Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A novel colorimetric sensing platform for the detection of S. aureus with high sensitivity and specificity†
Yun Zhang‡
 *a,
Shuyou Shi‡a,
Jiajia Xingb,
Wenqing Tana,
Chenguang Zhanga,
Lin Zhangc,
Huan Yuana,
Miaomiao Zhanga and
Jinjuan Qiaod
*a,
Shuyou Shi‡a,
Jiajia Xingb,
Wenqing Tana,
Chenguang Zhanga,
Lin Zhangc,
Huan Yuana,
Miaomiao Zhanga and
Jinjuan Qiaod
aHenan Key Laboratory of Immunology and Targeted Therapy, Henan Collaborative Innovation Center of Molecular Diagnosis and Laboratory Medicine, School of Laboratory Medicine, Xinxiang Medical University, Xinxiang 453003, PR China. E-mail: zhangyun0126@126.com; Fax: +86 373 3029977; Tel: +86 373 3029977
bSchool of International Education, Xinxiang Medical University, Xinxiang 453003, PR China
cSchool of Innovation and Entrepreneurship, Xinxiang Medical University, Xinxiang 453003, PR China
dDepartment of Medical Laboratory, Weifang Medical University, Weifang 261053, PR China
First published on 18th October 2019
Abstract
In this study, a novel colorimetric sensing platform was developed for the detection of S. aureus using dog immunoglobulin G (IgG) as the capture antibody and chicken anti-protein A immunoglobulin Y labeled with horseradish peroxidase (HRP-IgY) as the detection antibody. Dog IgG labeled with magnetic beads was used to capture S. aureus through the interaction between the Fc region of dog IgG and Staphylococcal protein A (SPA). HRP-IgY was introduced to recognize the residual SPA on the surface of S. aureus and to create a sandwich format, after which a soluble 3,3′,5,5′-tetramethylbenzidine (TMB) substrate was added. A stop solution was utilized to cease the enzymatic chromogenic reaction, and then optical density was read at 450 nm. Under optimal conditions, the proposed method displayed a low detection limit of 1.0 × 103 CFU mL−1 and a wide linear range of 3.1 × 103 to 2.0 × 105 CFU mL−1. This detection method exhibited high specificity against other foodborne bacteria. The recovery rates ranged from 95.2% to 129.2%. To our knowledge, this is the first report to employ dog IgG and chicken IgY as an antibody pair to detect S. aureus. This technique exhibits high application potential for S. aureus monitoring in various kinds of samples.
Introduction
Staphylococcus aureus (S. aureus) is one of the most common food-borne pathogens, and can cause various infections ranging from scalded skin syndrome to life-threatening septicemia.1–3 Annually, S. aureus causes about 241![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cases of food-borne illnesses, which pose a significant threat to public health and food safety.4,5 It is therefore important to develop effective detection strategies for S. aureus.
000 cases of food-borne illnesses, which pose a significant threat to public health and food safety.4,5 It is therefore important to develop effective detection strategies for S. aureus.
To date, several detection strategies have been developed to monitor S. aureus. Among them, the conventional culture-based method is considered as the gold standard. This method relies on multiple steps to obtain a sufficient number of bacterial cells prior to biochemical assays.6,7 Although the culture-based method is simple, the total detection procedure time required is longer than 2 days, making it unsuitable for rapid diagnosis of S. aureus. Polymerase chain reaction (PCR)-based approaches have been utilized to detect specific genes of S. aureus with high sensitivity and specificity.8,9 However, they are limited by the complex matrix, long-term bacterial enrichment, low bacterial abundance and sample cross-contamination.10 Enzyme linked immunosorbent assay (ELISA) can couple the amplification effect of enzymatic reactions with selective antigen–antibody binding and increase the sensitivity and specificity of the assay. Due to its low-cost, high sensitivity and high throughput application, ELISA is the most frequently used method for antigen detection.11,12 Nevertheless, few ELISA-based methods have been used to detect S. aureus cells. This is likely due to the lack of suitable antibody pairs, which limit the development of specific immunoassays for S. aureus.13 Since mammalian monoclonal or polyclonal antibodies against S. aureus can also bind to protein G producing Streptococcus by interactions between the Fc region of mammalian Immunoglobulin G (IgG) and protein G,14 the specificity of detection is not guaranteed. Non-specific interactions of the bio-recognition element with target analytes lead to inaccurate results in the detection process.15 Thus, it is of particular significance to select a feasible antibody pair for an ELISA-based detection of S. aureus.
In the present study, a novel colorimetric sensing platform was developed using dog IgG and chicken IgY as an antibody pair to detect S. aureus. Primarily, Staphylococcal protein A (SPA) was used as an ideal detection marker of S. aureus, since SPA is only expressed in the cell wall of 99% S. aureus isolates and the number of SPA molecules is estimated to approximately 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in each bacterium.16–19 Dog IgG was utilized as the capture antibody that binds to S. aureus based on the strong affinity between the Fc region of dog IgG and SPA. Since dog IgG possesses strong binding capacity towards protein A and low binding capacity towards protein G,20,21 it can eliminate the interference from protein G producing Streptococcus to a certain extent. Moreover, chicken anti-protein A IgY was adopted as the detection antibody to further increase the specificity, because chicken anti-protein A IgY does not exhibit non-specific binding to protein G.22–25 Therefore, the proposed antibody pair could be used to improve the selectivity of ELISA.
000 in each bacterium.16–19 Dog IgG was utilized as the capture antibody that binds to S. aureus based on the strong affinity between the Fc region of dog IgG and SPA. Since dog IgG possesses strong binding capacity towards protein A and low binding capacity towards protein G,20,21 it can eliminate the interference from protein G producing Streptococcus to a certain extent. Moreover, chicken anti-protein A IgY was adopted as the detection antibody to further increase the specificity, because chicken anti-protein A IgY does not exhibit non-specific binding to protein G.22–25 Therefore, the proposed antibody pair could be used to improve the selectivity of ELISA.
Immunomagnetic beads based ELISA (IMBs-ELISA) is developed by combining ELISA with magnetic separation.26–28 IMBs can be easily separated from the reaction mixture with a magnet and re-dispersed immediately following removal of the magnet, which allow for a nearly “in solution” reaction.12,29 These characteristics render IMBs-ELISA more efficient and sensitive. Therefore, the aim of the present study was to establish an IMBs-ELISA method that could combine with the high sensitivity of IMBs-ELISA and the high specificity of the novel antibody pair. Dog IgG coated on magnetic beads (MBs) and horseradish peroxidase (HRP) conjugated chicken anti-protein A IgY were utilized to bind with SPA on the surface of S. aureus in a sandwich structure (as illustrated in Fig. 1). Following enzymatic amplification, the optical density was measured and a standard curve was generated for quantitative analysis of S. aureus. Moreover, the proposed method was applied to detect the number of bacteria in spiked samples and validate the potential application of this method in real samples.
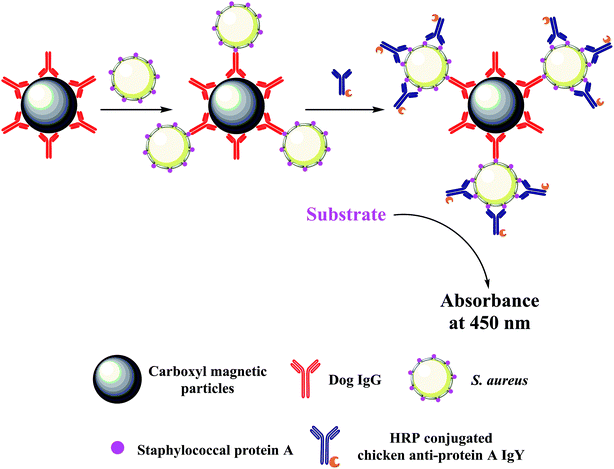 | ||
| Fig. 1 Schematic illustration of the novel colorimetric sensing platform for the detection of S. aureus (not to scale). | ||
Experimental
Reagents and materials
S. aureus (ATCC 12598), S. aureus (ATCC 25923), S. aureus (ATCC 139843), E. coli O157:H7 (EDL 933), Salmonella (ATCC 14028) and Listeria monocytogenes (ATCC 19115) were obtained from BeNa Culture Collection (Beijing, China). Streptococcus agalactiae (isolated from raw milk) was gifted from Prof. Hongping Wei, Wuhan Institute of Virology, Chinese Academy of Science. Carboxylated MBs with a 0.82 μm diameter (25 mg mL−1) were purchased from Spherotech (USA). N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) were bought from Aladdin Industrial Corporation (Shanghai, China). Dry powder of dog IgG (10 mg, dissolved in 10 mL of phosphate-buffered saline (PBS) (10 mmol L−1, pH 7.4)), streptavidin-Cy5.5 (1 mg mL−1) and Tween-20 were supplied by Solarbio (Beijing, China). HRP conjugated chicken anti-protein A IgY (HRP-IgY) (1 mg mL−1) was purchased from Abcam (England). Soluble 3,3′,5,5′-tetramethylbenzidine (TMB) substrate was procured from Biodragon Immunotechnologies (Beijing, China). Bovine serum albumin (BSA) and skim milk powder were purchased from Biosharp (Hefei, China) and BD Difco (USA), respectively. 96 well flat-bottomed microplates were obtained by Jet bio-filtration (Guangzhou, China). The aqueous solutions were prepared in ultrapure water (18.2 MΩ) obtained from Ulupure UPR-II-10T (Sichuan, China). All other chemicals and reagents purchased from commercial sources were of the highest purity grade. Orange juice and spring water were procured from a local supermarket, and human urine samples were collected from healthy volunteers. All experiments were performed in compliance with the relevant laws of China and institutional guidelines of Xinxiang medical university. Volunteers signed an informed consent approved by the ethics committee of Xinxiang medical university.Preparation and culture bacteria
S. aureus, E. coli O157:H7 and Salmonella were prepared by growing the stock cultures in Luria–Bertani broth medium, and Streptococcus agalactiae and Listeria monocytogenes were cultured in the brain heart infusion broth medium. Following incubation at 37 °C for 12 to 16 h, the bacteria were collected by centrifugation at 3000 rpm for 10 min and subsequently washed twice by sterile PBS (10 mmol L−1, pH 7.4). The bacterial precipitate was re-suspended in sterilized PBS (10 mmol L−1, pH 7.4) containing 20% glycerol and the bacterial solutions were stored at −20 °C until further use. The concentration of bacteria was determined by conventional colony counting method. The bacterial solutions were diluted 10-fold with sterile PBS and 100 μL of these samples were incubated with the corresponding medium agar at 37 °C overnight. The number of colonies formed on the plates was counted to calculate the bacterial concentration.Preparation of IMBs
Bio-conjugation of carboxyl groups of the MBs with the amino-groups of the dog IgG was achieved via a classic EDC/NHS amidization reaction.30 Briefly, 1 mg of MBs was separated using magnetic separation racks (Goldmag Biotech, China) to remove the storage buffer. The MBs were subsequently washed twice with PBS (pH 6.8) by a magnetic separation process. A total of 2 mg of EDC and 1 mg of NHS were dissolved in 200 μL of PBS (10 mmol L−1, pH 6.8) and were utilized to activate the carboxyl groups of the MBs. Following 30 min of activation, 100 μL of dog IgG (1 mg mL−1) was added to the MBs. The samples were incubated on a rotating platform (180 rpm) at 37 °C for 4 h to form MBs-dog IgG conjugates. Afterwards, 1 mL of 5% BSA and 15% skim milk was added to block non-specific sites on MBs for an additional 2 h and the samples were washed twice. Finally, the IMBs were re-suspended in 1 mL of PBS containing 1% BSA and stored at 4 °C.Detection procedure
A typical sandwich assay was performed in Eppendorf (EP) tubes fitted to the magnetic separation racks. Each incubation step was processed under shaking conditions (180 rpm) at 37 °C. 20 μL of IMBs (1 mg mL−1) was pipetted into each tube and then 100 μL of S. aureus solution was added. The mixture was incubated for 30 min and the beads were subsequently secured by placing the EP tube on the magnetic separation racks. Following removal of the supernatant, the beads were rinsed five times with PBST (PBS with 0.05% Tween-20). Subsequently, the MBs-dog IgG-S. aureus complex was dispersed in 100 μL of HRP-IgY (0.5 μg mL−1) and the entire mixture was rotated for 30 min. HRP-IgY formed a sandwich structure with S. aureus and MBs-dog IgG by an immune reaction. The immune sandwich complex was separated magnetically and the supernatant was discarded. The immune complex was rinsed five times with PBST to effectively remove unbound HRP-IgY. Thereafter, 100 μL of soluble TMB substrate was added into each EP tube, followed by incubation for 10 min. The reaction was terminated by the addition of 100 μL of stop solution (2 mol L−1 sulfuric acid). Following magnetic separation, 100 μL of the supernatant was transferred to the 96 well plate and the absorbance at 450 nm was determined by the Varioskan Flash multimode reader (Thermo Scientific, USA).Results and discussion
Optimization of experimental components
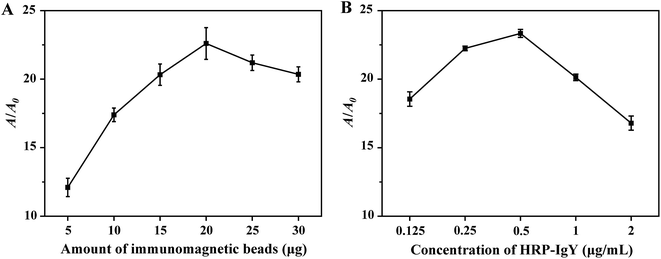
Initially, fluorescence microscopy was used to characterize the binding of dog IgG towards S. aureus. The latter was stained with fluorescence using streptavidin-Cy5.5 linked with biotin-dog IgG. The biotin–streptavidin reaction was used in order to demonstrate whether dog IgG could bind to S. aureus cells. The blue colour of the precipitate was noted at the bottom of the EP tubes following centrifugation, indicating the attachment of dog IgG to S. aureus (as shown in ESI Fig. S1A†). In addition, the stained bacteria were observed by fluorescence microscope (Pannoramic DESK P-MIDI, 3DHISTECH), and the fluorescence images illustrated strong red fluorescent emission from Cy5.5 on the cells of S. aureus, demonstrating the binding of dog IgG to S. aureus (as illustrated in ESI Fig. S1B†). Subsequently, the experimental components were optimized. In IMBs-ELISA, the dose of IMBs was a key determinant of the detection sensitivity of the proposed assay. Therefore, it was important to optimize the amount of IMBs added to the IMBs-ELISA. In Fig. 2, the parameters A and A0 represented the absorbance intensity at 450 nm for 5.0 × 104 CFU mL−1 S. aureus and for a blank of PBS, respectively. As shown in Fig. 2A, A and A0 were gradually increased with increasing amounts of IMBs. This may be attributed to non-specific binding of HRP-IgY onto IMBs caused by the increase in the amount of IMBs. Therefore, in order to obtain an optimal balance between high detection sensitivity and low nonspecific signal, optimal A/A0 was achieved at 20 μg of IMBs addition (as illustrated in ESI Fig. S2A†). 20 μg of IMBs was selected for the subsequent tests.The process of the colorimetric assay is mainly derived from HRP-catalyzed oxidation of TMB. For this reason, the concentration of HRP-IgY in the proposed method should be optimized. As illustrated in Fig. 2B, A and A0 were increased at higher HRP-IgY concentration. The increase in A0 was faster than that of A when the concentration of HRP-IgY was higher than 0.5 μg mL−1. This result suggested that an HRP-IgY concentration higher than 0.5 μg mL−1 exceeded the amount required for target capture, which led to nonspecific absorption of HRP-IgY onto IMBs, thereby increasing the background signal. As presented in ESI Fig. S2B,† a strong signal intensity with relatively lower background signal was recorded when the HRP-IgY concentration was 0.5 μg mL−1. Therefore, 0.5 μg mL−1 was determined to be the optimal concentration of HRP-IgY.
Detection of S. aureus in PBS
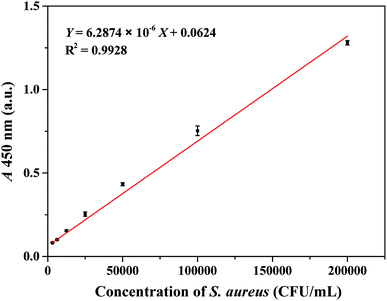
Under optimal conditions, a serial concentration of S. aureus was tested ranging from 3.1 × 103 to 2.0 × 105 CFU mL−1. As shown in Fig. 3, a calibration curve was developed based on the relationship between the different concentrations of S. aureus and the absorbance intensity at 450 nm. The linear regression equation was the following: Y = 6.2874 × 10−6X + 0.0624 (R2 = 0.9928), where Y and X represented the absorbance intensity (450 nm) of the reaction solution and the concentration of S. aureus (CFU mL−1), respectively. The detection limit (3σ, where σ is the standard deviation of PBS processed similarly as the S. aureus samples, n = 10, data were provided in ESI Table S1†) was calculated to be 1.0 × 103 CFU mL−1. The total detection procedure was completed in less than 90 min. The reproducibility of the colorimetric immunoassay was tested using 2.0 × 105 CFU mL−1 S. aureus. The variability data of 11 replicates were obtained. As listed in ESI Table S2,† the relative standard deviation (RSD) was 4.12%, which demonstrated that the assay exhibited optimal reproducibility and reliability for S. aureus detection.In order to test the robustness of the proposed method, the IMBs capturing S. aureus were performed in different pH values and PBS concentrations corresponding to the buffer solutions. As shown in ESI Fig. S3 and S4,† the IMBs exhibited ideal performance in the buffer solution over a wide range of the pH values (pH 4–9), and the PBS concentrations of the buffer solutions did not significantly affect the experimental results.
The satisfactory detection sensitivity was attributed to the following factors. MBs functioning as the immobile phase exhibited a large surface area, which resulted in the immobilization of a high number of antibodies and the efficient capture of S. aureus. In addition, S. aureus was captured by MBs through the interaction between SPA and dog IgG during the primary incubating step. A limited number of SPA molecules were occupied by antibodies due to steric hindrance between magnetic particles (∼0.82 μm) and S. aureus (∼0.8 μm). Given that one cell of S. aureus contains several SPA molecules and that the HRP-IgY antibodies exhibit relatively small molecular size, many residual SPA molecules could be recognized by HRP-IgY, thereby amplifying the colorimetric signal. The proposed IMBs-ELISA indicated comparable sensitivity with previous colorimetric methods for S. aureus detection.13,31–35 It possessed the advantages of not utilizing rare metals, complicated nanomaterial preparations, trained staff or sophisticated instruments, and all reagents were cheap and commercially available.
Detection of S. aureus in spiked samples
The effectiveness of the proposed detection system was evaluated on spiked samples. Briefly, stock solutions of S. aureus were diluted with orange juice, spring water and human urine, respectively. Standard curves were generated (as shown in ESI Fig. S5–S7†). The detection limits for orange juice, spring water and human urine were all estimated to 1.5 × 103 CFU mL−1, indicating that the sensitivity of this assay was sufficient to detect low concentration of bacteria in real samples.13,36,37 The results of the spike-recovery study were provided in Table 1. As seen from this table, the recovery percentage of S. aureus tested by the assay ranged from 95.2% to 129.2%. As listed in ESI Table S3,† the results obtained from the present method were consistent with those obtained from the ELISA method, which further demonstrated the suitability of the quantitative assay for testing various types of samples. These results demonstrated that the proposed method was robust and was effective in detecting S. aureus in real samples.| Samples | Added amount (CFU mL−1) | Found amount (CFU mL−1) | RSD (%) | Recovery (%) |
|---|---|---|---|---|
| a The sample was 5 times diluted.b The sample was 10 times diluted. | ||||
| Orange juice 1a | 6.3 × 103 | 8.1 × 103 | 5.0 | 129.2 |
| Orange juice 2a | 2.5 × 104 | 3.0 × 104 | 2.9 | 119.0 |
| Orange juice 3a | 1.0 × 105 | 1.1 × 105 | 7.3 | 106.4 |
| Spring water 1 | 6.3 × 103 | 6.3 × 103 | 0.8 | 100.3 |
| Spring water 2 | 2.5 × 104 | 2.4 × 104 | 1.9 | 95.2 |
| Spring water 3 | 1.0 × 105 | 1.1 × 105 | 2.0 | 111.4 |
| Human urine 1b | 6.3 × 103 | 6.5 × 103 | 7.9 | 103.6 |
| Human urine 2b | 2.5 × 104 | 2.6 × 104 | 3.4 | 104.0 |
| Human urine 3b | 1.0 × 105 | 1.0 × 105 | 4.7 | 101.9 |
Specificity
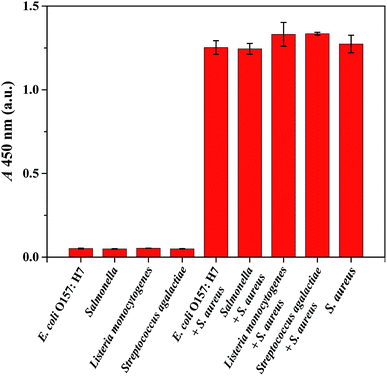
The specificity of the proposed IMBs-ELISA in detecting S. aureus was evaluated against various non-target pathogenic bacteria (E. coli O157:H7, Salmonella, Listeria monocytogenes and Streptococcus agalactiae). As illustrated in Fig. 4, S. aureus at 2.0 × 105 CFU mL−1 and its mixture with non-target pathogenic bacteria (all at 2.0 × 106 CFU mL−1) displayed a high intensity signal, while non-target pathogenic bacteria produced negative results since they lack protein A in their cell wall. Negligible interference from Streptococcus was observed since dog IgG has weak binding affinity towards protein G, and chicken anti-protein A IgY reacts solely with protein A. These results suggested high specificity of the assay for S. aureus as a result of the highly specific recognition between the novel antibody pair and S. aureus.SPA has been utilized as a detection marker for S. aureus in the agglutination test.38 Recently, several methods relying on the recognition of SPA have been put forwarded for the detection of S. aureus.17,32,39,40 For example, a chemiluminescent method for S. aureus detection was established by utilizing SPA-modified magnetic beads as competitive bacterial analogues and HRP labeled IgG as tracer, which could be completed in 50 min with a low detection limit of 6 CFU mL−1,17 while the presence of protein G producing Streptococcus would also result in positive detection signals. Dual-recognition strategies for S. aureus detection were based on vancomycin-affinity, utilizing IgG as the second recognition agent to bind with SPA.32,39 Vancomycin is a type of broad-spectrum antibiotics that can bind with multiple Gram-positive bacteria. Therefore these methods were expected to be prone to interference from Gram-positive bacteria. Chicken anti-protein A IgY was previously utilized as an antibody pair for the quantitative detection of S. aureus with high sensitivity and selectivity.40 Despite its desired selectivity, the consumption of chicken anti-protein A IgY that served as capture antibody was relatively high, resulting in higher cost of the test. To lower the test cost, dog IgG was introduced as a capture agent to detect S. aureus in the present study. However, there is a major limitation of these proposed SPA recognition-based methods is caused by the different expression levels of SPA genes noted among different strain types (as shown in ESI Fig. S8†). In addition, the different growth phases of S. aureus must be taken into consideration.41 In some cases, these SPA recognition based methods could be only used to detect S. aureus qualitatively.
Conclusion
Owing to the large number of SPA molecules on one S. aureus, dog IgG and HRP-IgY were used as the capture antibody and detection antibody, respectively, to develop a novel colorimetric sensing platform for highly sensitive and specific detection of S. aureus in this study. A low detection limit of 100 CFU of S. aureus in 100 μL of PBS without enrichment, with a linear range from 3.1 × 103 to 2.0 × 105 CFU mL−1 was obtained. If the enrichment of magnetic beads is considered, the detection limit can be much lower (10 CFU in 100 μL can be determined if the detection volume is 1 mL). The whole detection procedure was completed within 90 min. Moreover, the method built here circumvented the interference of protein G generating Streptococcus due to the advantage of the novel antibody pair being highly selective and no cross-reactivity with other food-borne bacteria. The proposed IMBs-ELISA method presents an important platform that can be applied to develop a commercial kit for S. aureus screening.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful for the financial support from the National Natural Science Foundation of China (Grant No. 81702099). We appreciated the collaboration of other members of our research group.References
- C. Zi, D. Zeng, N. Ling, J. Dai, F. Xue, Y. Jiang and B. Li, BMC Microbiol., 2018, 18, 132 CrossRef CAS.
- B. Esteban-Fernandez de Avila, M. Pedrero, S. Campuzano, V. Escamilla-Gomez and J. M. Pingarron, Anal. Bioanal. Chem., 2012, 403, 917–925 CrossRef CAS.
- R. H. Deurenberg and E. E. Stobberingh, Infect., Genet. Evol., 2008, 8, 747–763 CrossRef CAS.
- J. Kadariya, T. C. Smith and D. Thapaliya, BioMed Res. Int., 2014, 2014, 827965 Search PubMed.
- T. L. Harris, R. J. Worthington and C. Melander, Angew. Chem., 2012, 51, 11254–11257 CrossRef CAS.
- O. D. Hendrickson, V. D. Nikitushkin, A. V. Zherdev and B. B. Dzantiev, Arch. Microbiol., 2019, 201, 313–324 CrossRef CAS.
- J. Qiao, X. Meng, Y. Sun, Q. Li, R. Zhao, Y. Zhang, J. Wang and Z. Yi, J. Microbiol. Methods, 2018, 153, 92–98 CrossRef CAS.
- Y. M. Park, S. Y. Lim, S. J. Shin, C. H. Kim, S. W. Jeong, S. Y. Shin, N. H. Bae, S. J. Lee, J. Na, G. Y. Jung and T. J. Lee, Anal. Chim. Acta, 2018, 1027, 57–66 CrossRef CAS.
- A. Huletsky, R. Giroux, V. Rossbach, M. Gagnon, M. Vaillancourt, M. Bernier, F. Gagnon, K. Truchon, M. Bastien, F. J. Picard, A. van Belkum, M. Ouellette, P. H. Roy and M. G. Bergeron, J. Clin. Microbiol., 2004, 42, 1875–1884 CrossRef CAS.
- H. Zheng, F. Han, H. Lin, L. Cao, T. R. Pavase and J. Sui, Food Chem., 2019, 294, 468–476 CrossRef CAS.
- E. Engvall and P. Perlmann, J. Immunol., 1972, 109, 129–135 CAS.
- T. Alefantis, P. Grewal, J. Ashton, A. S. Khan, J. J. Valdes and V. G. Del Vecchio, Mol. Cell. Probes, 2004, 18, 379–382 CrossRef CAS.
- J. Yu, Y. Zhang, Y. Zhang, H. Li, H. Yang and H. Wei, Biosens. Bioelectron., 2016, 77, 366–371 CrossRef CAS PubMed.
- S. Yang, M. Wang, L. Wang and Z. Fu, Sens. Actuators, B, 2017, 252, 1003–1009 CrossRef CAS.
- M. Rubab, H. M. Shahbaz, A. N. Olaimat and D. H. Oh, Biosens. Bioelectron., 2018, 105, 49–57 CrossRef CAS.
- L. H. Xiong, R. Cui, Z. L. Zhang, X. Yu, Z. Xie, Y. B. Shi and D. W. Pang, ACS Nano, 2014, 8, 5116–5124 CrossRef CAS PubMed.
- J. Xiong, W. Wang, Y. Zhou, W. Kong, Z. Wang and Z. Fu, Microchim. Acta, 2016, 183, 1507–1512 CrossRef CAS.
- W. Hu, Y. Zhang, H. Yang, J. Yu and H. Wei, J. Microbiol. Methods, 2015, 115, 47–53 CrossRef CAS.
- J. Sjoquist, B. Meloun and H. Hjelm, Eur. J. Biochem., 1972, 29, 572–578 CrossRef CAS.
- D. D. Richman, P. H. Cleveland, M. N. Oxman and K. M. Johnson, J. Immunol., 1982, 128, 2300–2305 CAS.
- Millipore Sigma, https://www.sigmaaldrich.com/china-mainland/zh/technical-documents/articles/biology/affinity-chromatography-antibodies/protein-a-g-binding.html, accessed October 2019.
- N. Mizutani, Y. Sugita-Konishi, K. Omoe, K. Shinagawa, H. Kawakami, S. Kanno, K.-i. Sugiyama and Y. Kamata, Int. J. Food Sci. Technol., 2012, 47, 155–159 CrossRef CAS.
- P. K. Reddy, A. Shekar, J. J. Kingston, M. H. Sripathy and H. Batra, J. Immunol. Methods, 2013, 391, 31–38 CrossRef CAS.
- P. Reddy, S. Ramlal, M. H. Sripathy and H. V. Batra, J. Immunol. Methods, 2014, 408, 114–122 CrossRef CAS.
- W. Jin, K. Yamada, M. Ikami, N. Kaji, M. Tokeshi, Y. Atsumi, M. Mizutani, A. Murai, A. Okamoto, T. Namikawa, Y. Baba and M. Ohta, J. Microbiol. Methods, 2013, 92, 323–331 CrossRef CAS.
- Y. Zhang, C. Yan, H. Yang, J. Yu and H. Wei, Food Chem., 2017, 234, 332–338 CrossRef CAS PubMed.
- F. Zhao, Y. Tian, Q. Shen, R. Liu, R. Shi, H. Wang and Z. Yang, Talanta, 2019, 195, 55–61 CrossRef CAS.
- D. Cheng, M. Yu, F. Fu, W. Han, G. Li, J. Xie, Y. Song, M. T. Swihart and E. Song, Anal. Chem., 2015, 88, 820–825 CrossRef.
- S. Akbari Nakhjavani, B. Khalilzadeh, P. Samadi Pakchin, R. Saber, M. H. Ghahremani and Y. Omidi, Biosens. Bioelectron., 2018, 122, 8–15 CrossRef CAS.
- Y. Zhang, X. Si, M. Zhang, X. Yang, H. Yuan, X. Wang, Y. Zhang and H. Wang, Anal. Lett., 2019, 52, 602–612 CrossRef CAS.
- Y. J. Sung, H. J. Suk, H. Y. Sung, T. Li, H. Poo and M. G. Kim, Biosens. Bioelectron., 2013, 43, 432–439 CrossRef CAS.
- H. Gao, S. Yang, J. Han, J. Xiong, W. Kong, C. Li, G. Liao and Z. Fu, Chem. Commun., 2015, 51, 12497–12500 RSC.
- C. Yan, Y. Zhang, H. Yang, J. Yu and H. Wei, Talanta, 2017, 170, 291–297 CrossRef CAS.
- C. Z. Li, K. Vandenberg, S. Prabhulkar, X. Zhu, L. Schneper, K. Methee, C. J. Rosser and E. Almeide, Biosens. Bioelectron., 2011, 26, 4342–4348 CrossRef CAS.
- N. Verdoodt, C. R. Basso, B. F. Rossi and V. A. Pedrosa, Food Chem., 2017, 221, 1792–1796 CrossRef CAS.
- B. Hill, B. Smythe, D. Lindsay and J. Shepherd, Int. J. Food Microbiol., 2012, 157, 305–308 CrossRef.
- G. Walcher, M. Gonano, J. Kummel, G. C. Barker, K. Lebl, O. Bereuter, M. Ehling-Schulz, M. Wagner and B. Stessl, J. Dairy Res., 2014, 81, 462–470 CrossRef CAS.
- M. Wilkerson, S. McAllister, J. M. Miller, B. J. Heiter and P. P. Bourbeau, J. Clin. Microbiol., 1997, 35, 148–151 CAS.
- S. Yang, H. Ouyang, X. Su, H. Gao, W. Kong, M. Wang, Q. Shu and Z. Fu, Biosens. Bioelectron., 2016, 78, 174–180 CrossRef CAS.
- Y. Zhang, W. Tan, L. Zhang, S. Shi, Y. Niu, X. Yang, J. Qiao and H. Wang, Anal. Methods, 2019, 11, 3665–3670 RSC.
- J. J. Iangone, Adv. Immunol., 1982, 32, 157–252 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra05304b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |