke 88 exchange functional and correlation functional of Lee, Yang and Parr.32,33 This is the most widely used functional and is able to reproduce experimental results very accurately. The basis set employed here is adequate for the elements which are present in the current reaction system. To ensure that all the structures are minima, analytical regularity calculation has been done at the same level of theory. All the energy minima have been found to have only positive frequencies. Same set of analytical frequencies has been used to calculate zero point vibrational energy corrections. Further, electronic energies of all the structures has been corrected by performing calculations at single point with CCSD (T)/6-311++G (d, p) level of theory.34,35 No transition state (TS) has been observed in this work. The barrier less nature of the reactions has been confirmed by performing constraint optimizations along proposed reaction coordinate. Gaussian 09 was used to perform all calculations of electronic structure.36Analytical grade reagents
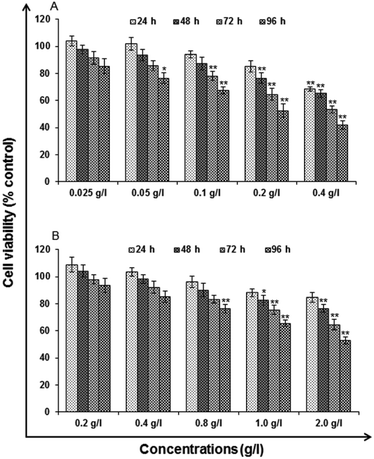
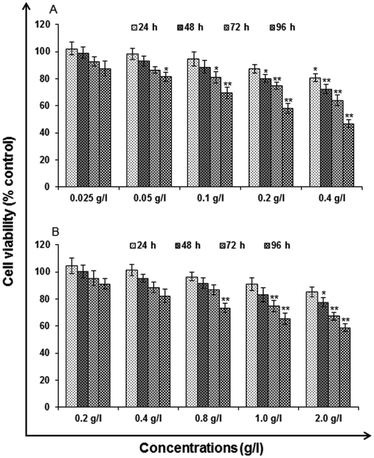
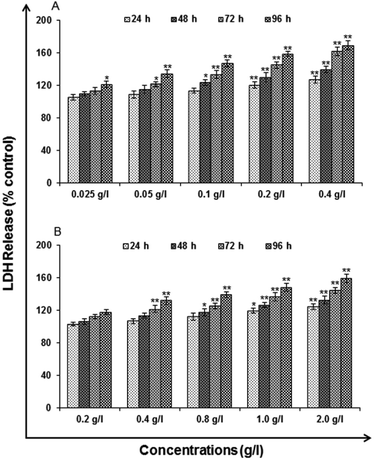
Analytical grade reagents and chemicals were used in the experiments unless otherwise stated. Milli Q water (water deionized and double distilled) was used in every part of the experiments. Zinc sulphate heptahydrate (ZnSO4·7H2O), manganese(II) chloride tetrahydrate (MnCl2·4H2O), L-cysteine (97% purity), imidazole and ethanol be procured from Sigma Aldrich. Sodium sulphide (Na2S·XH2O) was procured from SD Fine Chemicals (Mumbai). Milli-Q water was used to prepare the stock solutions and stored at 4 °C until their usage. All the particular biological assay chemicals and reagents be purchased from Sigma (Sigma St. Louis, MO, USA) unless otherwise stated. Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F-12) was used as culture medium, antibiotics, fetal bovine serum (FBS) and trypsin–EDTA were procured from Gibco BRL, USA. The plastic wares, Culture wares and other material used in the study were purchased commercially from Nunc, Denmark.MTT assay. Following the protocol of Tripathi V. K. et al. 2014, Cytotoxicity assessment was done using standard endpoint i.e., tetrazolium bromide MTT assay. In concise, A549 cells (1 × 104 cells per well) were incubated in the CO2 incubator for 24 h at 37 °C and seeded in 96-well tissue culture plates. Cells were then exposed to medium under high humid conditions containing uncapped QDs (0.025, 0.05, 0.1, 0.2 and 0.4 g l−1) and capped QDs (0.2, 0.4, 0.8, 1.0 and 2.0 g l−1) for 24–96 h at 37 °C in 5% CO2-95% atmosphere. Prior to the completion of respective incubation periods, tetrazolium salt (10 μl per well; 5 mg ml−1 of stock in phosphate-buffered saline [PBS]) was added 4 h. At the completion of the incubation period, 200 μl of culture grade dimethyl sulfoxide was added to each well. The content was mixed well until dissolved completely. Using multi-well microplate reader (Synergy HT; Bio-Tek, USA) plates were then incubated for 10 min at room temperature and color was read at 550 nm. To maintain basal control the unexposed sets were also run parallel under identical conditions.
Lactate dehydrogenase release assay
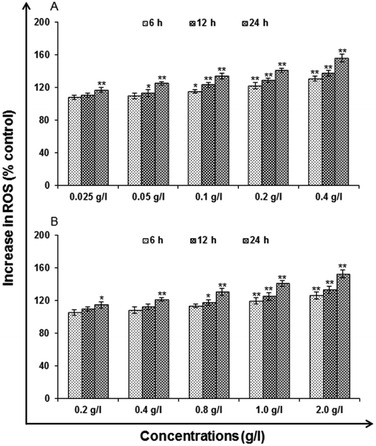
LDH release assays a reliable, nonradiative technique for analysis of cytotoxic lytic activity. LDH release assay is a method to measure the membrane integrity as a function of the amount of cytoplasmic LDH released into the medium. Using ready-made commercially available LDH assay kit for in vitro cytotoxicity evaluation (TOX-7, Sigma St. Louis, MO, USA) the assay was carried out. By the action of LDH, reduction of NAD take place in the method. In the stoichiometric conversion of a tetrazolium dye the reduced NAD (NADH+) was utilized. Using the multi-well plate reader the colored compound was measured at wavelengths 490 and 690 nm. The cells were processed for LDH release assay similar to MTT assay as the cells were exposed to 0.025, 0.05, 0.1, 0.2 and 0.4 g l−1 of uncapped QDs and 0.2, 0.4, 0.8, 1.0 and 2.0 g l−1 capped QDs for different time periods after the completion of the respective time periods. As per the experimental schedule culture plates were removed from CO2 incubator and centrifuged at 250×g for 4 min. Then supernatant of each well was transferred to a flat bottom 96 well culture plate and processed further for enzymatic analysis as per the instructions.Reactive oxygen species (ROS) generation. ROS are highly reactive molecules to oxidize cell. In this analysis, according to method of Kashyap M. P. et al. 2011, uncapped and capped QDs induced ROS generation was assessed in A549 cells using 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA, Sigma Aldrich) dye as fluorescence agent. Through fluorometric analysis ROS generation was studied.
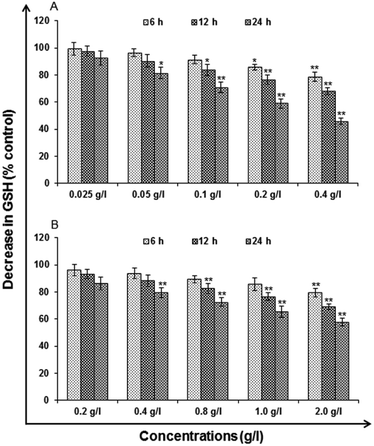
Glutathione (GSH) levels. Glutathione (GSH) levels deficiency put the cell at the risk for oxidative damage so this method were assessed using commercially available kit (ApoGSH Glutathione Colorimetric Assay Kit, Catalog No. #K261-100, Biovision, USA). In brief, using multi-well microplate Reader (Synergy HT, Bio-Tek, USA) the deproteinated samples were transferred to 96 well plates (50 μl per well), mixed with 150 μl freshly prepared assay cocktail and read at 405 nm at 5 min intervals up to 30 min. To calculate the experimental values standard curve was plotted using the glutathione standard supplied in the kit.
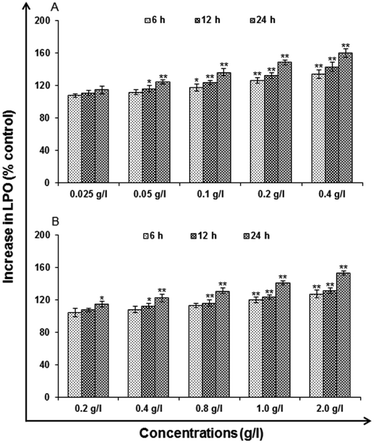
Lipid peroxidation (LPO). LPO is the oxidative degradation of lipids resulting in cell damage. For analysis, commercially available kit (Lipid Peroxidation Assay Kit, catalog no. 705002; Cayman Chemicals, USA) as per the manufacturer's protocol used for the exposure of uncapped QDs (0.025, 0.05, 0.1, 0.2 and 0.4 g l−1) and (0.2, 0.4, 0.8, 1.0 and 2.0 g l−1) capped QDs for 6, 12 and 24 h. The cells were harvested by centrifugation at 1000 rpm for 10 min and processed for the estimation of lipid peroxidation.
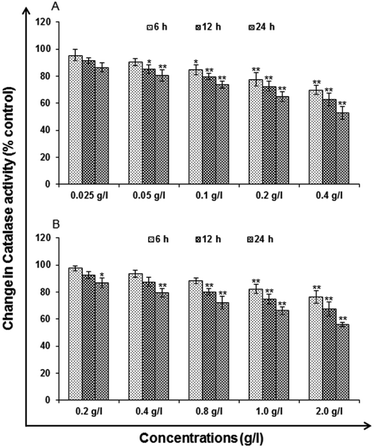
Catalase (CAT) levels. Catalase is an enzyme, protein that catalyzes chemical reactions and protect cell from oxidative damage by ROS. Activity was similar to the LPO for catalase analysis. Activity was measured using commercially available kit for catalase activity (Catalog no. 707002; Cayman Chemicals, USA).
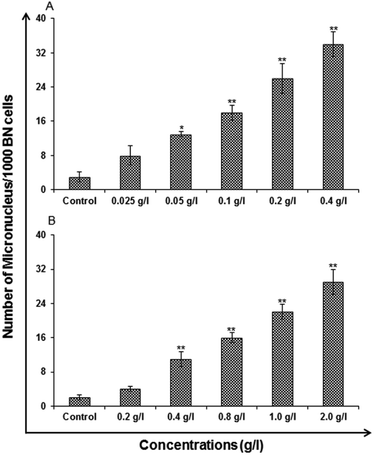
Micronucleus (MN) assay. In vitro micronucleus assay was carried out using standard protocols Kashyap M. P. et al. 2010 for toxicological screening for genotoxic compounds. The micronucleus (MNs) analysis, was performed by 1 × 105 A549-cells which were seeded in the each well of a six well tissue culture plates (Nunc.) and was further allowed to adhere for 24 h in CO2 incubator at 37 °C. The exposure given to the cells were of varied concentration for 24 h. The uncapped QDs were exposed to concentration 0.025, 0.05, 0.1, 0.2 and 0.4 (g l−1) and capped were exposed to 0.2, 0.4, 0.8, 1.0 and 2.0 (g l−1). Washing and supplementing of the cells was done with cytochalasin B (3 μg ml−1, Sigma) containing medium. Incubation time was kept as 24 h. A hypotonic buffer (0.075 M KCl) for 5–10 min at 37 °C was used for harvesting and Carnoy's fixative (methanol/acetic acid, 3
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was used for fixation of the cells. To get good imaging cells were dropped in a proper orientation onto the slides and stained with 5% Giemsa in phosphate buffer for which pH: 6.8 was stabilized for 15–20 min. The mounting with DPX was finally done for microscopic examination. Using a Nikon Eclipse 80i upright microscope attached to a Nikon digital CCD cool camera (Model DS-Ri1 of 12.7 megapixel) a minimum of 1000 binucleated cells with well-defined cytoplasm in each slide was scored for the presence of micronucleus. Mean of threeslides was evaluated for presenting the data.
1) was used for fixation of the cells. To get good imaging cells were dropped in a proper orientation onto the slides and stained with 5% Giemsa in phosphate buffer for which pH: 6.8 was stabilized for 15–20 min. The mounting with DPX was finally done for microscopic examination. Using a Nikon Eclipse 80i upright microscope attached to a Nikon digital CCD cool camera (Model DS-Ri1 of 12.7 megapixel) a minimum of 1000 binucleated cells with well-defined cytoplasm in each slide was scored for the presence of micronucleus. Mean of threeslides was evaluated for presenting the data.
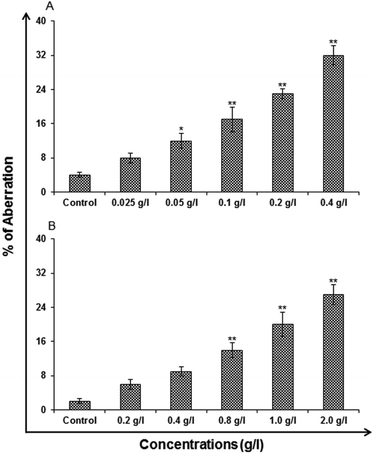
Chromosomal aberration (CA) assay. The CA assay was carried out using standard protocols Kapoor E et al. 2014 to identify the toxicological screening of genotoxicity. The cells that were cultured for the assay was A549 cells. The cells were exposed to different concentration of uncapped and capped QDs for 24 h. To harvest the cells, colcemid (0.15 μg ml−1 final concentration) was added and then kept for 4 h. The cells were washed with Hank's balanced salt solution. Later the cells were given a hypotonic shock in potassium citrate (0.8%) for 30 min. Cold fixative (methanol
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetic acid, 5
acetic acid, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratios) was used for fixing the cells. Cells were stained in 5% Gurr's Giemsa on a clear slide. Using a Nikon Eclipse 80i upright microscope attached to a Nikon digital CCD cool camera (Model DS-Ri1 of 12.7 Megapixel) the cells were scored for the presence of CA. Mean of three slides was taken for presenting the final data.
2 ratios) was used for fixing the cells. Cells were stained in 5% Gurr's Giemsa on a clear slide. Using a Nikon Eclipse 80i upright microscope attached to a Nikon digital CCD cool camera (Model DS-Ri1 of 12.7 Megapixel) the cells were scored for the presence of CA. Mean of three slides was taken for presenting the final data.
Statistical analysis. For statistical analysis at least three experiments were conducted and the data was expressed as mean and standard error of means (mean ± SE). Thus,*P < 0.05 was taken to indicate significant differences. Softwares like ANOVA which was also followed by post hoc Dunnett's test was employed to detect differences between the groups of treated and control.
Results and discussions
Experimental work
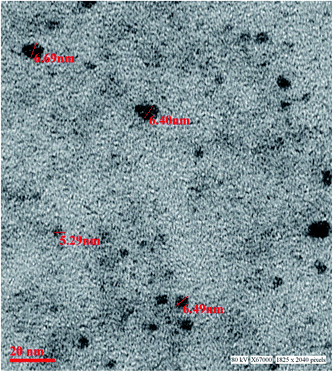
The aqueous route of synthesis was involved to obtain the luminescent manganese doped zinc sulphide QDs. The precursors used in the study was zinc sulphate salt, manganese salt while sodium sulphide was used to provide sulphide ions for the Mn doped ZnS nanomaterial. The imidazole molecule has two nitrogen atom one having N–H bond and other having lone pair. The synthesis was performed in water and further change to pH was done with NaOH. The base attacks on the H atom attached to the nitrogen atom and this result in the formation of nitrogen metal bond by the influence of negative charge on the first nitrogen atom. The deprotonation of imidazole was done to obtain highly basic anion. The capping agent was added in appropriate amount to reduce the formation of only metal ligand complex. The imidazole bonded zinc was further reacted with sulphide ion. There is always a competition between capping agent and anion hence controlling this aspect is crucial. To overcome this, fast addition of sulphide ion was done to form zinc sulphide precipitate immediately. After addition of sulphide ion the temperature of the reaction mixture was increased for 1 hour. As the temperature increases the rate of ZnS formation increases as well. Synthesis of ZnS nanoparticle as a result involves another step where the formed precipitate undergoes ageing. The ageing of the nanomaterial has significant role in its crystallinity. Here room temperature was chosen for growing the particles. Subsequent to 18 h of ageing the material was washed with ethanol and water several times and final mixture was kept on oven for drying. The synthesis resulted in imidazole capped Mn doped zinc sulphide quantum dots following aqueous route of synthesis.The as synthesized material obtained was characterized further by TEM for particle size determination as shown in the Fig. 1. The particles in the size range 5.5–6.5 nm were obtained. The TEM results show that homogeneous particles have been synthesized. The typical particle size of the material is below 10 nm hence it is inferred that QD have been obtained. The dark image of the material is due to the zinc metal which is classified as heavy metal. The strong transmission of the material confirms the constituent of the material under investigation. There is no clump or cluster in the image which proves that imidazole acts as good capping agent and controls the particle size effectively.
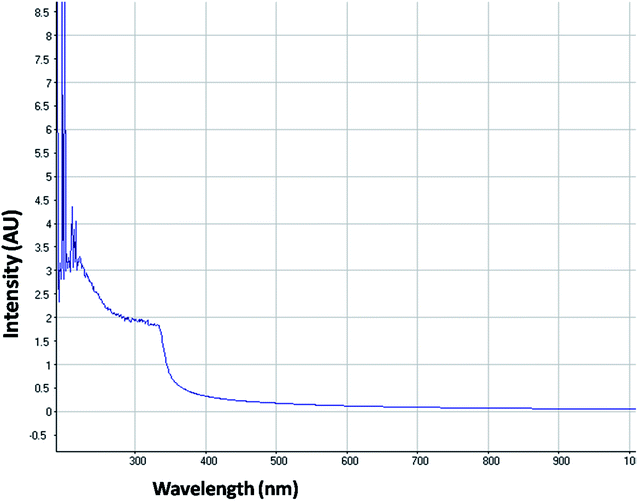
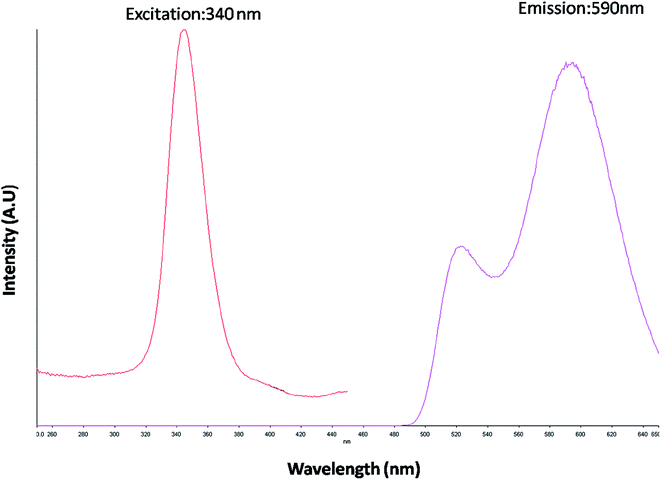
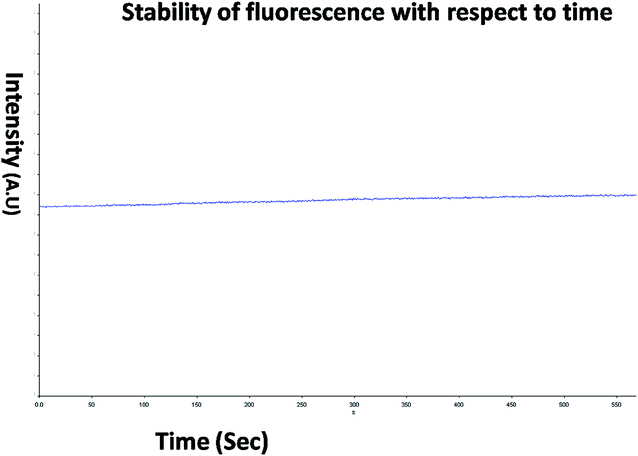
The optical property of the as synthesized QDs was performed by UV and spectrofluorometer. The absorbance at 310 nm and cut off at 340 nm as shown in Fig. 2, proves the blue shift. The cutoff at 340 nm also supports the photoluminescence spectra (Fig. 3). The photoluminescence spectra show that the excitation of the QD is 340 nm and emission is at 590 nm. The strong quantum confinement is there in the band gap due to small sized QD. The emission at 590 nm confirms that manganese ion has been doped in the crystal lattice. The trap states created due to the manganese ion results in higher emission wavelength. Thus the material with highly enhanced optical property has been synthesized. Both UV and photoluminescence spectra support the band gap (3.6 eV), excitation (340 nm) and emission (590 nm) parameter. Hence imidazole molecule had not altered the optical property of the synthesized quantum dots. The optical stability of the synthesized QD was evaluated as well, which shows a stable photoluminescence for a significant 550 seconds (Fig. 4). Thus the luminescence intensity of the imidazole capped QD do not decrease with respect to time and hence highly efficient probe was synthesized which can be used for a longer time in imaging.
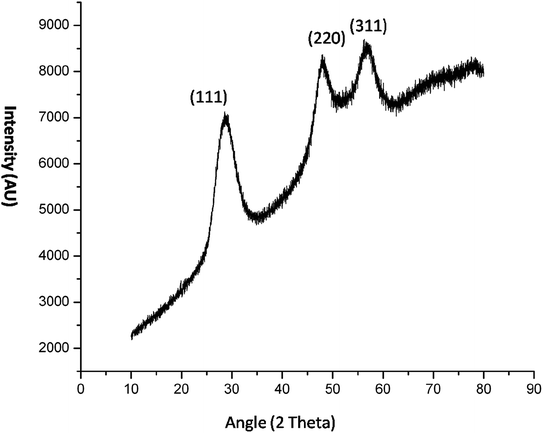
The crystal property of the material was evaluated by XRD measurement. It was observed in the diffraction pattern that high intensity peaks were observed at 28.47 and 56. The XRD pattern (Fig. 5) shows miler planes at (111), (220) and (311) which is in accordance with JCPDS no. 05-0566. The lattice parameters of the QD synthesized and library is similar which confirms the zinc sulphide lattice of the type cubic zinc blende. The purity of the material can be acknowledged by having no additional peaks in the diffraction pattern. The peaks in the diffraction pattern shows that they are of high intensity which attribute to the high crystalline property of the QDs. The dimension of the imidazole capped QD was 9 nm. The smaller size has been also confirmed by the XRD data since the peaks are broaden. The size of the material was calculated by Scherrer equation β1/2 = 0.94λ/d![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
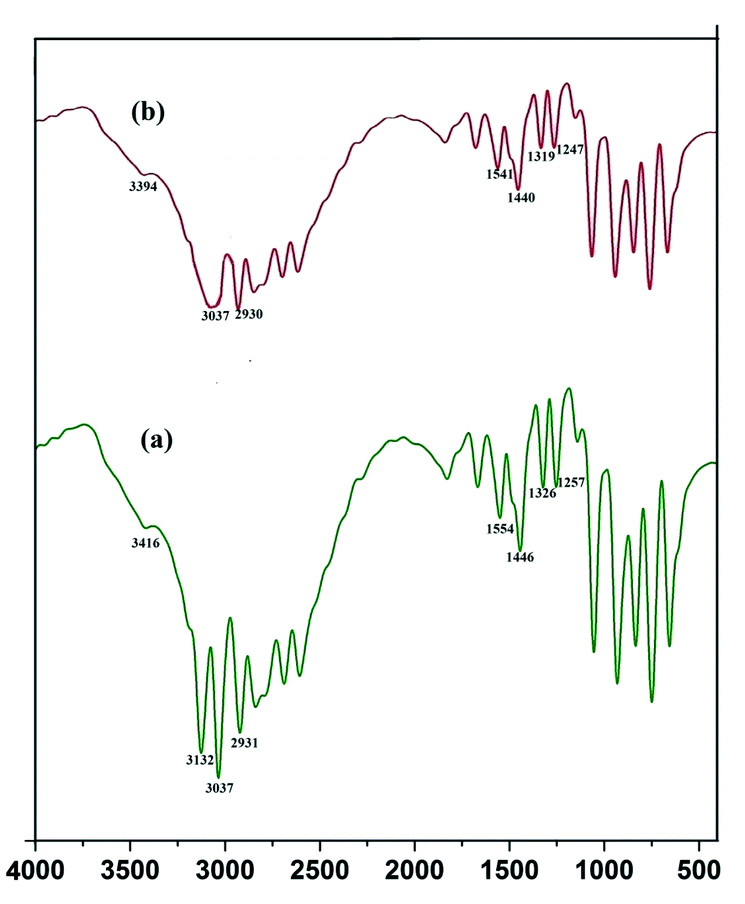
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ (Liu et al. 2010).12 The peak width shows the characteristics of ZnS QDs. Further the capping of imidazole with Mn doped ZnS QDs were confirmed by analyzing FTIR bands. The decrease in intensity of N–H band (Fig. 16b) (3416 cm−1) shows strong interaction of N–H bond with the zinc ion. Although other bands like C–H stretching (2931–3132 cm−1) shows lower intensity in the imidazole capped QDs (Fig. 16b).
θ (Liu et al. 2010).12 The peak width shows the characteristics of ZnS QDs. Further the capping of imidazole with Mn doped ZnS QDs were confirmed by analyzing FTIR bands. The decrease in intensity of N–H band (Fig. 16b) (3416 cm−1) shows strong interaction of N–H bond with the zinc ion. Although other bands like C–H stretching (2931–3132 cm−1) shows lower intensity in the imidazole capped QDs (Fig. 16b).
Hence high quality manganese doped zinc sulphide QD have been synthesized. It is inferred that the imidazole controls the size of the material without affecting the chemical property and material property. Here we have also evaluated the biological applicability of the capped QD with respect to uncapped one.
Computational insight to explain imidazole–zinc bonding
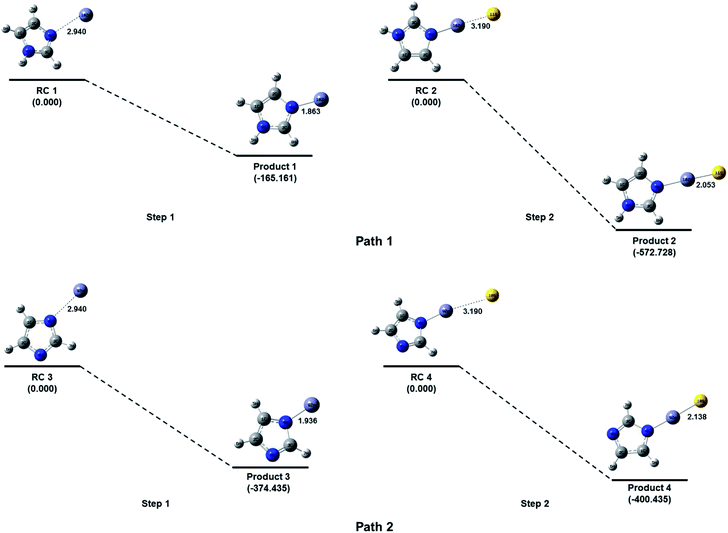
Imidazole is a five membered ring with both basic and acidic nature due to the presence of lone pair on the nitrogen atom and NH group. Here in the quantum chemical study with the help of computational simulation following density functional theory we have studied the formation of zinc imidazole complex following two approaches. In the first approach we have modeled the reaction pathway of zinc ion with imidazole molecule. To model the reaction, Zn ion is placed in the plane of imidazole ring at 2.940 Å (sum of van der Walls radius of Zn and N) from the nitrogen atom (other than NH). This initial structure is optimized to minimum energy and we obtain the Zn–imidazole complex (hereafter product 1). A lone pair of electrons is present on this nitrogen atom of the imidazole molecule which is donated to zinc ion resulting in the formation of this complex (path 1, Fig. 6). This step is barrier less and exothermic with the enthalpy of formation (ΔH) is −165.1605 kcal mol−1 with respect to the separated reactants. Product 1 now carries a positive charge and is susceptible to the nucleophilic attack. The S2− which is added after the formation of product 1 in the reaction system, now quickly react and form a bond with Zn–imidazole complex producing imidazole capped ZnS (product 2). This reaction is exothermic with enthalpy of formation being −572.7279 kcal mol−1.Further, in the similar way the addition of Zn2+ to the NH group of imidazole was also attempted by placing Zn2+ at 109.5° with imidazole plane and at 2.940 Å from the nitrogen atom of the NH group. However, the zinc ion was not approaching towards the NH bond for the bond formation during optimization of the reaction system. The probable reason for this phenomenon could be the unavailability of electrons on the nitrogen of NH functional group as compared to the other nitrogen of the imidazole ring. The addition of Zn2+ on this nitrogen (i.e. of NH) can be feasible if a high electron density could be possible on the group which can interact with the zinc ion. Here in we have removed the H of N–H bond by reacting imidazole molecule with base as described in the experimental section which results in the formation of the imidazole anion. The imidazole anion is a symmetric heterocyclic ring with a uni-negative charge which is distributed among all the atoms in the ring. To study the interaction of imidazole anion with Zn2+, it is placed at 2.940 Å from nitrogen of the ring in the same plane as imidazole (Fig. 6). Optimization of this initial structure results in the product 3 which forms barrierlessly with enthalpy of formation −374.4349 kcal mol−1. It will further react barrierlessly with sulfide ion (S2−) to produce product 4 (with ΔH = −400.7276 kcal mol−1).
The above results shows that when reaction of imidazole and Zn2+ occur within the basic medium, its efficiency increases, (ΔH = −165.1610 kcal mol−1 without basic medium, and ΔH = −374.4351 kcal mol−1 in basic medium). The first step in path 1 is the ion-molecule interaction which is less efficient compared to first step in path 2 where the interaction is between imidazole ion and Zn2+ ion, which is an ionic interaction with respect to both of the reactants therefore this should be spontaneous. The product 4 has 313.3782 kcal mol−1 less energy than product 2. Due to symmetric behavior of imidazolium anion there is equal probability of attack of zinc ion towards both of the nitrogen of the imidazolium anion. The pathway 2 is more efficient thermodynamically than pathway 1 (total ΔH = −737.8884 kcal mol−1 without basic medium, and ΔH = −775.1625 kcal mol−1 in basic medium). Hence it can be argued that pathway 2 is suitable to produce imidazole capped—ZnS quantum dots.
In vitro toxicity and genetic toxicology was evaluated by various parameters. Cellular toxicity was evaluated by MTT, NRU, LDH, ROS, catalase, GSH and LPO. The toxicity at genetic level was performed by chromosomal aberration and micronucleus test. In vitro toxicity and genetic toxicology was evaluated by various parameters. Cellular toxicity was evaluated by MTT, NRU, LDH, ROS, catalase, GSH and LPO. The toxicity at genetic level was performed by chromosomal aberration and micronucleus test.
Cytotoxicity assessment
Genotoxicity studies
Conclusion
Mn doped ZnS QD is synthesized by employing aqueous route of synthesis. The imidazole is used as capping agent which does not alter the optical property or chemical structure of the synthesized material as is evident by the characterization using TEM, XRD and spectroscopy. Quantum chemical and experimental studies have been performed to understand the capping mechanism of ZnS quantum dot via imidazole. It is observed that most efficient capping is observed in the basic medium where deprotonation of the molecule leads to the formation of anionic species that forms a strong covalent bond with the zinc ion. The imidazole capped QD was further analysed and tested by toxicity parameters. Toxicity profiling of the synthesized QD was done at both cellular and genetic level. The toxicity study shows that uncapped QD shows toxic behavior as compared to imidazole capped QD.Conflicts of interest
This work does not involve any conflict of interest and all the authors have given their consent.Acknowledgements
This work is fully supported by SERB-DST, New Delhi. Vivek Pandey is indebted to SERB-DST for providing N-PDF fellowship. Authors acknowledge Mr Vivek Mishra for technical support.References
- K. T. Vadiraj and S. L. Belagali, Synthesis and characterization of Mn: ZnS quantum dots for photovoltaic applications, Nano-Struct. Nano-Objects, 2018, 14, 118–124 CrossRef CAS.
- V. Chantada and P. Maria, Development and application of molecularly imprinted polymer–Mn-doped ZnS quantum dot fluorescent optosensing for cocaine screening in oral fluid and serum, Talanta, 2018, 181, 232–238 CrossRef.
- H. Elmizadeh, M. Soleimani, F. Faridbod and G. R. Bardajee, Fabrication and optimization of a sensitive tetracycline fluorescent nano-sensor based on oxidized starch polysaccharide biopolymer-capped CdTe/ZnS quantum dots: Box–Behnken design, J. Photochem. Photobiol., A, 2018, 367, 188–199 CrossRef CAS.
- E. Hofman, R. J. Robinson, Z. J. Li, B. Dzikovski and W. Zheng, Controlled dopant migration in CdS/ZnS core/shell quantum dots, J. Am. Chem. Soc., 2017, 139, 8878–8885 CrossRef CAS.
- S. V. Bozrova, In Vitro Cytotoxicity of CdSe/ZnS Quantum Dots and Their Interaction with Biological Systems, KnE Energy & Physics, 2018, 3, 58–63 Search PubMed.
- B. A. Rzigalinski and J. S. Strobl, Cadmium-containing nanoparticles: perspectives on pharmacology and toxicology of quantum dots, Toxicol. Appl. Pharmacol., 2009, 238, 280–288 CrossRef CAS.
- M. Bruchez, M. Moronne, P. Gin, S. Weiss and A. P. Alivisatos, Semiconductor Nanocrystals as Fluorescent Biological Labels, Science, 1998, 281, 2013–2016 CrossRef CAS.
- H. Elmizadeh, M. Soleimani, F. Faridbod and G. R. Bardajee, Fabrication and optimization of a sensitive tetracycline fluorescent nano-sensor based on oxidized starch polysaccharide biopolymer-capped CdTe/ZnS quantum dots: Box–Behnken design, J. Photochem. Photobiol., A, 2018, 367, 188–199 CrossRef CAS.
- T. D. Talantikite, H. Merzouk, H. Haddad and A. Tounsi, Effect of dopant concentration on structural and optical properties Mn doped ZnS films prepared by CBD method, Optik, 2017, 136, 362–367 CrossRef.
- A. Fontes, R. B. de Lira, M. A. B. L. Seabra, T. G. da Silva, A. G. de Castro and B. S. Santos, Quantum dots in biomedical research, Biomedical Engineering-Technical Applications in Medicine, 2012, pp. 269–290 Search PubMed.
- S. Roy, M. Manna and A. Chattopadhyay, Complex Transfer Reaction from ZnO to ZnS Quantum Dots Driven by Surface Anions, J. Phys. Chem., 2018, 122, 9939–9946 CAS.
- S. Pradeep, S. Raghuram, M. G. Chaudhury and S. Mazumder, Synthesis and Characterization of Fe3+ and Mn2+ Doped ZnS Quantum Dots for Photocatalytic Applications: Effect of 2-Mercaptoethanol and Chitosan as Capping Agents, J. Nanosci. Nanotechnol., 2017, 17, 1125–1132 CrossRef CAS.
- B. Demirhan, B. E. Demirhan, N. Ertas and H. E. Kara, Rapid Detection of Acrylamide in Food Using Mn-Doped ZnS Quantum Dots as a Room Temperature Phosphorescent Probe, Food Anal. Methods, 2018, 11, 1367–1373 CrossRef.
- P. Yang and G. Zhou, Phase transfer of hydrophobic QDs for water-soluble and biocompatible nature through silanization, Mater. Res. Bull., 2011, 46, 2367–2372 CrossRef CAS.
- N. Gaponik, D. V. Talapin, A. L. Rogach, K. Hoppe, E. V. Shevchenko, A. Kornowski, A. Eychmüller and H. Weller, Thiol-capping of CdTe nanocrystals: an alternative to organometallic synthetic routes, J. Phys. Chem. B, 2002, 106, 7177–7185 CrossRef CAS.
- L. Chang, X. He, L. Chen and Y. Zhang, Mercaptophenylboronic acid-capped Mn-doped ZnS quantum dots for highly selective and sensitive fluorescence detection of glycoproteins, Sens. Actuators, B, 2017, 243, 72–77 CrossRef CAS.
- C. F. Peng, Y. Y. Zhang, Z. J. Qian and Z. J. Xie, Fluorescence sensor based on glutathione capped CdTe QDs for detection of Cr3+ ions in vitamins, Food Science and Human Wellness, 2018, 7, 71–76 CrossRef.
- W. C. W. Chan and S. M. Nie, Quantum Dot Bioconjugates for Ultrasensitive Nonisotopic Detection, Science, 1998, 281, 2016–2018 CrossRef CAS.
- S. Coe, W. K. Woo, M. G. Bawendi and V. Bulovi, Electroluminescence from single monolayers of nanocrystals in molecular organic devices, Nature, 2003, 420, 800 CrossRef PubMed.
- L. E. Brus, A simple model for the ionization potential, electron affinity, and aqueous redox potentials of small semiconductor crystallites, J. Chem. Phys., 1983, 79, 5566–5571 CrossRef CAS.
- A. I. Ekimov and A. A. Onushchenko, Size quantization of the electron energy spectrum in a microscopic semiconductor crystal, JETP Lett., 1984, 40, 1136–1139 Search PubMed.
- R. Rossetti, S. Nakahara and L. E. Brus, Quantum size effects in the redox potentials, resonance Raman spectra, and electronic spectra of CdS crystallites in aqueous solution, J. Chem. Phys., 1983, 79, 1086–1088 CrossRef CAS.
- M. G. Bawendi, W. L. Wilson, L. Rothberg, P. J. Carroll, T. M. Jedju, M. L. Steigerwald and L. E. Brus, Electronic structure and photoexcited-carrier dynamics in nanometer-size CdSe clusters, Phys. Rev. Lett., 1990, 65, 1623–1626 CrossRef CAS.
- P. Reiss, S. Carayon, J. Bleuse and A. Pron, Low polydispersity core/shell nanocrystals of CdSe/ZnSe and CdSe/ZnSe/ZnS type: preparation and optical studies, Synth. Met., 2003, 649–652 CrossRef CAS.
- M. Darawsheh, H. Abu Ali, A. Abuhijleh, E. Rappocciolo, M. Akkawi, S. Jaber, S. Maloul and Y. Hussein, Synthesis, crystallographic, spectroscopic studies and biological activity of new cobalt(II) complexes with bioactive mixed sulindac and nitrogen-donor ligands, Eur. J. Med. Chem., 2014, 82, 152–163 CrossRef CAS.
- A. Dołęga, A. Farmas, K. Baranowska and A. Herman, Novel zinc complexes with acetyloacetonate, imidazole and thiolate ligands: Crystal structure of a zinc complex of relevance to farnesyl transferase, Inorg. Chem. Commun., 2009, 12, 823–827 CrossRef.
- S. Leon, Comprehensive Pharmacy Review, Lippincott Williams and Wilkins, 6th edn, 2007, p. 930, 0781774004 Search PubMed.
- B. Martinez-Perez, D. Quintanar-Guerrero, M. T.-T. R. Cisneris-Tamayo, M. L. Zambrano-Zaragoza, S. Alcala-Alcala, N. Mendoza-Munoz and S. Pinon, Controlled release biodegradable nanoparticles: From preparations to vaginal applications, Eur. J. Pharm. Sci., 2018, 115, 185–195 CrossRef CAS.
- B. Sun, K. Liu, J. Han, L. Y. Zhao, X. Su, B. Lin, D. M. Zhao and M. S. Cheng, Design, synthesis, and biological evaluation of amide imidazole derivatives as novel metabolic enzyme CYP26A1 inhibitors, Bioorg. Med. Chem., 2015, 15, 6763–6773 CrossRef.
- B. Zhang, F. Han, L. Wang, D. Li, D. Yang, X. Yang, J. Yang, X. Li, D. Zhao and R. Wang, Catalytic Asymmetric Michael Reaction of 5H-Oxazol-4-Ones with α, β-Unsaturated Acyl Imidazoles, Chem.–Eur. J., 2015, 21, 17234–17238 CrossRef CAS.
- D. W. Appleton and B. Sarkar, Studies of Zn(II) and Co(II) complexes of imidazole and n-methylimidazole with regard to the activity related ionization in carbonic anhydrase, Bioinorg. Chem., 1977, 7, 211–224 CrossRef CAS.
- A. D. Becke, Density-functional exchange-energy approximation with correct asymptotic behavior, Phys. Rev. A: At., Mol., Opt. Phys., 1988, 38, 3098 CrossRef CAS PubMed.
- C. Lee, W. Yang and R. G. Parr, Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density, Phys. Rev. A: At., Mol., Opt. Phys., 1988, 37, 785 CrossRef CAS.
- K. Raghavachari, G. W. Trucks, J. A. Pople and G. M. Head, A fifth-order perturbation comparison of electron correlation theories, Chem. Phys. Lett., 1989, 157, 479–483 CrossRef CAS.
- J. D. Watts, J. Gauss and J. R. J. Bartlell, Chem. Phys., 1993, 98, 9718 Search PubMed.
- F. Trani, G. Scalmani, G. Zheng, I. Carnimeo, M. J. Frisch and V. Barone, Time-dependent density functional tight binding: new formulation and benchmark of excited states, J. Chem. Theory Comput., 2011, 7, 3304–3313 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery, J. E. Peralta Jr, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski, and D. J. Fox, Gaussian 09, Gaussian, Inc., Wallingford CT, 2009, https://gaussian.com/glossary/g09/ Search PubMed.
- R. Arunan, P. Joseph, M. Sivakumar and S. C. G. K. Daniel, One Pot Aqueous Synthesis of LHistidine Amino Acid Capped Mn: ZnS Quantum Dots for Dopamine Sensing, Curr. Nanosci., 2019 DOI:10.2174/1573413715666190520093625.
Footnote |
| † Equal first authors. |
| This journal is © The Royal Society of Chemistry 2019 |