Open Access Article
Open Access ArticleA highly selective fluorescent probe for human NAD(P)H:quinone oxidoreductase 1 (hNQO1) detection and imaging in living tumor cells†
Ya Zhu‡
a,
Jialing Han‡a,
Qian Zhanga,
Zhou Zhaoa,
Jin Wanga,
Xiaowei Xu *a,
Haiping Hao*a and
Jun Zhang*b
*a,
Haiping Hao*a and
Jun Zhang*b
aSchool of Pharmacy, China Pharmaceutical University, 210009, Nanjing, China. E-mail: xw@cpu.edu.cn; haipinghao@cpu.edu.cn
bSchool of Pharmacy, Nanjing Medical University, 211166, Nanjing, China. E-mail: zhangjun@njmu.edu.cn
First published on 27th August 2019
Abstract
Human NAD(P)H:quinone oxidoreductase (hNQO1) can be used as a biomarker for the early diagnosis of cancer. In this article, a novel fluorescent probe 1 for detecting hNQO1 was designed and synthesized by using 2-dicyanomethylene-3-cyano-4,5,5-trimethyl-2,5-dihydrofuran (TCF) derivative (TCF-OH) as a fluorophore and quinone propionic acid (QPA) as a recognition group. The probe, which has high selectivity for hNQO1 and a Stokes shift of about 117 nm, shows a linear relationship with hNQO1 concentrations in the range of 0.25–3 μg ml−1 with the color changes from yellow to red and was successfully applied to intracellular hNQO1 imaging.
Introduction
Human NAD(P)H:quinone oxidoreductase 1 (hNQO1) is a flavin protease that catalyzes a two-electron reduction of various quinones directly to the hydroquinone by utilizing NAD(P)H as a cofactor.1–5 Hydroquinone can be easily combined with glutathione or glucuronic acid and excreted from cells, so hNQO1 plays a crucial detoxification role in organisms.6 hNQO1 is expressed in various mammalian tissues and organs, and hNQO1 is mainly present in the cytoplasm and has a low expression level in the nucleus.4 Studies have shown that hNQO1 is overexpressed in a variety of tumor cells, including colon cancer,7 breast cancer,8,9 pancreatic cancer,10–12 lung cancer,13,14 cholangioma,15 melanoma,16 liver cancer,17 etc. hNQO1 is expressed 2–50 times more in cancer cells than in normal cells, so hNQO1 can be used as a biomarker for cancer diagnosis, which is significant for the early diagnosis of cancer.18–222-Dicyanomethylene-3-cyano-4,5,5-trimethyl-2,5-dihydrofuran (TCF) has three conjugated systems of strong electron-withdrawing cyano and its derivative TCF-OH contains a donor–π–acceptor (D–π–A) structure, which is widely used to construct novel near-infrared fluorescent probes and non-linear optic chromophores and avoid autofluorescence interference by tissue.23–27 Recently, a variety of fluorescent probes for biological imaging have been reported based on TCF structures.23,24,28–31 Herein, we designed and synthesized a novel fluorescent probe 1 for detection of hNQO1 based on TCF-OH as a chromophore and quinone propionic acid (QPA) as a recognition group. QPA annihilates the fluorescence of TCF-OH as an electron withdrawing group before probe 1 is not reduced by hNQO1, so the probe 1 has weaker fluorescence. When probe 1 responds to hNQO1 and NAD(P)H, hNQO1 selectively catalyzes the reduction of QPA to trimethylhydroquinone, then forms a lactone structure to separate from the probe and opens the lactonestructure of probe 1 to release the fluorophore TCF-OH, thereby realizing the detection of hNQO1, and this process is accompanied by the color of the solution changing from yellow to red. Probe 1 has a Stokes shift of about 117 nm, which better eliminates background interference. The probe 1 has the advantages of simple synthetic route, low cytotoxicity, good cell permeability, visual and stable detection, and may be applied to sense intracellular hNQO1.
