Open Access Article
Open Access ArticleKnockdown of USP14 inhibits PDGF-BB-induced vascular smooth muscle cell dedifferentiation via inhibiting mTOR/P70S6K signaling pathway†
Hongbin Liua,
Xiaoliang Lia,
Guobei Yan *a and
Ruihua Lunb
*a and
Ruihua Lunb
aDepartment of Laboratory, The First Affiliated Hospital of Henan Polytechnic University, The Second People's Hospital of Jiaozuo, No. 17 of Minzhu South Road, Jiaozuo 454001, Henan, P. R. China. E-mail: yan_guobei3@126.com; Tel: +86-0391-2758507
bDepartment of Laboratory, Jiaozuo Maternal and Children's Hospital, Jiaozuo 454001, Henan, P. R. China
First published on 11th November 2019
Abstract
Atherosclerosis is a chronic progressive cardiovascular disease, which may result in many clinical consequences. Ubiquitin-specific protease 14 (USP14), a member of the USP family, has been found to be involved in cardiovascular disease. In the present study, we aimed to explore the role of USP14 in atherosclerosis. The results showed that USP14 expression was markedly increased in atherosclerotic tissues as compared to control tissues. Then we next examined the role of USP14 in primary human aortic smooth muscle cells (HASMCs) in response to PDGF-BB stimulation. The results demonstrated that PDGF-BB induced the USP14 expression in a dose- and time-dependent manner. Knockdown of USP14 in HASMCs suppressed PDGF-BB-induced proliferation and migration of HASMCs. The expressions of VSMCs markers including α-SMA, calponin and SM-MHC were markedly increased by knockdown of USP14, indicating that USP14 knockdown suppressed phenotypic modulation of HASMCs. However, USP14 overexpression exhibited the opposite effects. Furthermore, PDGF-BB-induced phosphorylation of mTOR and P70S6K in HASMCs was prevented by knockdown of USP14. In addition, MHY-1485, an activator of mTOR signaling, reversed the effects of USP14 knockdown on PDGF-BB-induced HASMCs. These data suggested that knockdown of USP14 prevented PDGF-BB-induced proliferation, migration, and phenotypic modulation of HASMCs via inhibiting the mTOR/P70S6K signaling pathway.
1. Introduction
Atherosclerosis is a chronic progressive cardiovascular disease that can result in many clinical consequences, such as myocardial infarction, ischaemic stroke and peripheral arterial disease.1,2 Therefore, atherosclerosis is considered as one of the major leading causes of death worldwide. Vascular smooth muscle cells (VSMCs), the major constituents in the media layer of medium- and large-sized arteries, are critical to maintain the integrity of the arterial wall.3 Studies on the mechanisms of atherogenesis have demonstrated that VSMCs are involved in the disease progression of atherogenesis.4,5 VSMCs participate in arterial wall remodeling in order to maintain blood flow in affected vessels due to atherosclerotic changes.Mature VSMCs is quiescent and characterized by a ‘contractile’ phenotype. VSMCs stably express several smooth muscle (SM) contractile proteins such as SMα-actin, SM22α, SM myosin heavy chain (MHC), H1-calponin and smoothelin, which are recognized as selective VSMCs markers.6 However, VSMCs has been found to exhibit phenotypic and functional plasticity in respond to vascular injury.7 VSMCs switch to a dedifferentiated, proliferative, and migratory phenotype. This ‘phenotypic switching’ has been considered of fundamental importance to the development of atherosclerosis.7 Calcium (Ca2+) signaling pathway is responsible for VSMC phenotype switch. Injury to the vessel wall is accompanied by VSMC phenotype switch from a contractile quiescent to a synthetic phenotype and by alteration of many components of VSMC Ca2+ signaling pathways.8 Therefore, preventing the generation of a pro-atherogenic phenotype in VSMCs is beneficial for the treatment of atherosclerosis.
Deubiquitinating enzymes (DUBs) play vital roles in the ubiquitin-specific proteases (USPs) by removing ubiquitin from substrate proteins and regulating their proteasomal degradation and sub-localization.9,10 The USPs family represents the majority of DUBs, containing at least 50 members. USP14, a member of USPs family, could induce deubiquitination of targeted proteins to stabilize the substrate protein.11 A previous study showed that USP14 modulates cancer cell motility by deubiquitinating the chemokine receptor CXC chemokine receptor 4 (CXCR4).12 In addition, it was reported that USP14 plays an important role in cardiovascular disease.13 However, the role of USP14 in atherosclerosis has not been reported.
In the present study, we investigated the role of USP14 in atherosclerosis in vitro. The results showed that USP14 is up-regulated in human atherosclerotic lesions and exerted a pro-atherosclerotic role in atherosclerosis.
2. Results
2.1 USP14 is highly expressed in atherosclerotic tissues
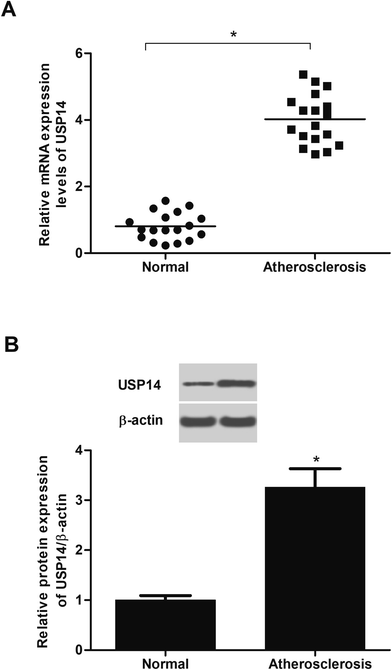
To identify the changes of USP14 expression in atherosclerotic tissues, the qRT-PCR and western blot assays were performed. The data in Fig. 1A showed that the RNA expression levels USP14 were significantly upregulated in atherosclerotic tissues compared with control tissues. Similarly, the protein expression levels were also increased (3.27 fold) in atherosclerosis tissues (Fig. 1B), which indicated a potential correlation between atherosclerosis and USP14.2.2 USP14 expression is increased in PDGF-BB-stimulated HASMCs
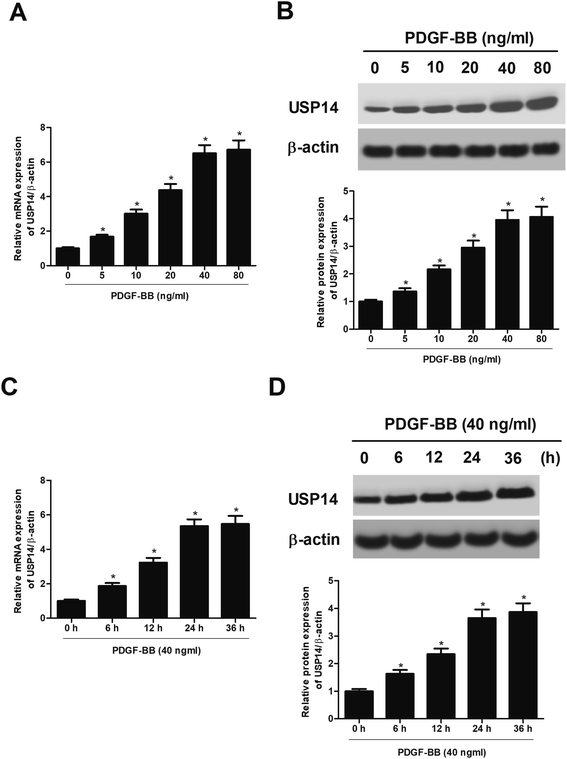
Then we sought to explore the cellular function of USP14 in HASMCs. The HASMCs were incubated with increasing concentrations of PDGF-BB (0, 5, 10, 20, 40, 80 ng ml−1) for 24 h. As indicated in Fig. 2A and B, treatment with 5–80 ng ml−1 of PDGF-BB caused gradual increase in USP14 expression at both mRNA and protein levels in HASMCs. Then the HASMCs were incubated with 40 ng ml−1 of PDGF-BB for a series of time periods (0, 6, 12, 24, 36 h). The data indicated that PDGF-BB induced USP14 expression at both mRNA and protein levels in HASMCs in a time-dependent manner (Fig. 2C and D). The concentration of 40 ng ml−1, and the time period of 24 h were selected for the following experiments.2.3 USP14 regulates the proliferation and migration of HASMCs exposed to PDGF-BB
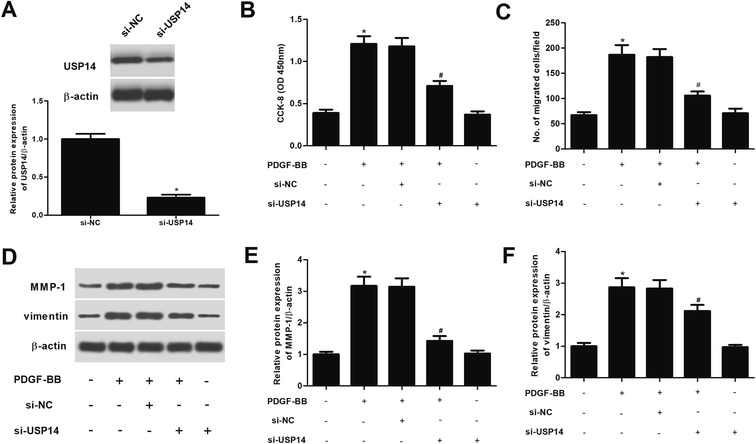
We therefore investigated the roles of USP14 in PDGF-BB- treated HASMCs. USP14 was knocked down in HASMCs through transfection with si-USP14. Western blot analysis showed that the si-USP14 significantly reduced USP14 protein levels to 23.4% of the si-NC group (Fig. 3A). Then CCK-8 and transwell assays were performed to evaluate cell proliferation and migration, respectively. The results proved that downregulation of USP14 significantly inhibited the proliferation and migration in PDGF-BB-induced HASMCs (Fig. 3B and C). In addition, we showed that knockdown of USP14 greatly suppressed the protein expression levels of MMP-1 and vimentin induced PDGF-BB in HASMCs (Fig. 3D). Furthermore, we observed that si-USP14 only had no significant change on cell proliferation/migration, MMP-1 and vimentin expressions in HASMCs (Fig. 3B–F). Then, we also examined the effects of USP14 overexpression on cell proliferation and migration of HASMCs, and the results indicated that overexpression of USP14 significantly promoted the proliferation and migration of HASMCs exposed to PDGF-BB (ESI Fig. 1†).2.4 USP14 regulates expression of VSMCs markers in PDGF-BB-stimulated HASMCs
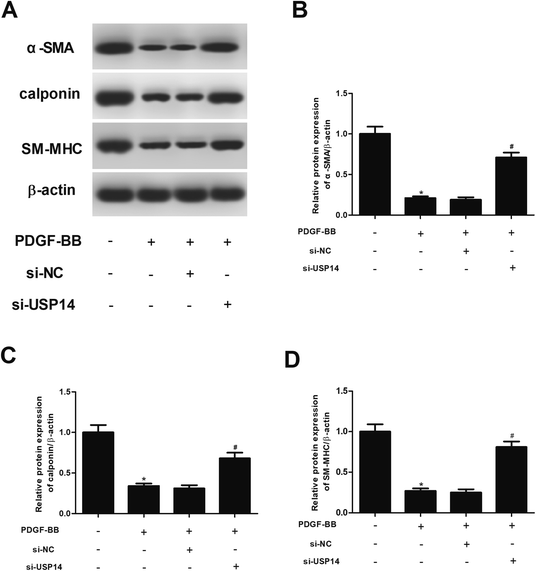
Then the expression levels of VSMCs markers including α-SMA, calponin and SM-MHC were measured using qRT-PCR and western blot analysis. As shown in Fig. 4A–D, the expressions of α-SMA, calponin and SM-MHC were significantly decreased after PDGF-BB treatment. However, the inhibitory effects of PDGF-BB on expressions of α-SMA, calponin and SM-MHC were mitigated by knockdown of USP14. In addition, these proteins expressions were not effect by si-USP14 only (ESI Fig. 2†). Furthermore, we found that overexpression of USP14 significantly inhibited the protein expressions of α-SMA, calponin and SM-MHC in HASMCs exposed to PDGF-BB (ESI Fig. 3†).2.5 USP14 regulates the activation of mTOR/P70S6K signaling in HASMCs in response to PDGF-BB
It has been demonstrated that mTOR/P70S6K plays an important role in the progress of atherosclerosis. We then evaluated the effect of USP14 on mTOR/P70S6K signaling pathway. Western blot analysis illustrated that PDGF-BB markedly induced the phosphorylation of mTOR and P70S6K in HASMCs, while USP14 knockdown suppressed the phosphorylation of mTOR and P70S6K in PDGF-BB-stimulated HASMCs (Fig. 5A–C). Moreover, si-USP14 only had no obvious effect on mTOR/P70S6K pathway in HASMCs (ESI Fig. 4†). Furthermore, we showed that USP14 knockdown also inhibited the phosphorylation of Akt in PDGF-BB-stimulated HASMCs (ESI Fig. 5†). Besides, overexpression of USP14 could induce the activation of mTOR/P70S6K signaling in HASMCs in response to PDGF-BB (ESI Fig. 6†). Previous studies reported that PSMD7 is a substrate of USP14,14 and PSMD7 downregulation induces apoptosis and suppresses tumorigenesis of esophageal squamous cell carcinoma via the mTOR/p70S6K pathway,15 thus, we speculated USP14 regulates mTOR/p70S6K pathway via the interaction with PSMD7. Co-IP assay verified the overlap between USP14 and PSMD7 proteins (Fig. 5D).2.6 Upregulation of mTOR signaling by MHY-1485 reversed the effects of si-USP14 on HASMCs
To further explore the role of mTOR/P70S6K signaling pathway in the effects of USP14, HASMCs were treated with MHY-1485 to activate the mTOR/P70S6K signaling. The results in Fig. 6A and B revealed that MHY-1485 treatment blocked the inhibitory effects of USP14 knockdown on proliferation and migration in HASMCs. In addition, the increased expression levels of α-SMA, calponin and SM-MHC caused by USP14 knockdown were reversed by MHY-1485 (Fig. 6C).2.7 Inhibitors of USP14 and mTOR/P70S6K pathway aggravated the effects of si-USP14 on HASMCs
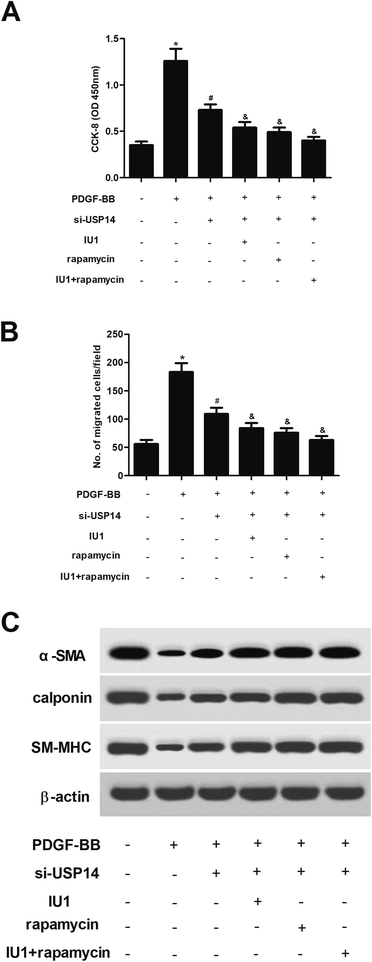
Next, HASMCs were treated with IU1 (a small-molecule inhibitor of USP14) or/and rapamycin (an mTOR inhibitor). The results showed that IU1 or/and rapamycin enhanced the inhibitory effects of USP14 knockdown on proliferation and migration in HASMCs (Fig. 7A and B). Moreover, the increased expression levels of α-SMA, calponin and SM-MHC caused by USP14 knockdown were enhanced by IU1 or/and rapamycin (Fig. 7C).3. Discussion
Ubiquitination is crucial process for protein degradation and thereby regulates a plethora of physiological processes, such as cell survival, differentiation, innate and adaptive immunity.16 Ubiquitin-proteasome system (UPS), the main cytosolic proteolytic system for ubiquitination, includes ubiquitin, ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2), ubiquitin ligase (E3), 26S proteasome and DUBs.17 It has been demonstrated that deregulation of the UPS is implicated in the pathogenesis of many human diseases, including cancer, cardiovascular disease and neurodegenerative disorders.18–20 USPs represent the major family of DUBs which remove ubiquitin from specific protein substrates, allowing the protein salvage from proteasome degradation, thereby regulating protein localization or activation.21 Previous studies have identified their involvement in several pathologies, thus, USPs are considered as potential target sites for drug discovery.22–24It has been demonstrated that USP14 is up-regulated in left ventricular tissues from abdominal aorta constriction (AAC) rat models.13 In addition, USP14 expression is elevated throughout the various cardiac hypertrophy stages, implying that USP14 may be associated with the pathogenesis of cardiac hypertrophy. Further in vitro investigations proved that USP14 expression is significantly increased in primary cardiac muscle cells after angiotensin II (Ang-II) treatment. USP14 overexpression in cardiomyocytes causes cardiomyocyte hypertrophy, while knockdown of USP14 in vitro alleviates the hypertrophic response.13 These findings suggested that USP14 is involved in cardiovascular disease. Therefore, we aimed to explore whether USP14 was implicated in atherogenesis. Firstly, we found that USP14 expression was markedly increased in atherosclerotic tissues. Then we next examined the role of USP14 in atherogenesis in vitro. According to the previous studies, phenotypic plasticity of VSMCs plays a crucial role in atherosclerosis.25 The capacity of VSMCs to switch the phenotype is regulated by various external signals. PDGF-BB is a member of PDGFs family, which can regulate cell proliferation, migration, extracellular matrix (ECM) accumulation, as well the production of inflammatory mediators.26 PDGF-BB is primarily released by vascular endothelial cells and platelets at the sites of vascular injury. PDGF-BB is considered to be one of the most potent mitogens and chemoattractant stimulants for the regulation of VSMCs proliferation, migration and phenotypic modulation.27,28 Therefore, PDGF-BB is usually applied for the induction of VSMCs in vitro.
Our study demonstrated that PDGF-BB significantly induced HASMCs proliferation, migration and phenotypic modulation. Moreover, the USP14 expression was also elevated after PDGF-BB treatment. Knockdown of USP14 in HASMCs suppressed PDGF-BB-induced proliferation, migration and phenotypic modulation of HASMCs, indicating that knockdown of USP14 might alleviate atherosclerosis. It is well established that PDGF-BB induces VSMCs dedifferentiation through several transcription factors and key molecular signaling pathways. The mTOR signaling pathway has been found to impact most major cellular functions.29 This signaling is observed to be activated in many diseases, including atherosclerosis.30 Pan et al.31 reported that PDGF-BB induces VSMCs dedifferentiation, which is associated with the mTOR signaling and its effectors P70S6K. Lu et al.32 demonstrated that chicoric acid prevents PDGF-BB-induced VSMCs dedifferentiation, which is attributed to the inhibition of mTOR/P70S6K signaling cascade. Therefore, we assessed the role of mTOR/P70S6K signaling in the effect of USP14 on PDGF-BB-induced HASMCs dedifferentiation. The results showed that PDGF-BB induced the phosphorylation of mTOR and P70S6K in HASMCs, indicating that PDGF-BB caused the activation of mTOR/P70S6K signaling. However, the PDGF-BB-induced activation of mTOR/P70S6K signaling was prevented by knockdown of USP14. In addition, upregulation of mTOR signaling by MHY-1485 reversed the effects of USP14 knockdown on HASMCs. These data suggested that suppression of mTOR/P70S6K signaling contributed to the effects of USP14 knockdown on PDGF-BB-induced HASMCs.
In conclusion, we identified USP14 as a pro-atherosclerotic factor through controlling the HASMCs proliferation, migration and phenotypic modulation. The effects of USP14 were mediated by the mTOR/P70S6K signaling pathway (ESI Fig. 7†). Collectively, USP14 might be a potential target for the treatment of atherosclerosis.
4. Materials and methods
4.1 Tissue specimens
All experiments were performed in accordance with the Guidelines of the First Affiliated Hospital of Henan Polytechnic University, the Second People's Hospital of Jiaozuo (Jiaozuo, China), and experiments were approved by the ethics committee at the First Affiliated Hospital of Henan Polytechnic University, the Second People's Hospital of Jiaozuo (approval ID: 00031). Informed consents were obtained from human participants of this study. Fresh atherosclerosis tissues were collected from 18 patients undergoing carotid endarterectomy at the Department of Vascular Surgery, the Second People's Hospital of Jiaozuo. 18 normal left internal mammary artery obtained from patients undergoing coronary artery bypass surgery were used as non-atherosclerotic control arteries. The atherosclerotic lesions and control tissues were frozen after collection and stored at −80 °C for future preparation of total RNA.4.2 Cell culture
Primary human aortic smooth muscle cells (HASMCs; ScienCell Research Laboratories, San Diego, CA, USA) were cultured in smooth muscle cell medium (SMCM; ScienCell Research Laboratories) containing 5% fetal bovine serum (FBS; Hyclone, Logan, UT, USA), 1% smooth muscle cell growth supplement (ScienCell Research Laboratories), 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (Sigma-Aldrich, St. Louis, MO, USA). Cells were incubated at 37 °C in 5% CO2/95% mixed ambient air.4.3 SiRNA-USP14 and cell transfection
USP14-target-specific siRNA (si-USP14) and negative-control siRNA (si-NC) were synthesized by OriGene (Rockville, MD, USA). Transfections were performed using Lipofectamine-2000 transfection system (Invitrogen, Carlsbad, CA, USA) as described in the instruction.4.4 Construction of the pcDNA3.1-USP14 vector and cell transfection
The full-length USP14 open reading frame was amplified from HASMCs by RT-PCR, and cloned into the pcDNA3.1 expression vector to construct the pcDNA3.1-USP14 recombinant expression vector. HASMCs were transfected with pcDNA3.1-USP14 or pcDNA3.1 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions.4.5 Cell proliferation assay
Totally of 2 × 103 cells per well were seeded in 96-well plates and incubated for 48 h. Subsequently, 10 μl of CCK-8 reagent was supplied and incubated for 4 h. The absorbance of each sample was determined at 450 nm with a multi-mode plate reader (Bio-Tek Instruments, Winooski, VT, USA).4.6 Cell migration assay
Cell migration ability was measured using transwell assay with Transwell chambers (8 μm pore size; Corning Inc., Corning, NY, USA). Briefly, 4 × 104 cells diluted in 100 μl serum-free medium were seeded into the upper transwell chamber. To induce chemotaxis, the lower chamber was filled with 800 μl medium containing 10% FBS. After incubation at 37 °C for 24 h, cells were fixed in 4% paraformaldehyde and stained with 1% crystal violet. The numbers of mitigated cells in five randomly selected fields were counted using an inverted microscope (Olympus, Tokyo, Japan).4.7 Real-time Quantitative PCR
Total RNA was isolated using the TRIzol reagent (Invitrogen). After determining the concentration, purity, and integrity of RNA, the RNA (1 μg) was reverse transcribed into cDNA using QuantiTect Reverse Transcription Kit (Qiagen, Hilde, Germany) according to the manufacturer's instructions. The mRNA levels of target genes were analyzed by qPCR using SYBR Green Real-time PCR Master Mix Kit (Bio-Rad, Hercules, CA, USA) on an iQ5 Real-time PCR System (Bio-Rad). The following primer pairs were used to PCR amplification: USP14, forward 5′- TGATGTGATGCAATCTGTG-3′, reverse 5′-ATCCTGCCCATTCTATTC-3’; β-actin, forward 5′-AAATCGTGCGTGACATCAAAGA-3′, and reverse 5′-GGC CATCTCCTGCTCGAA-3’. Relative levels of gene expression were calculated using the 2−ΔΔCT method.4.8 Western blot analysis
Total proteins were prepared with RIPA buffer (Thermo Fisher Scientific, Waltham, MA, USA) and then the protein concentration was determined using the BCA protein assay kit (Thermo). The western blot analysis was performed as previously described. Briefly, the proteins were incubated with primary antibodies (anti-USP14, anti-MMP-1, anti-vimentin, anti-α-SMA, anti-calponin, anti-SM-MHC, anti-mTOR, anti-p-mTOR, anti-P70S6K, anti-p-P70S6K, anti-p-Akt, anti-Akt or anti-β-actin) for 12 h at 4 °C, followed by incubation in HRP-conjugated secondary antibody. All antibodies were obtained from Abcam (Cambridge, MA, USA). The bands were enhanced with ECL chemiluminescent detection system reagents (Thermo) and detected using the chemidoc XRS imaging system (Bio-Rad).4.9 Coimmunoprecipitation (co-IP) assay
After washing with phosphate buffer saline (PBS), cells were lysed in RIPA buffer. Lysates were centrifuged, and then the supernatant was collected and incubated with primary antibody pre-absorbed protein A and G sepharose beads at 4 °C for 6 h. Then, the sediments were washed with the RIPA buffer and boiled in 1× loading buffer for 10 min. Protein expression was detected by western blot.4.10 Statistical analysis
Data were expressed as the mean ± standard of the mean (SEM) of three independent experiments (each performed in triplicate). The unpaired two-tailed Student's t-test and ANOVA with Bonferroni post hoc test were applied to identify the differences between two groups or multiple comparisons using SPSS 23.0 software (SPSS, Inc., Chicago, IL, USA). A p-value less than 0.05 was considered statistically significant.Conflicts of interest
The authors declare that there is no conflict of interest regarding the publication of this paper.Acknowledgements
I'd like to express my sincere thanks to all those who have lent me hands in the course of my writing this paper.References
- G. Liuzzo, Atherosclerosis: an inflammatory disease, Rays, 2001, 26, 221–230 CAS.
- J. D. Marsh, Pathobiology of atherosclerosis – A brief review, Semin. Thromb. Hemostasis, 2004, 30, 665–672 CrossRef.
- A. Frismantiene, M. Philippova, P. Erne and T. J. Resink, Smooth muscle cell-driven vascular diseases and molecular mechanisms of VSMC plasticity, Cell. Signalling, 2018, 52, 48–64 CrossRef CAS.
- T. Ira, G. C. A. Guillermo and G. K. Owens, Recent insights into the cellular biology of atherosclerosis, J. Cell Biol., 2015, 209, 13–22 CrossRef.
- M. R. Bennett, S. Sinha and G. K. Owens, Vascular Smooth Muscle Cells in Atherosclerosis, Circ. Res., 2016, 118, 692–702 CrossRef CAS.
- M. J. Zhang, Y. Zhou, L. Chen, Y. Q. Wang, X. Wang, Y. Pi, C. Y. Gao, J. C. Li and L. L. Zhang, An overview of potential molecular mechanisms involved in VSMC phenotypic modulation, Histochem. Cell Biol., 2016, 145, 119–130 CrossRef CAS.
- D. A. Chistiakov, A. N. Orekhov and Y. V. Bobryshev, Vascular smooth muscle cell in atherosclerosis, Acta Physiol., 2015, 214, 33–50 CrossRef CAS.
- S. J. House, M. Potier, J. Bisaillon, H. A. Singer and M. Trebak, The non-excitable smooth muscle: calcium signaling and phenotypic switching during vasculardisease, Pflügers Arch., 2008, 456, 769–785 CrossRef CAS.
- A. Wang, F. Zhu, R. Liang, D. Li and B. Li, Regulation of T cell differentiation and function by ubiquitin-specific proteases, Cell. Immunol., 2019, 103922 CrossRef CAS.
- M. J. Lee, B. H. Lee, J. Hanna, R. W. King and D. Finley, Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes, Mol. Cell. Proteomics, 2011, 10, R110 Search PubMed.
- R. K. Mialki, J. Zhao, J. Wei, D. F. Mallampalli and Y. Zhao, Overexpression of USP14 protease reduces I-kappaB protein levels and increases cytokine release in lung epithelial cells, J. Biol. Chem., 2013, 288, 15437–15441 CrossRef CAS.
- M. A. Mines, J. S. Goodwin, L. E. Limbird, F. F. Cui and G. H. Fan, Deubiquitination of CXCR4 by USP14 is critical for both CXCL12-induced CXCR4 degradation and chemotaxis but not ERK ativation, J. Biol. Chem., 2009, 284, 5742–5752 CrossRef CAS.
- N. Liu, R. Chai, B. Liu, Z. Zhang, S. Zhang, J. Zhang, Y. Liao, J. Cai, X. Xia, A. Li, J. Liu, H. Huang and S. Liu, Ubiquitin-specific protease 14 regulates cardiac hypertrophy progression by increasing GSK-3beta phosphorylation, Biochem. Biophys. Res. Commun., 2016, 478, 1236–1241 CrossRef CAS.
- B. Liu, S. Jiang, M. Li, X. Xiong, M. Zhu, D. Li, L. Zhao, L. Qian, L. Zhai, J. Li, H. Lu, S. Sun, J. Lin, Y. Lu, X. Li and M. Tan, Proteome-wide analysis of USP14 substrates revealed its role in hepatosteatosis via stabilization of FASN, Nat. Commun., 2018, 13, 4770–4781 CrossRef.
- K. Shi, J. Z. Zhang, R. L. Zhao, L. Yang and D. Guo, PSMD7 downregulation induces apoptosis and suppresses tumorigenesis of esophageal squamous cell carcinoma via the mTOR/p70S6K pathway, FEBS Open Bio, 2018, 7, 533–543 CrossRef PubMed.
- D. Popovic, D. Vucic and I. Dikic, Ubiquitination in disease pathogenesis and treatment, Nat. Med., 2014, 20, 1242–1253 CrossRef CAS PubMed.
- T. Nam, J. H. Han, S. Devkota and H. W. Lee, Emerging Paradigm of Crosstalk between Autophagy and the Ubiquitin-Proteasome System, Mol. Cells, 2017, 40, 897–905 CAS.
- C. W. Olanow and K. S. Mcnaught, Ubiquitin-proteasome system and Parkinson's disease, Mov. Disord., 2006, 21, 1806–1823 CrossRef.
- F. Wang, A. Lerman and J. Herrmann, Dysfunction of the ubiquitin-proteasome system in atherosclerotic cardiovascular disease, Am. J. Cardiovasc. Dis., 2015, 5, 83–100 CAS.
- M. Shen, S. Schmitt, D. Buac and Q. P. Dou, Targeting the ubiquitin-proteasome system for cancer therapy, Cancer Sci., 2013, 17, 1091–1108 CAS.
- H. Zhou, J. Zhao, J. Cai and S. B. Patil, Ubiquitin-specific proteases function in plant development and stress responses, Plant Mol. Biol., 2017, 94, 565–576 CrossRef CAS PubMed.
- L. Daviet and F. Colland, Targeting ubiquitin specific proteases for drug discovery, Biochimie, 2008, 90, 270–283 CrossRef CAS.
- A. Pal and N. J. Donato, Ubiquitin-specific proteases as therapeutic targets for the treatment of breast cancer, Breast Cancer Res., 2014, 16, 461–468 CrossRef.
- A. Pal, M. A. Young and N. J. Donato, Emerging Potential of Therapeutic Targeting of Ubiquitin-Specific Proteases in the Treatment of Cancer, Cancer Res., 2014, 74, 4955–4966 CrossRef CAS PubMed.
- M. Davies, J. Harman, H. Yu, M. Bennett and H. Jørgensen, The epigenetic phenotypic switch of vascular smooth muscle cells involved in atherosclerosis, Lancet, 2013, 381, S34 CrossRef.
- C. Betsholtz, Biology of platelet-derived growth factors in development, Birth Defects Res. C. Embryo Today, 2003, 69, 272–285 CrossRef CAS.
- E. S. Park, K. P. Lee, S. H. Jung, D. Y. Lee, K. J. Won, Y. P. Yun and B. Kim, Compound K, an intestinal metabolite of ginsenosides, inhibits PDGF-BB-induced VSMC proliferation and migration through G1 arrest and attenuates neointimal hyperplasia after arterial injury, Atherosclerosis, 2013, 228, 53–60 CrossRef CAS.
- K. Kingsley, J. L. Huff, W. L. Rust, K. Carroll, A. M. Martinez, M. Fitchmun and G. E. Plopper, ERK1/2 mediates PDGF-BB stimulated vascular smooth muscle cell proliferation and migration on laminin-5, Biochem. Biophys. Res. Commun., 2002, 293, 1000–1006 CrossRef CAS.
- Q. Yang and K. L. Guan, Expanding mTOR signaling, Cell Res., 2007, 17, 666–681 CrossRef CAS.
- A. Soltani, A. Bahreyni, N. Boroumand, M. K. Roshan, M. Khazaei, M. Ryzhikov, S. Soleimanpour, A. Avan and S. M. Hassanian, Therapeutic potency of mTOR signaling pharmacological inhibitors in the treatment of proinflammatory diseases, current status and perspectives, J. Cell. Physiol., 2017, 233, 4783–4790 CrossRef.
- S. Pan, H. Lin, H. Luo, F. Gao, L. Meng, C. Zhou, C. Jiang, Y. Guo, Z. Ji, J. Chi and H. Guo, Folic acid inhibits dedifferentiation of PDGF-BB-induced vascular smooth muscle cells by suppressing mTOR/P70S6K signaling, Am. J. Transl. Res., 2017, 9, 1307–1316 CAS.
- Q. B. Lu, M. Y. Wan, P. Y. Wang, C. X. Zhang, D. Y. Xu, X. Liao and H. J. Sun, Chicoric acid prevents PDGF-BB-induced VSMC dedifferentiation, proliferation and migration by suppressing ROS/NFkappaB/mTOR/P70S6K signaling cascade, Redox Biol., 2018, 14, 656–668 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra04726c |
| This journal is © The Royal Society of Chemistry 2019 |