Experimental section
Materials and instruments
hNQO1 and NADPH were purchased from Sigma and Aladdin, respectively. All chemicals and solvents were purchased commercially and used without further purification unless noted otherwise. All experimental cells were from ATCC. Dulbecco's Modified Eagle's Medium (DMEM), fetal bovine serum (FBS), penicillin, streptomycin, and trypsin–EDTA were obtained from Gibco. LM-CS was performed on waters instrument. Fluorescence spectra were recorded using a Shimadzu RF6000 with both excitation and emission slit widths of 5 nm. Fluorescence images were captured by using an Zeiss LSM700 laser confocal fluorescence microscope.The synthesis of compound 3
3,3-Dimethylacrylate (1.1 ml, 15.6 mmol) was added to a mixture of 2,3,5-trimethyl-1,4-benzenediol (2 g, 13 mmol) and methane sulfonic acid (16 ml), and the mixture was condensed and refluxed at 70 °C for 3 h. The reaction mixture was cooled to room temperature and then extracted three times with dichloromethane (DCM). The extracts were washed with a saturated NaHCO3 solution and a NaCl solution, dried over anhydrous Na2SO4 for 0.5 h, and the organic solvent was evaporated to give a pale yellow solid (3.49 g). Purified by column chromatography (petroleum ether (PE)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DCM = 2
DCM = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give a white solid (2.24 g, 64% yield). 1H NMR (500 MHz, chloroform-d) δ: 4.62 (s, 1H), 2.60 (s, 2H), 2.40 (s, 3H), 2.27 (s, 3H), 2.23 (s, 3H), 1.50 (s, 6H).
1) to give a white solid (2.24 g, 64% yield). 1H NMR (500 MHz, chloroform-d) δ: 4.62 (s, 1H), 2.60 (s, 2H), 2.40 (s, 3H), 2.27 (s, 3H), 2.23 (s, 3H), 1.50 (s, 6H).
The synthesis of compound 4
Compound 3 (2.2 g, 9.4 mmol) was dissolved in acetonitrile (111 ml) and water (47 ml), and N-bromosuccinimide (NBS) (1.8 g, 10.3 mmol) was added. The reaction was stirred at room temperature for 1 h and then extracted three times with DCM. The extracts were washed with a NaCl solution, dried over anhydrous Na2SO4 for 0.5 h, and the organic solvent was evaporated to give a pale yellow solid (2.43 g). Purified by column chromatography (PE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate (EA) = 8
ethyl acetate (EA) = 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give a white solid (1.86 g, 76% yield). 1H NMR (300 MHz, chloroform-d) δ: 3.04 (s, 2H), 2.16 (s, 3H), 1.97 (s, 3H), 1.95 (s, 6H), 1.46 (s, 6H). 13C NMR32 (100 MHz, CDCl3) δ (ppm): 190.88, 187.47, 178.33, 152.04, 142.99, 139.04, 138.41, 47.21, 37.94, 28.80, 14.32, 12.52, 12.12.
1) to give a white solid (1.86 g, 76% yield). 1H NMR (300 MHz, chloroform-d) δ: 3.04 (s, 2H), 2.16 (s, 3H), 1.97 (s, 3H), 1.95 (s, 6H), 1.46 (s, 6H). 13C NMR32 (100 MHz, CDCl3) δ (ppm): 190.88, 187.47, 178.33, 152.04, 142.99, 139.04, 138.41, 47.21, 37.94, 28.80, 14.32, 12.52, 12.12.
The synthesis of compound 7
Compound 6 (92 mg, 0.75 mmol) was dissolved in anhydrous tetrahydrofuran (THF) (3 ml) and anhydrous EtOH (0.75 ml), then compound 5 (150 mg, 0.75 mmol) and ammonium acetate (59 mg, 0.75 mmol) were added, and the mixture was reacted under N2 for 6 h at room temperature. After the reaction, the mixture was extracted three times with DCM. The extracts were washed with a NaCl solution, masticated by methanol and filtered to obtain orange-red solid powder (181.5 mg, 59% yield). 1H NMR (300 MHz, DMSO-d6) δ: 10.61 (s, 1H), 7.91 (d, J = 16.2 Hz, 1H), 7.81 (d, J = 7.7 Hz, 2H), 7.03 (d, J = 16.2 Hz, 1H), 6.91 (d, J = 7.8 Hz, 2H), 1.78 (s, 6H). 13C NMR33 (125 MHz, DMSO-d6): δ/ppm 177.9, 176.5, 162.9, 148.9, 132.9 (2C), 126.3, 117.0 (2C), 113.5, 112.7, 112.3, 111.8, 99.6, 97.3, 53.8, 25.9 (2C).The synthesis of probe 1
Compound 7 (61 mg, 0.2 mmol) and compound 4 (60 mg, 0.24 mmol), EDC (76 mg, 0.4 mmol) and DMAP (49 mg, 0.4 mmol) were added to the reaction flask, the oil pump was evacuated and replaced with N2, then anhydrous DCM (1 ml) was added, and the reaction was stirred overnight at room temperature under N2. After the reaction, the mixture was extracted three times with DCM. The extracts were washed with a NaCl solution, dried over anhydrous Na2SO4 for 0.5 h, and the organic solvent was evaporated to give a pale yellow solid (166 mg). Purified by column chromatography (PE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DCM = 4
DCM = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give a solid (53.2 g). Chromatography on a thin layer of silica gel plate (developing agent PE
1) to give a solid (53.2 g). Chromatography on a thin layer of silica gel plate (developing agent PE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EA = 1
EA = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), squeegee, rinse with ethyl acetate, and control the temperature within 40 °C, remove the organic solvent by rotary evaporation to give a yellow solid (30.8 mg, 57.8% yield). 1H NMR (300 MHz, chloroform-d) δ: 7.67 (m, 2H), 7.63 (d, J = 11.1 Hz, 2H), 7.16 (d, J = 8.5 Hz, 1H), 6.99 (d, J = 16.4 Hz, 1H), 3.31 (s, 2H), 2.21 (s, 3H), 1.96 (s, 6H), 1.81 (s, 6H), 1.56 (s, 6H). 13C NMR (126 MHz, CDCl3) δ: 190.78, 187.35, 175.14, 173.53, 170.78, 153.82, 151.44, 145.99, 142.70, 139.63, 138.86, 131.43, 130.27, 122.84, 115.01, 111.51, 110.77, 110.11, 100.37, 97.75, 47.74, 38.43, 29.04, 26.44, 14.48, 12.67, 12.20. HRMS m/z: calculated for C32H29N3NaO5 558.2005 (M + Na)+, found 558.2007.
1), squeegee, rinse with ethyl acetate, and control the temperature within 40 °C, remove the organic solvent by rotary evaporation to give a yellow solid (30.8 mg, 57.8% yield). 1H NMR (300 MHz, chloroform-d) δ: 7.67 (m, 2H), 7.63 (d, J = 11.1 Hz, 2H), 7.16 (d, J = 8.5 Hz, 1H), 6.99 (d, J = 16.4 Hz, 1H), 3.31 (s, 2H), 2.21 (s, 3H), 1.96 (s, 6H), 1.81 (s, 6H), 1.56 (s, 6H). 13C NMR (126 MHz, CDCl3) δ: 190.78, 187.35, 175.14, 173.53, 170.78, 153.82, 151.44, 145.99, 142.70, 139.63, 138.86, 131.43, 130.27, 122.84, 115.01, 111.51, 110.77, 110.11, 100.37, 97.75, 47.74, 38.43, 29.04, 26.44, 14.48, 12.67, 12.20. HRMS m/z: calculated for C32H29N3NaO5 558.2005 (M + Na)+, found 558.2007.
Spectrophotometric experiments
The probe 1 was dissolved in DMSO to prepare a 10 mM stock solution. In all spectrum measurements, the stock solution was diluted to a working concentration of 2.5 μM with PBS (10 mM, pH = 7.4) containing 0.025% DMSO. The NADPH (50 μM) and the hNQO1 solutions (0.0625 μg ml−1, 0.25 μg ml−1, 0.5 μg ml−1, 1 μg ml−1, 2 μg ml−1, 3 μg ml−1, 4 μg ml−1, 8 μg ml−1 and 16 μg ml−1) were prepared, respectively. Then the probe 1, NADPH and different concentrations of hNQO1 were mixed uniformly and then placed in a water bath at 37 °C for 10 min. For all fluorescence spectroscopy studies, a fluorescence emission spectrum at 460 to 720 nm was recorded at a maximum excitation light wavelength of 444 nm, excitation and emission slits are 5 nm.Selectivity of probe 1 towards hNQO1
Free amino acids, common ions and biothiols present in the human body may limit the use of probe 1. To investigate the selectivity of probe 1 for hNQO1 substrates, the probe 1 (2.5 μM) and various interfering reactants including NADPH (50 μM), GSH (250 μM), glucose (250 μM), Cys (250 μM), Ala (250 μM), Tyr (250 μM), Arg (250 μM), His (250 μM), Na+ (250 μM), K+ (250 μM), Ca2+ (250 μM), Mg2+ (250 μM) were added to several test vials in a water bath for 10 minutes. The fluorescence emission spectrum at 460 to 720 nm was recorded at a maximum excitation light wavelength of 444 nm, excitation and emission slits are 5 nm.Cytotoxicity and bioimaging application
Prior to exploring the imaging ability of probe 1 in live cells, the cytotoxicity of probe 1 was assessed using a CCK-8 assay. After incubating the MDA-MB-231 (hNQO1−) cells for 24 h, the cells were incubated for 20 h with fresh medium containing serial concentration of probe 1 (0, 2, 4, 6, 8, 10, 12, 14, 16 μM). The cells were then washed with warm PBS, and the cells were further incubated with medium containing 10% CCK-8, and then the absorbance was measured at 490 nm. Then we evaluated the ability of probe 1 to image intracellular hNQO1 in live cancer cells and cultured MDA-MB-231 (hNQO1+) cells transfected with the hNQO1 gene and hNQO1 negative expression cell MDA-MB-231 (hNQO1−), and used MDA-MB-231 (hNQO1+) cells to observe the effect of hNQO1 inhibitor (dicoumarin) on the imaging of probe 1. The cells were incubated with DMEM medium containing 10% FBS and 1% penicillin and streptomycin. After adhering the cells for 24 h in a cell culture incubator (37 °C, 5% CO2), the supernatant in the culture dish was discarded, and the cells were further incubated with fresh DMEM medium containing probe 1 (10 μM) for 30 min, then washed three times with warm PBS and images were collected by confocal laser scanning microscopy. The inhibitor group was incubated with fresh DMEM medium containing dicoumarol (50 μM) for 6 h and then incubated with fresh DMEM medium containing the probe (10 μM) for 30 min and then imaged under a confocal laser scanning microscopy.Results and discussion
Synthesis route of the probe 1
Scheme 1 summarizes the synthetic route of probe 1 and all the synthesized structures were confirmed by 1H NMR, 13C NMR and HRMS spectra (Fig. S1–S6†). The structure of probe 1 includes a fluorescent response unit TCF-OH and an hNQO1 recognition unit QPA, which are linked by an ester bond. After the probe 1 reacts with hNQO1, QPA is reduced to trimethylhydroquinone, and then automatically detaches from the probe 1 structure, the fluorophore lactone structure is opened, and the fluorescence of TCF-OH is recovered to achieve fluorescence enhancement. Fig. 1A described the possible interaction mechanism of the probe 1 with hNQO1. Furthermore, the results of LM-CS also demonstrated the interaction mechanism of the probe 1 with hNQO1 (Fig. 1B). It was found that the reaction products of probe 1 (2.5 μM) in the presence of hNQO1 (16 μg ml−1) and NADPH (50 μM) exhibited chromatographic peaks at tR = 4.96 min and 4.65 min, respectively, corresponding perfectly to the compound 7 and 8, indicating the removal of QPA moiety from the probe 1 and the formation of TCF-OH. Absence of the peak (tR = 8.21 min) of probe 1 in the reaction mixture indicated complete reaction of probe 1 with hNQO1.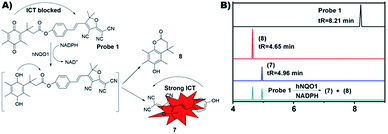 | ||
| Fig. 1 (A) The reaction of probe 1 with hNQO1. (B) LM-CS analysis of probe 1 activation. LM-CS spectra of probe 1, compound 7, compound 8 and the reaction mixture of probe 1 and hNQO1. | ||
Spectroscopic response of probe 1 to hNQO1
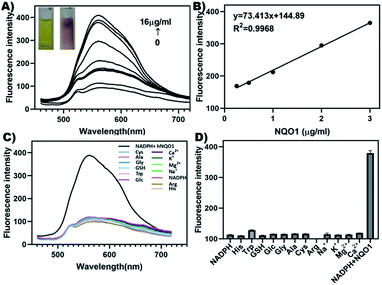
As shown in Fig. 2A, probe 1 exhibited only weak fluorescence at excitation light with a maximum excitation wavelength of 444 nm. Then the probe 1, NADPH and different concentrations of hNQO1 were mixed uniformly and then placed in a water bath at 37 °C for 10 min. It was found that the fluorescence intensity at 561 nm increased gradually under the excitation of the same light, along with a change in color from yellow to red. In addition, when the concentration of hNQO1 was 16 μg ml−1, the fluorescence intensity of the solution increased by about 4 folds. Furthermore, the concentration-dependent curve is as shown in (Fig. 2B). There was a good linear proportional relationship between probe 1 and hNQO1, and the concentrations of hNQO1 in the range of 0.25 to 3 μg ml−1.Selectivity evaluation
The emission spectra of probe 1 and NADPH, GSH, glucose, Cys, Ala, Tyr, Arg, His, Na+, K+, Ca2+, Mg2+ were measured separately and the fluorescence emission spectrum at 460 to 720 nm was recorded. The results implied that the fluorescence intensity of probe 1 hardly changed upon addition of other interfering substances, but the fluorescence intensity was significantly enhanced after the addition of hNQO1 (Fig. 2C). In addition, bars represent the average final fluorescence intensity between probe 1 and hNQO1 or interfering reactants at 561 nm after 10 min incubation (Fig. 2D). It was shown that the probe 1 only produced fluorescence enhancement in the presence of hNQO1, indicating a high selectivity of probe 1 for hNQO1.Cytotoxicity evaluation and bio-imaging of probe 1 in cancer cells
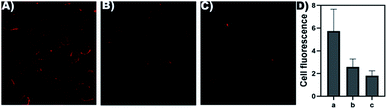
The cytotoxicity of probe 1 for MDA-MB-231 (hNQO1−) cells was measured using a CCK-8 assay. As shown in (Fig. 3), when the concentration of probe 1 was 14 μM, the cell viability of MDA-MB-231 (hNQO1−) was about 83%, illustrating low toxicity and good biocompatibility of probe 1. Then we evaluated the ability of probe 1 to image intracellular hNQO1 in MDA-MB-231 (hNQO1+) cells and hNQO1 negative expression cell MDA-MB-231 (hNQO1−). As shown in (Fig. 4A), fluorescence mainly appeared in the cytoplasm of MDA-MB-231 (hNQO1+), indicating that probe 1 undergoes electron transfer induced by NADPH in the cell, and reacted with hNQO1 to form the fluorophore TCF-OH and emit strong fluorescence. After MDA-MB-231 (hNQO1+) was incubated with dicoumarin, the imaging ability of the probe was significantly reduced, which indicated the high selectivity of the probe to hNQO1 (Fig. 4B). As shown in (Fig. 4C), hNQO1 negative expression cell MDA-MB-231 (hNQO1−) only showed weak fluorescence compared to MDA-MB-231 (hNQO1+), which indicating that the probe 1 could monitor the intracellular hNQO1 level in biological systems. In addition, the fluorescence data obtained from the images were used to calculate average fluorescence intensities by ImageJ (Fig. 4D). Intracellular imaging demonstrates the sensitivity of the probe 1 to hNQO1 and can be used for early judgment of cancer.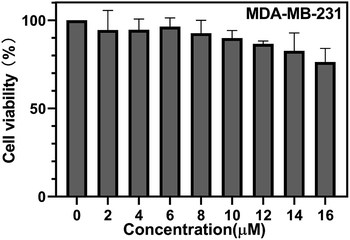 | ||
| Fig. 3 The cell viability of MDA-MB-231 (hNQO1−) after incubation with serial concentrations of probe 1. | ||
Conclusion
In summary, a novel fluorescent probe 1 for detecting hNQO1 was designed and synthesized by using TCF-OH as a fluorophore and QPA as a recognition group. Probe 1 was weakly excited by excitation light at 444 nm, and there was no obvious emission peak. The probe solution was incubated with hNQO1 at 37 °C for 10 min, and a significant emission peak appeared at 561 nm with the same wavelength of light excitation, and the fluorescence intensity was correlated with the concentration of hNQO1, and the linear response range was 0.25–3 μg ml−1. The probe has high selectivity for hNQO1 and is stable under physiological pH conditions. The result that the fluorescence of the probe is significantly enhanced in MDA-MB-231 (hNQO1+) cells compared to MDA-MB-231 (hNQO1−) cells shows that the probe can be successfully used for imaging detection of living cells. These results indicate that probe 1 has potential application value for detection and biological imaging of cancer biology.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by National Natural Science Foundation of China (grants 21602254 to X. X., 81430091, 81421005, and 81720108032 to H. H., 81773986 to J. Z.), Overseas Expertise Introduction Center for Discipline Innovation (111 project to H. H., B17047), the Natural Science Foundation of Jiangsu Province (grant BK20160767 to X. X.) and the project for Major New Drugs Innovation and Development (grant 2015ZX09501010 to H. H.).Notes and references
- J. J. Pink, S. M. Planchon, C. Tagliarino, M. E. Varnes, D. Siegel and D. A. Boothman, J. Biol. Chem., 2000, 275, 5416 CrossRef CAS PubMed.
- H. Zhu and Y. B. Li, Cardiovasc. Toxicol., 2012, 12, 39 CrossRef CAS.
- D. Siegel, C. Yan and D. Ross, Biochem. Pharmacol., 2012, 83, 1033 CrossRef CAS PubMed.
- S. L. Winski, Y. Koutalos, D. L. Bentley and D. Ross, Cancer Res., 2002, 62, 1420 CAS.
- M. Nakamura and T. Hayashi, J. Biochem., 1994, 115, 1141 CrossRef CAS.
- V. Vasiliou, D. Ross and D. W. Nebert, Hum. Genomics, 2006, 2, 329 CrossRef CAS PubMed.
- L. L. Ji, Y. Z. Wei, T. Jiang and S. Y. Wang, Int. J. Clin. Exp. Pathol., 2014, 7, 1124 Search PubMed.
- M. S. Bentle, K. E. Reinicke, Y. Dong, E. A. Bey and D. A. Boothman, Cancer Res., 2007, 67, 6936 CrossRef CAS PubMed.
- Y. Yang, Y. Zhang, Q. Y. Wu, X. L. Cui, Z. H. Lin, S. P. Liu and L. Chen, J. Exp. Clin. Cancer Res., 2014, 33, 14 CrossRef CAS PubMed.
- N. S. Awadallah, D. Dehn, R. J. Shah, N. S. Russell, Y. K. Chen, D. Ross, J. S. Bentz and K. R. Shroyer, Appl. Immunohistochem. Mol. Morphol., 2008, 16, 24 CAS.
- A. M. Lewis, M. Ough, J. Du, M. S. Tsao, L. W. Oberley and J. J. Cullen, Mol. Carcinog., 2017, 56, 1825 CrossRef CAS PubMed.
- P. Khare, A. Bose, P. Singh, S. Singh, S. Javed, S. K. Jain, O. Singh and R. Pal, Mol. Carcinog., 2017, 56, 359 CrossRef CAS PubMed.
- J. J. Schlager and G. Powis, Int. J. Cancer, 1990, 45, 403 CrossRef CAS PubMed.
- D. Siegel, W. A. Franklin and D. Ross, Clin. Cancer Res., 1998, 4, 2065 CAS.
- B. Buranrat, A. Chaawan, A. Puapairoj, A. Prawan, P. Zeekpudsa and V. Kukongviriyapan, Asian Pac. J. Cancer Prev., 2012, 13, 131 CrossRef.
- M. Garate, A. A. Wani and G. Li, Free Radicals Biol. Med., 2010, 48, 1601 CrossRef CAS PubMed.
- T. Cresteil and A. K. Jaiswal, Biochem. Pharmacol., 1991, 42, 1021 CrossRef CAS PubMed.
- S. Danson, T. H. Ward, J. Butler and M. Ranson, Cancer Treat. Rev., 2004, 30, 437 CrossRef CAS PubMed.
- M. Fiorillo, F. Sotgia, D. Sisci, A. R. Cappello and M. P. Lisanti, Oncotarget, 2017, 8, 20309 CrossRef.
- P. Sutton, J. Evans, R. Jones, H. Malik, D. Vimalachandran, D. Palmer, C. Goldring and N. Kitteringham, Lancet, 2015, 385, S95 CrossRef.
- A. M. Malkinson, D. Siegel, G. L. Forrest, A. F. Gazdar, H. K. Oie, D. C. Chan, P. A. Bunn, M. Mabry, D. J. Dykes and S. D. Harrison, et al., Cancer Res., 1992, 52, 4752 CAS.
- S. R. Punganuru, H. R. Madala, V. Arutla and K. S. Srivenugopal, Cancers, 2018, 10, 470 CrossRef PubMed.
- A. C. Sedgwick, H. H. Han, J. E. Gardiner, S. D. Bull, X. P. He and T. D. James, Chem. Commun., 2017, 53, 12822 RSC.
- B. C. Zhu, H. Kan, J. K. Liu, H. Q. Liu, Q. Wei and B. Du, Biosens. Bioelectron., 2014, 52, 298 CrossRef CAS PubMed.
- Y. L. Meng, Z. H. Xin, Y. J. Jia, Y. F. Kang, L. P. Ge, C. H. Zhang and M. Y. Dai, Spectrochim. Acta, Part A, 2018, 202, 301 CrossRef CAS PubMed.
- Z. Q. Guo, S. Park, J. Y. Yoon and I. Shin, Chem. Soc. Rev., 2014, 43, 16 RSC.
- Y. J. Wang, Y. Shi, Z. Wang, Z. Zhu, X. Zhao, H. Nie, J. Qian, A. Qin, J. Z. Sun and B. Z. Tang, Chemistry, 2016, 22, 9784 CrossRef CAS PubMed.
- Y. P. Dai, T. Z. Xue, X. X. Zhang, S. Misal, H. F. Ji and Z. J. Qi, Spectrochim. Acta, Part A, 2019, 216, 365 CrossRef CAS PubMed.
- X. Y. Wang, J. Min, W. J. Wang, Y. Wang, G. Yin and R. Y. Wang, Analyst, 2018, 143, 2641 RSC.
- L. Gwynne, A. C. Sedgwick, J. E. Gardiner, G. T. Williams, G. Kim, J. P. Lowe, J. Y. Maillard, A. T. A. Jenkins, S. D. Bull, J. L. Sessler, J. Yoon and T. D. James, Front. Chem., 2019, 7, 255 CrossRef PubMed.
- M. Y. Wu, K. Li, C. Y. Li, J. T. Hou and X. Q. Yu, Chem. Commun., 2013, 50, 183 RSC.
- D. Pan, F. Luo, X. Liu, W. Liu, W. Chen, F. Liu, Y. Q. Kuang and J. H. Jiang, Analyst, 2017, 142, 2624 RSC.
- M. Ipuy, C. Billon, G. Micouin, J. Samarut, C. Andraud and Y. Bretonnière, Org. Biomol. Chem., 2014, 12, 3641 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra05650e |
| ‡ These authors contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |