Open Access Article
Open Access ArticleAstragali radix total flavonoid synergizes cisplatin to inhibit proliferation and enhances the chemosensitivity of laryngeal squamous cell carcinoma†
Jiajia Cui‡
ah,
Xiwang Zheng‡ah,
Dongli Yang‡ab,
Yinghuan Huah,
Changming Anc,
Yunfeng Bod,
Huizheng Lif,
Yuliang Zhangah,
Min Niuah,
Xuting Xueah,
Yan Lug,
Yemei Tangab,
Hongyu Yinab,
Zhenyu Li *eh,
Wei Gao
*eh,
Wei Gao *abh and
Yongyan Wu
*abh and
Yongyan Wu *abh
*abh
aShanxi Key Laboratory of Otorhinolaryngology Head and Neck Cancer, First Hospital of Shanxi Medical University, Taiyuan 030001, Shanxi, China. E-mail: wuyongyan@sxent.org; gaoweisxent@sxent.org
bDepartment of Otolaryngology Head & Neck Surgery, First Hospital of Shanxi Medical University, Taiyuan 030001, Shanxi, China
cDepartment of Head and Neck Surgery, Cancer Hospital, National Cancer Center, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing 100021, China
dDepartment of Pathology, Shanxi Cancer Hospital, Shanxi Medical University, Taiyuan 030001, Shanxi, China
eModern Research Center for Traditional Chinese Medicine, Shanxi University, Taiyuan 030001, Shanxi, China. E-mail: lizhenyu@sxu.edu.cn
fDepartment of Otolaryngology Head & Neck Surgery, Dalian Municipal Friendship Hospital, Dalian 116001, Liaoning, China
gDepartment of Otolaryngology Head & Neck Surgery, The First Hospital, Jinzhou Medical University, Jinzhou 121001, Liaoning, China
hThe Key Scientific and Technological Innovation Platform for Precision Diagnosis and Treatment of Head and Neck Cancer, Shanxi Province, Taiyuan 030001, Shanxi, China
First published on 7th August 2019
Abstract
Laryngeal squamous cell carcinoma (LSCC) is the most common head and neck cancer. Astragali radix extracts play crucial roles in the regulation of cancer progression. However, the role of Astragali radix extracts in LSCC and the related mechanisms remains unclear. Here, we evaluated the inhibitory effects of the combined use of Astragali radix total flavonoid (TFA) and cisplatin (CDDP) on an LSCC mouse model by pharmacodynamics. Ultra-high-performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS) was employed to define the prototype of TFA in vivo. The potential drug targets were identified through the integrative analysis of LSCC microarrays, RNA sequencing data and the main bioactive component of TFA. Furthermore, a protein–protein interaction network, compound–target network and target–pathway network were constructed based on the prototype and potential drug targets to identify the main targets and pathways. Animal experiments showed that TFA has significant synergistic antitumor activity with cisplatin and attenuates the nephrotoxicity caused by CDDP chemotherapy, improving the survival of LSCC-bearing mice. Using UPLC-MS/MS, we identified 8 constituents of TFA in experimental mice serum: formononetin, ononin, calycosin, calycosin-7-O-β-D-glucoside, 7,2′-dihydroxy-3′,4′-dimethoxyisoflavan, 7,2′-dihydroxy-3′,4′-dimethoxyisoflavaneglucoside, 3-hydroxy-9,10-dimethoxypterocarpan and 9,10-dimethoxyptercarpan-3-O-β-D-glucoside. Integrative analysis predicted 19 target genes for TFA constituents, and the target genes were mainly involved in the EGFR-related cancer signaling, metabolism and oxidative stress. Collectively, these findings highlight the role of TFA in the regulation of LSCC and provide potential targets for a high-efficiency and low-toxicity therapeutic strategy of LSCC.
1 Introduction
LSCC is the most common head and neck squamous cell carcinoma (HNSCC).1 Despite recent advances in oncological and surgical treatments, the prognosis of advanced LSCC remains extremely poor.2 Hence, chemotherapy becomes an ideal adjuvant treatment for LSCC. Cisplatin (CDDP) as a first-line agent or after being combined with paclitaxel or carboplatin has been used for LSCC treatment.3,4 However, the severe adverse reactions of systemic chemotherapy are unavoidable because of poor chemosensitivity and increased cytotoxicity effects due to multidrug combinations.5 Therefore, the key point to improve the prognosis of LSCC is the development of a therapeutic strategy with enhanced tumor sensitivity to chemotherapy and low toxicity to normal cells.Medicinal plants and extracts have been used as chemotherapeutic agents, chemopreventive agents or chemosensitizer agents in cancer.6–8 Astragali radix (AR) has been used for centuries as a reinforcing vital energy herb in Chinese medicine.9 Recent studies link AR with cancer proliferation, anti-oxidation, relief of complications in cardiovascular diseases and chemopreventive efficiency.9–11 AR extracts show protective effects during oxaliplatin-induced lipid peroxidation, protein carbonylation, and DNA oxidation.12 Flavonoids are the main types of bioactive substances in AR extracts. Flavonoids inhibit multidrug resistance and have excellent safety and tolerability.13,14 Hence, we proposed that Astragali radix total flavonoid (TFA) can sensitize LSCC to chemotherapy.
In this study, we evaluated the effects of TFA and CDDP on an LSCC xenograft model. UPLC-MS/MS identified bioactive ingredients in the serum of TFA-treated mice. The target genes of TFA and the related signaling pathways were investigated by the integrative analysis of the TFA bioactive ingredients, LSCC microarrays and RNA sequencing data. In addition, we constructed a protein–protein interaction (PPI) network, compound–target (C–T) network and target–pathway (T–P) network to reveal the possible mechanisms of the TFA synergistic anti-tumor effect.
2 Methods
2.1 Extract preparation
Astragali Radix was purchased from Shanxi Datong Materials Market (Hunyuan, China) on September 2016 and identified as the roots of A. membranaceus var. mongholicus by Professor Zhenyu Li. The preparation and chemical composition characterization of the TFA extract have been reported in our previous study.15 Briefly, 2.5 kg of Astragali Radix was smashed and soaked in distilled water at room temperature and then extracted by refluxing (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, w/v) first for 2 h and then (1
10, w/v) first for 2 h and then (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8, w/v) for 2 h. The filtrates were combined and concentrated in a rotary evaporator. The concentrated solution was further separated using an AB-8 macroporous resin, which was eluted sequentially with distilled water, 10% ethanol, and 35% ethanol. The 35% ethanol eluate was concentrated and portioned three times with ethyl acetate to afford TFA. Next, nuclear magnetic resonance and LC-MS were performed to determine the amount of flavonoids in TFA, and the content of flavonoids in TFA used in this study was found to be 40–60%.
8, w/v) for 2 h. The filtrates were combined and concentrated in a rotary evaporator. The concentrated solution was further separated using an AB-8 macroporous resin, which was eluted sequentially with distilled water, 10% ethanol, and 35% ethanol. The 35% ethanol eluate was concentrated and portioned three times with ethyl acetate to afford TFA. Next, nuclear magnetic resonance and LC-MS were performed to determine the amount of flavonoids in TFA, and the content of flavonoids in TFA used in this study was found to be 40–60%.
2.2 Cell culture and reagents
The human LSCC cell line HEp-2 (China Center for Type Culture Collection, Wuhan, China) was maintained in DMEM supplemented with 10% fetal bovine serum (Biological Industries, CT), 100 U mL−1 penicillin, and 0.1 mg mL−1 streptomycin in a humidified incubator with 5% CO2 at 37 °C.Cisplatin (Selleck, Houston, TX) was dissolved in 0.9% saline to make a 0.5 mM stock solution. The reference standards formononetin, calycosin-7-O-β-D-glucoside, 7,2′-dihydroxy-3′,4′-dimethoxyisoflavan, 7,2′-dihydroxy-3′,4′-dimethoxyisoflavaneglucoside, 3-hydroxy-9,10-dimethoxypterocarpan and 9,10-dimethoxyptercarpan-3-O-β-D-glucoside were obtained from Shanghai Everlasting Biotechnology Co. Ononin and calycosin were obtained from Chengdu Manst Co. LC-MS-grade acetonitrile and formic acid were obtained from Thermo Fisher (USA). Ultrapure water (18.2 MΩ) was supplied with a Milli-Q system (Millipore, France). Other reagents were of analytical grade.
2.3 Animal experiments
Animal experiments followed the Health Guide for the Care and Use of Laboratory Animals and were approved by the medical ethics committee of Shanxi Medical University. Specific pathogen-free (SPF)-grade female BALB/C nude mice (6–8 week-old) were purchased from Beijing Vital River Laboratory Animal Technology corporation. All mice were housed in isolation and ventilation cages in a climate-controlled room with 12 h light–dark illumination cycles at 23 ± 1.5 °C and 45 ± 15% humidity and were maintained in the laboratory for 1 week and used for further experiments if no abnormal behaviors were observed.2.4 Therapeutic studies in xenografts
For xenograft experiments, 2 × 106 HEp-2 cells were subcutaneously injected into the axilla of the right forelimb (0.2 mL per mouse) of 36 nude mice. The control group ([C], n = 6) had free access to normal chow and water. Tumor sizes were measured every 2 days by using a caliper, and the volume was calculated by L × W2 × 0.5, where L and W represent long diameter and short diameter, respectively. When tumors reached 350 mm2, the mice were randomly divided into 5 groups (n = 6) for treatment: (1) low-dose group (12 mg per kg per day TFA + 1 mg per kg per day CDDP, intraperitoneally [ip]); (2) medium-dose group (24 mg per kg per day TFA + 1 mg per kg per day CDDP, ip); (3) high-dose group (48 mg per kg per day TFA + 1 mg per kg per day CDDP, ip); (4) positive control group (CDDP, 1 mg per kg per day for 20 days); and (5) the model (M) group (given equal amounts of saline). Body weights were measured every 2 days during the experimental period. Mice were killed, and the tumors were harvested, weighed, and photographed at endpoint. The excised organs including liver, kidneys, spleen, lungs and heart of each mouse were trimmed of any adherent tissue and weighed. Gross necropsy was performed for all dissected organs and tissues.2.5 Prototype identification of TFA in vivo
Three SPF-grade subcutaneous tumor-bearing nude mice were randomly numbered 1, 2 and 3. The mice were allowed free access to water and fasted for 12 h before the experiment. The control serum samples were taken from the eyeballs of mice 1 and 2 after anesthesia. We intraperitoneally injected mouse 3 with TFA combined with CDDP (48 mg kg−1 TFA and 1 mg kg−1 CDDP, ip). At 30 min after administration, serum was taken from eyeballs. The supernatants were stored at −80 °C.2.6 Assessment of tumor response
The growth curve and tumor growth inhibition rate [R = (1 − T/C) × 100%] were used to evaluate the effects of each treatment, where T and C represent the mean tumor weights (mg) of treated groups and the model group (M), respectively. R ≥ 40% is considered significant antitumor activity.162.7 Hematoxylin–eosin (HE) staining
All tissues were fixed with 4% paraformaldehyde for 24 h; then, they were embedded in paraffin, sectioned at 3 μm, and stained with HE. Finally, the sections were reviewed by a pathologist to confirm the diagnosis.2.8 Immunohistochemistry (IHC) analysis
Tissue sections were de-paraffinized and heated and antigen retrieval with citrate buffer solution (pH 7.4) was carried out using high pressure. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide, followed by blocking with normal goat serum for 40 min at room temperature. Anti-Ki67 (MAX, Ready-to-use) antibody were incubated overnight at 4 °C and then incubated with biotin-conjugated goat anti-rabbit/mouse IgG secondary antibody for 30 min at room temperature. Visualization was performed with a DAB detection system. Slides were scanned on a Panoramic digital slide scanner.2.9 TFA prototype identification by UPLC-MS/MS
2.10 Chromatography and MS conditions
The Thermo Fisher U3000 system was fitted with a Waters HSS T3 column (2.1 × 100 mm, 1.7 μm, Waters, USA) with an ESI ion source. The elution procedure used was previously reported by Zhang WN.15 The Z-spray ionization source was maintained at 300 °C with spray voltage of 3.5 kV in the positive ionization mode. Capillary temperature 320 °C, lens voltage 55 kPa, mass resolution 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, and mass scan range 80–1500 m/z in the full-scan mode were maintained. The exact mass of the compounds was calculated by using Qual browser in Xcalibur 2.3.l (Thermo Fisher).
000, and mass scan range 80–1500 m/z in the full-scan mode were maintained. The exact mass of the compounds was calculated by using Qual browser in Xcalibur 2.3.l (Thermo Fisher).
2.11 System pharmacology analysis
2.12 Statistical analysis
Data are presented as mean ± SD. Differences were determined by two-tailed Student's t-test or ANOVA test by SPSS 22.0 (Chicago, IL, USA), with two-tailed P < 0.05 considered significant.3 Results
3.1 TFA synergizes with CDDP to inhibit tumor growth in LSCC mouse xenografts
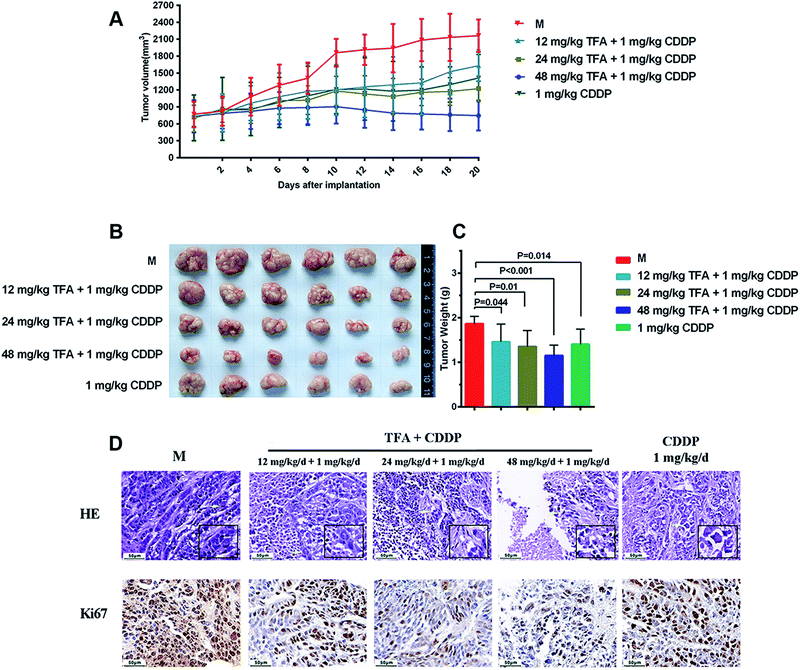
The effect of TFA combined with CDDP in vivo was evaluated using a xenograft model. After a combined treatment with TFA and CDDP, the volumes of the HEp-2 tumors dose-dependently decreased as compared with that for the model group (M) (Fig. 1A). In addition, as compared with CDDP alone, the co-administration of 48 mg kg−1 TFA and CDDP remarkably inhibited tumor growth.Compared with the results for the model group, the mean tumor size and weight for the TFA- and CDDP-treated groups or for the group treated with only CDDP were reduced remarkably (Fig. 1B and C). The tumor inhibition rate was higher with 24 mg kg−1 TFA + 1 mg kg−1 CDDP and 48 mg kg−1 TFA + 1 mg kg−1 CDDP compared to that with CDDP alone (29.3% and 40.2% vs. 26.8%, respectively) (Table 1).
| Group | Mean tumor weight (g) | R |
|---|---|---|
| a Data are mean ± SD. Compared with M group, *P < 0.05, **P < 0.01. TFA, Astragali radix total flavonoid; CDDP, cisplatin. | ||
| Model group (M) | 1.881 ± 0.179 | — |
| 12 mg kg−1 TFA + 1 mg kg−1 CDDP | 1.445 ± 0.38* | 23.1% |
| 24 mg kg−1 TFA + 1 mg kg−1 CDDP | 1.329 ± 0.337* | 29.3% |
| 48 mg kg−1 TFA + 1 mg kg−1 CDDP | 1.108 ± 0.284** | 40.2% |
| 1 mg kg−1 CDDP | 1.378 ± 0.321* | 26.8% |
The HE staining results revealed that the combination of TFA and CDDP induced higher tumor cell necrosis as compared with CDDP alone (Fig. 1D). Moreover, we noted that Ki67 positive cells substantially decreased in 48 mg kg−1 TFA and 1 mg kg−1 CDDP-treated groups as compared to that in the group treated with CDDP alone. Taken together, we inferred that TFA enhanced LSCC sensitivity to CDDP in vivo in a dose-dependent manner.
3.2 TFA improved the survival state of CDDP-treated mice
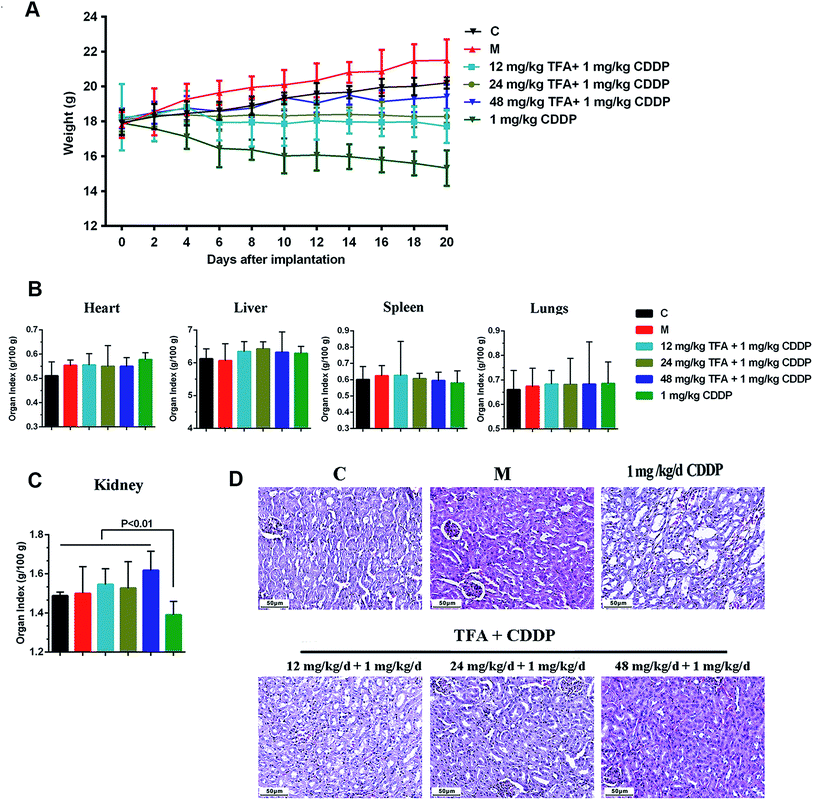
To investigate the safety of TFA and CDDP combination, we assessed general behavior, body weight, organ coefficients and pathological morphology. Strikingly, 6 groups showed significant differences in body weights although there was no animal death during the experimental period (Fig. 2A). The loss of body weight is a typical feature with CDDP chemotherapy. In the co-administration group, TFA improved the mouse body weight in a dose-dependent manner.Next, we investigated the toxicity of drugs on the animals. The renal index in the group treated with CDDP alone was significantly lower than that with other treatments (P < 0.01), and the addition of TFA significantly improved the renal index (Fig. 2C). However, the organ indices of heart, liver, spleen and lung showed no significant change among different groups (Fig. 2B). The HE staining results showed that the kidney of the mouse treated with CDDP alone had serious tubular damage, mainly in the corticomedullary junction, characterized by tubular dilation, necrosis, vacuolization and formation of hyaline casts, with no effect on glomeruli (Fig. 2D). The co-treatment with TFA reduced the number of hyaline casts and the extent of tubular necrosis induced by CDDP in the corticomedullary area (Fig. 2D). Thus, TFA attenuated the nephrotoxicity caused by CDDP chemotherapy.
3.3 Identification of prototype ingredients of TFA
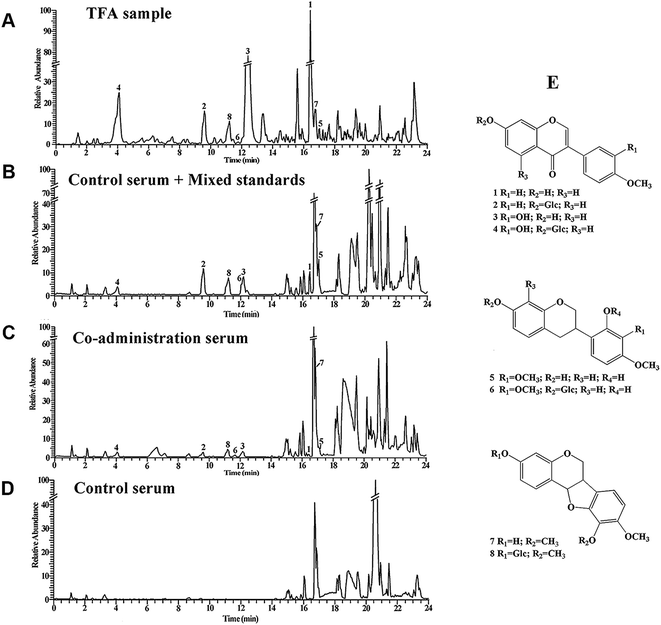
We further detected the potential bioactive ingredients in the serum of TFA-treated mice. Fig. 3A–D show the base peak intensity chromatograms of the TFA samples (Fig. 3A), control serum samples with mixed standard products (Fig. 3B), serum samples of 48 mg kg−1 TFA and 1 mg kg−1 CDDP (ip) (Fig. 3C) and control serum samples in positive ion modes (Fig. 3D). As a result, MS analysis identified 8 constituents in the serum of TFA-treated mice: formononetin (named 1), ononin (named 2), calycosin (named 3), calycosin-7-O-β-D-glucoside (named 4), 7,2′-dihydroxy-3′,4′-dimethoxyisoflavan (named 5), 7,2′-dihydroxy-3′,4′-dimethoxyisoflavaneglucoside (named 6), 3-hydroxy-9,10-dimethoxypterocarpan (named 7), and 9,10-dimethoxyptercarpan-3-O-β-D-glucoside (named 8). The detailed information for these constituents is listed in Table 2.| No. | Name | TR (min) | m/z | Fragments |
|---|---|---|---|---|
| a TR: retention time. | ||||
| 1 | Formononetin | 16.35 | 269.0808 | 254.0571, 237.0545 |
| 2 | Ononin | 9.60 | 431.1341 | 269.0810, 254.0573 |
| 3 | Calycosin | 12.40 | 285.0759 | 270.0529, 253.0494 |
| 4 | Calycosin-7-O-β-D-glucoside | 4.01 | 447.1287 | 285.0760, 279.0522 |
| 5 | 7,2′-Dihydroxy-3′,4′-dimethoxyisoflavane | 17.02 | 303.1219 | 193.0860, 181.0860, 167.0703 |
| 6 | 7,2′-Dihydroxy-3′,4′-dimethoxyisoflavaneglucoside | 12.19 | 465.1755 | 303.1225, 167.0703 |
| 7 | 3-Hydroxy-9,10-dimethoxypterocarpan | 16.70 | 301.1070 | 213.0907, 167.0704 |
| 8 | 9,10-Dimethoxypterocarpan-3-O-β-D-glucoside | 11.15 | 463.1596 | 301.1070 |
3.4 Potential targets of TFA can sensitize LSCC to CDDP
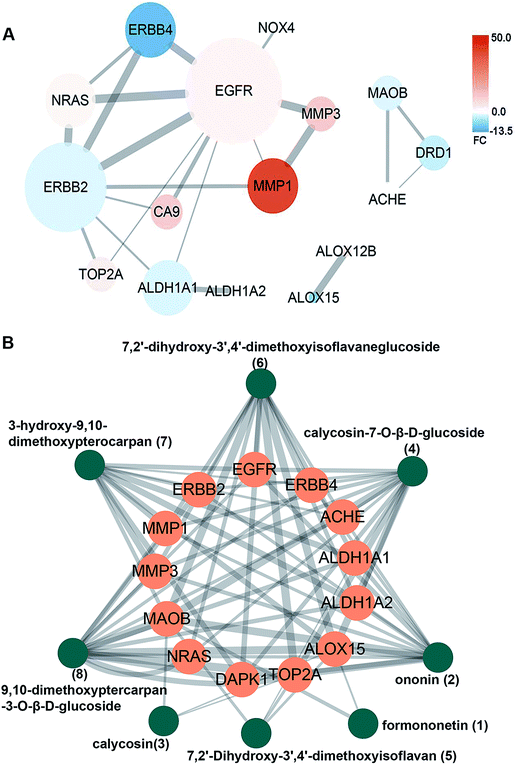
Next, a network pharmacology strategy was introduced to explore the mechanisms of multi-ingredient medicine. Consequently, 184 protein targets (drug targets) were predicted for the 8 primary TFA constituents. A total of 1096 LSCC-associated targets (disease targets) were collected from two datasets, of which 114 targets were from DisGeNet** and 996 targets were from the intersection of microarray data (6 LSCC and 6 paired adjacent normal margin) and RNA sequencing data (57 LSCC and 57 paired adjacent normal margin) (Fig. 4A). Venn analysis provided the intersection of drug targets and disease targets (Fig. 4B). Consequently, 19 candidate targets were identified: EGFR, ERBB2, ERBB4, MMP1, MMP3, NRAS, NOX4, TOP2A, DRD1, MAOB, ACHE, CA9, ALDH1A1, ALDH1A2, ALOX12B, ALOX15, DCT, CYP1B1 and DAPK1 (ESI Table S1†). We speculated that TFA may target these genes to achieve a synergistic effect with CDDP chemotherapy.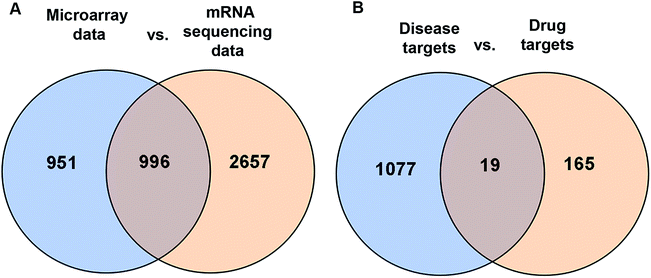 | ||
| Fig. 4 Identification of TFA potential therapeutic targets by Venn analysis. (A) 996 LSCC-associated targets stem from the intersection of microarray data and mRNA sequencing data. (B) 19 candidate targets stem from the intersection of disease targets and drug targets. Disease targets include 996 LSCC-associated targets and 114 targets from the network database DisGeNet.** Drug targets include 184 distinct protein targets of 8 primary constituents from TFA obtained by similarity ensemble approach§ and Swiss Target Prediction¶ and TCMSP database.|| | ||
3.5 Construction of PPI network with the 19 candidate targets
The interactions among proteins reflect the regulatory responses to the intrinsic or extrinsic perturbations of the pathway. We further constructed a PPI network of the 19 candidate targets of TFA. The highly connected nodes in the PPI network are called hubs. We noted that EGFR was a hub for TFA-treated LSCC, which interacted with ERBB2, ERBB4, MMP1, MMP3, NRAS, NOX4, TOP2A, CA9, and ALDH1A1. This suggested that TFA can function in LSCC through multiple mechanisms, and EGFR may be a core target of TFA.3.6 Interaction between TFA constituents and candidate targets
To further verify the quality and reliability of assessing protein-ligand interactions at the atomic level, molecular docking was used to model the interactions among the 8 compounds with potential molecular mechanisms in TFA and PPI network protein targets. Based on the molecular docking information, we constructed a compound–target (C–T) network. In the C–T network, 8 components of TFA could be docked with the candidate targets EGFR, ERBB2, ERBB4, MMP3, MMP1, ALDH1A, ALDH1A2, TOP2A, MAOB, ALOX15, NRAS, ACHE and DAPK1 with high docking scores (docking score > 4.52) (Fig. 5A, ESI Table S2†). Particularly, the flavonoid glycoside components ononin, calycosin-7-O-β-D-glucoside, 7,2′-dihydroxy-3′,4′-dimethoxyisoflavaneglucoside and 9,10-dimethoxyptercarpan-3-O-β-D-glucoside and 3-hydroxy-9,10-dimethoxypterocarpan (7) could bind to targets tightly, suggesting that they were the main contributors to the synergistic tumor inhibitory effect on LSCC by regulating these targets.3.7 Construction and analysis of target–pathway (T–P) network
To better understand the pharmacological mechanisms of the TFA synergistic effect with CDDP to inhibit LSCC, the 19 targets were annotated through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. GO analysis suggested that the target genes are involved in the process of oxidation-reduction, regulation of apoptosis, and cell proliferation (Fig. 6A). Furthermore, the KEGG pathway analysis results revealed that the 19 targets are enriched in PI3K-Akt, ErbB, and calcium signaling pathways (Fig. 6B). Interestingly, we found that EGFR is involved in half of these pathways (6 out of 12), followed by ERBB2 and ERBB4 (5 out of 12). The Kaplan–Meier analysis of TCGA cohorts revealed that the upregulation of EGFR, MMP1 and MMP3 was associated with poor outcomes with LSCC (Fig. 6C–E).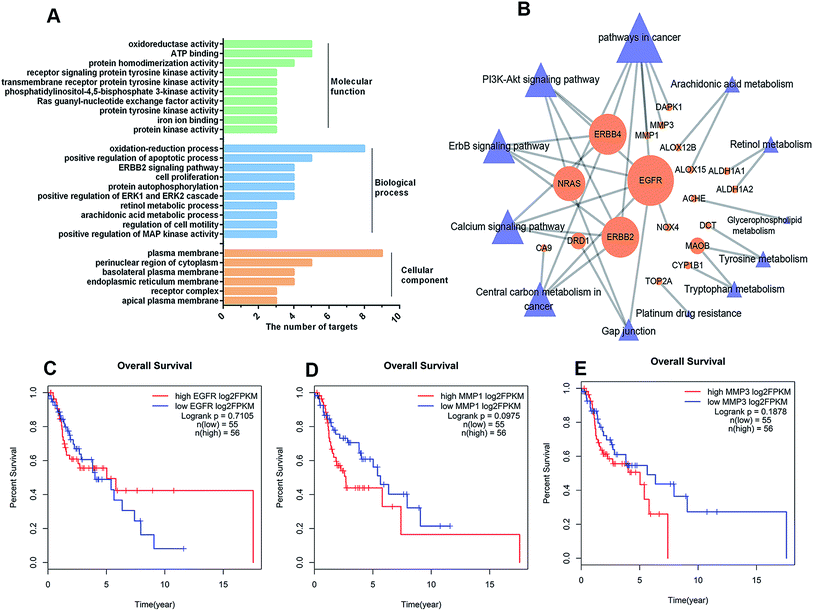 | ||
| Fig. 6 Functional annotation of predicted TFA targets. (A) GO analysis of the 19 candidate targets, including cellular component, molecular function, and biological process. (B) The target–pathway network (C–T network) by KEGG enrichment analysis. The orange circles are targets, and the blue triangles indicate the related pathway of TFA and cisplatin playing a synergistic anti-tumor effect. The size of all nodes is proportional to the degree value. Kaplan–Meier survival curves of LSCC patients with different EGFR (C), MMP1 (D) and MMP3 (E) levels in the TCGA|||| cohort. Upregulated EGFR, MMP1 and MMP3 were correlated with poor outcomes for patients with LSCC. | ||
Moreover, we found that the targets ALOX12B, ALOX15, ALDH1A1, ALDH1A2, DCT, MAOB, CYP1B1 and TOP2A were involved in metabolism pathways, including arachidonic acid metabolism, retinol metabolism, tyrosine metabolism, tryptophan metabolism, and platinum drug resistance (Fig. 6B). Taken together, our data indicated that the TFA constituents regulate cancer-associated pathways and metabolism in LSCC, thus inhibiting tumor growth and enhancing tumor sensitivity to chemotherapy.
4 Discussion
Laryngeal squamous cell carcinoma is the second highest incidence of HNSCC, especially in the northern area of China.2 Cisplatin (CDDP)-based chemotherapy is associated with significant chemoresistance and toxicity but has been adopted for LSCC. The low cure rate in LSCC is partly due to the lack of an effective chemotherapeutic strategy.3,4 The combination of traditional Chinese medicines with conventional chemotherapeutic drugs has become an alternative way to improve the outcome of cancer treatment.8 Particularly, emerging evidence has proved that the traditional Chinese medicine Astragali radix has anti-cancer functions.9,10 TFA is the main component of Astragali radix extracts, and the intake of TFA or flavonoid subclasses inhibits cancer growth.18,19 In the present study, for the first time, our data revealed that TFA has a significant anti-tumor effect in an LSCC preclinical model. The combined treatment of TFA and CDDP not only inhibited LSCC growth, but also attenuated the nephrotoxicity caused by CDDP chemotherapy.UPLC-MS/MS analysis identified 8 bioactive ingredients in the serum from the TFA-treated LSCC mouse model: formononetin, ononin, calycosin, calycosin-7-O-β-D-glucoside, 7,2′-dihydroxy-3′,4′-dimethoxyisoflavan, 7,2′-dihydroxy-3′,4′-dimethoxyisoflavan-7-O-β-D-glucoside, 3-hydroxy-9,10-dimethoxypterocarpan and 9,10-dimethoxyptercarpan-3-O-β-D-glucoside. Previous studies have revealed that formononetin20,21 and ononin22 inhibit cancer progression. Nevertheless, a further investigation of either single bioactive ingredient or in combination with these TFA ingredients for LSCC treatment is needed in future. Mechanistically, network pharmacology analysis screened 19 potential targets from the intersection data of drug targets and disease targets. We noted that the flavonoid glycosides ononin, calycosin-7-O-β-D-glucoside, 7,2′-dihydroxy-3′,4′-dimethoxyisoflavan-7-O-β-D-glucoside, and 9,10-dimethoxyptercarpan-3-O-β-D-glucoside have a higher binding degree with the candidate targets. Flavonoid glycosides maintain higher plasma concentrations and have a longer residence time in the blood than aglycones.23 Actually, previous studies revealed that ononin,24 calycosin-7-O-β-D-glucoside25,26 and 7,2′-dihydroxy-3′,4′-dimethoxyisoflavan-7-O-β-D-glucoside27 can inhibit cancer progression. Hence, we speculated that the flavonoid glycosides might be the main contributors to the synergistic tumor inhibitory effect on LSCC in TFA. However, considering TFA as a multi-component synergistic system that plays a therapeutic role through overall regulation on diseases, using the 4 flavonoid glycosides alone may not inhibit the LSCC growth effectively.
Moreover, we successfully constructed the PPI network, target–compound network, and target–pathway network; these networks had potential implications for understanding the pharmacological mechanism of the action and active substances of TFA and suggested that TFA ingredients interfere with the main hallmarks of LSCC. Notably, EGFR had the highest degree value and good docking score with 4 flavone glycoside components and 3-hydroxy-9,10-dimethoxypterocarpan, which highlighted that EGFR may play a key role in the TFA-mediated regulation of LSCC. The overexpression and mutation of EGFR have been frequently found in a variety of solid tumors, including those of head and neck, breast, and non-small cell lung cancers.28 The EGFR-targeted drug cetuximab in combination with radiotherapy has been used for locally advanced head and neck squamous cell carcinoma.29
Recent studies showed that the EGFR family members play a significant role in HNSCC.30 EGFR determination seems to be a very strong prognostic factor, especially for LSCC patients treated by induction chemotherapy followed by exclusive radiotherapy.31 Furthermore, simultaneously targeting EGFR, ERBB2, and ERBB4 by afatinib can help overcome the intrinsic and acquired cetuximab resistance in the HNSCC cell lines.32 Therefore, these data indicated that the EGFR family members EGFR, ERBB2, and ERBB4 are likely targets of TFA in our LSCC model.
GO analysis revealed that the potential TFA targets were involved in the oxidation–reduction process. Oxidative stress is a prominent feature of tumor cells and is involved in the whole process of tumor development.33 TFA has strong antioxidant activity,9,34 and studies have documented the beneficial effects of various natural products as antioxidants in CDDP-induced DNA damage and nephrotoxicity.35,36 Moreover, CDDP-induced oxidative stress has been linked to cisplatin resistance.37,38 Accordingly, TFA may enhance the LSCC sensitivity to CDDP by balancing the oxidative stress.
In the present study, the T–P network further showed that the EGFR family members contribute to multiple signaling pathways, including the PI3K-AKT signaling pathway, ErbB signaling pathway, calcium signaling pathway, central carbon metabolism in cancer and gap junction. These signaling pathways have already been testified and tightly linked to LSCC progression.2,39,40 Energy metabolism such as lipid and amino acid metabolism was closely associated with CDDP resistance.41,42 Moreover, metabolomics studies have demonstrated the significant changes in lipid and amino acids in cisplatin-induced nephrotoxicity.43,44 In our results, nearly half of the targets (9 out 19) were involved in lipid metabolism, amino acid metabolism and platinum resistance pathways. Hence, we speculated that TFA functions in LSCC via regulating the classical cancer-related pathways and energy metabolism.
In conclusion, TFA showed synergistic antitumor activity when combined with CDDP in LSCC. The TFA nephroprotective effect did not interfere with the anticancer activity of CDDP when evaluated in an LSCC mouse model. Network pharmacology analysis revealed that TFA functions through fine-tuning the oxidative stress, cancer-related pathways and metabolic levels. These findings provide basic data for the application of Astragali radix flavonoid extract as an adjuvant for LSCC chemotherapy.
Author contributions
WG, YW, and ZL conceived the study, supervised and approved the study design. JC, DY, YZ, MN, and YH performed most of the experiments and interpreted the results. CA, XX, and YL performed hematoxylin–eosin staining and immunohistochemistry experiments. YB and HL analyzed the HE staining and IHC results. JC and XZ performed bioinformatics analysis. YT and HY performed molecular biology experiments. JC, DY and YZ repeated the experiments. JC, WG, YW and ZL wrote and revised the paper. All authors read and approved the final manuscript.Conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.Abbreviations
| HNSCC | Head and neck squamous cell carcinoma |
| LSCC | Laryngeal squamous cell carcinoma |
| AR | Astragali Radix |
| TFA | Astragali radix total flavonoid |
| CDDP | Cisplatin |
| PBS | Phosphate buffered saline |
| HE | Hematoxylin–eosin |
| IHC | Immunohistochemistry |
| UPLC-MS/MS | Ultra-high-performance liquid chromatography tandem mass spectrometry |
| PPI | Protein–protein interaction |
| C–T network | Compound–target network |
| T–P network | Target–pathway network |
| OS | Over survival |
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grants: 81602394, 81802793, 81802948, 81872210), China Postdoctoral Science Foundation (grants: 2016M591412, 2017M610174), Shanxi Province Scientific and Technological Achievements Transformation Guidance Foundation (grants: 201604D131002 and 201604D132040), Key Technological Innovation Platform Foundation for Head and Neck Cancer Research of Shanxi Province (grants: 201605D151003), the Research Project of Shanxi Province Health and Family Planning Commission (grant: 201601038), the Excellent talent science and technology innovation project of Shanxi Province (grant: 201705D211018), Shanxi Scholarship Council of China (grant: 2017-017), Outstanding Youth Development Foundation of the First Hospital of Shanxi Medical University (grant: YR1601), Startup Foundation for Doctors of Shanxi Medical University (grant: BS03201624), and Fund of Shanxi “1331 Project”.References
- C. E. Steuer, M. El-Deiry, J. R. Parks, K. A. Higgins and N. F. Saba, Ca-Cancer J. Clin., 2017, 67, 31 CrossRef PubMed.
- W. Gao, C. Zhang, W. Li, H. Li, J. Sang, Q. Zhao, Y. Bo, H. Luo, X. Zheng, Y. Lu, Y. Shi, D. Yang, R. Zhang, Z. Li, J. Cui, Y. Zhang, M. Niu, J. Li, Z. Wu, H. Guo, C. Xiang, J. Wang, J. Hou, L. Zhang, R. F. Thorne, Y. Cui, Y. Wu, S. Wen and B. Wang, Mol. Ther., 2019, 27, 365 CrossRef CAS PubMed.
- R. I. Haddad, E. Massarelli, J. J. Lee, H. Y. Lin, K. Hutcheson, J. Lewis, A. S. Garden, G. R. Blumenschein, W. N. William, R. R. Pharaon, R. B. Tishler, B. S. Glisson, C. Pickering, K. A. Gold, F. M. Johnson, G. Rabinowits, L. E. Ginsberg, M. D. Williams, J. Myers, M. S. Kies and V. Papadimitrakopoulou, Ann. Oncol., 2019, 30, 471 CrossRef CAS PubMed.
- P. Szturz, K. Wouters, N. Kiyota, M. Tahara, K. Prabhash, V. Noronha, D. Adelstein, D. Van Gestel and J. B. Vermorken, Front. Oncol., 2019, 9, 86 CrossRef PubMed.
- J. M. Bauml, R. Vinnakota, Y. H. Anna Park, S. E. Bates, T. Fojo, C. Aggarwal, J. Di Stefano, C. Knepley, S. Limaye, R. Mamtani, J. Wisnivesky, N. Damjanov, C. J. Langer, R. B. Cohen and K. Sigel, Cancer, 2019, 125, 406 CrossRef CAS PubMed.
- Y. Matsui, J. Watanabe, M. Ikegawa, T. Kamoto, O. Ogawa and H. Nishiyama, Oncogene, 2008, 27, 4603 CrossRef CAS PubMed.
- R. Baena Ruiz and P. Salinas Hernández, Maturitas, 2016, 94, 13 CrossRef CAS PubMed.
- R. G. de Oliveira Júnior, A. F. Christiane Adrielly, J. da Silva Almeida, R. Grougnet, V. Thiéry and L. Picot, Fitoterapia, 2018, 129, 383 CrossRef PubMed.
- A. Gong, R. Duan, H. Y. Wang, X. P. Kong, T. Dong, K. Tsim and K. Chan, Medicines, 2018, 5, e46 CrossRef PubMed.
- Y. Jung, U. Jerng and S. Lee, Chin. J. Integr. Med., 2016, 22, 225 CrossRef CAS PubMed.
- Z. Guo, Y. Lou, M. Kong, Q. Luo, Z. Liu and J. Wu, Int. J. Mol. Sci., 2019, 20, 1463 CrossRef PubMed.
- L. Di CesareMannelli, M. Zanardelli, G. Bartolucci, A. Karioti, A. R. Bilia, A. Vannacci, A. Mugelli and C. Ghelardini, Planta Med., 2015, 81, 1045 CrossRef CAS PubMed.
- S. A. Hussain, A. A. Sulaiman, C. Balch, H. Chauhan, Q. M. Alhadidi and A. K. Tiwari, Nutr. Cancer, 2016, 68, 879 CrossRef CAS PubMed.
- M. Abotaleb, S. M. Samuel, E. Varghese, S. Varghese, P. Kubatka, A. Liskova and D. Büsselberg, Cancers, 2018, 11 Search PubMed.
- W. N. Zhang, A. P. Li, Y. S. Qi, X. M. Qin and Z. Y. Li, J. Pharm. Biomed. Anal., 2018, 158, 128 CrossRef CAS PubMed.
- Y. He, S. Peng, J. Wang, H. Chen, X. Cong, A. Chen, M. Hu, M. Qin, H. Wu, S. Gao, L. Wang, X. Wang, Z. Yi and M. Liu, Nat. Commun., 2016, 7, 13122 CrossRef CAS PubMed.
- C. Zhang, W. Gao, S. Wen, Y. Wu, R. Fu, D. Zhao, X. Chen and B. Wang, Neoplasma, 2016, 63, 888 CrossRef CAS PubMed.
- R. K. Lall, V. M. Adhami and H. Mukhtar, Mol. Nutr. Food Res., 2016, 60, 1396 CrossRef CAS PubMed.
- R. Zamora-Ros, D. K. Barupal, J. A. Rothwell, M. Jenab, V. Fedirko, I. Romieu, K. Aleksandrova, K. Overvad, C. Kyrø, A. Tjønneland, A. Affret, M. His, M. C. Boutron-Ruault, V. Katzke, T. Kühn, H. Boeing, A. Trichopoulou, A. Naska, M. Kritikou, C. Saieva, C. Agnoli, M. Santucci de Magistris, R. Tumino, F. Fasanelli, E. Weiderpass, G. Skeie, S. Merino, P. Jakszyn, M. J. Sánchez, M. Dorronsoro, C. Navarro, E. Ardanaz, E. Sonestedt, U. Ericson, L. Maria Nilsson, S. Bodén, H. B. Bueno-de-Mesquita, P. H. Peeters, A. Perez-Cornago, N. J. Wareham, K. T. Khaw, H. Freisling, A. J. Cross, E. Riboli and A. Scalbert, Int. J. Cancer, 2017, 140, 1836 CrossRef CAS PubMed.
- C. Kim, S. G. Lee, W. M. Yang, F. Arfuso, J. Y. Um, A. P. Kumar, J. Bian, G. Sethi and K. S. Ahn, Cancer Lett., 2018, 431, 123 CrossRef CAS PubMed.
- C. Yang, Q. Xie, X. Zeng, N. Tao, Y. Xu, Y. Chen, J. Wang and L. Zhang, Bioorg. Chem., 2019, 85, 445 CrossRef CAS PubMed.
- R. Zhou, H. Chen, J. Chen, X. Chen, Y. Wen and L. Xu, BMC Complementary Altern. Med., 2018, 18, 83 CrossRef PubMed.
- J. Xiao, Crit. Rev. Food Sci. Nutr., 2017, 57, 1874 CAS.
- R. Zhou, H. Chen, J. Chen, X. Chen, Y. Wen and L. Xu, BMC Complementary Altern. Med., 2018, 18, 83 CrossRef PubMed.
- M. Ren, X. Wang, G. Du, J. Tian and Y. Liu, Mol. Med. Rep., 2016, 13, 633 CrossRef CAS PubMed.
- S. Fu, Y. Gu, J. Q. Jiang, X. Chen, M. Xu, X. Chen and J. Shen, J. Ethnopharmacol., 2014, 155, 692 CrossRef CAS PubMed.
- W. I. Tou and C. Y. Chen, J. Biomol. Struct. Dyn., 2012, 30, 299 CrossRef CAS PubMed.
- K. Shostak and A. Chariot, Trends Mol. Med., 2015, 21, 385 CrossRef CAS PubMed.
- Y. Tian, J. Lin, Y. Tian, G. Zhang, X. Zeng, R. Zheng, W. Zhang and Y. Yuan, Int. J. Cancer, 2018, 142, 2198 CrossRef CAS PubMed.
- E. Henson, Y. Chen and S. Gibson, Cancers, 2017, 9, 27 CrossRef PubMed.
- X. Pivot, N. Magné, E. Guardiola, G. Poissonnet, O. Dassonville, M. Francoual, J. L. Formento, R. J. Bensadoun, F. Demard, M. Schneider and G. Milano, Oral Oncol., 2005, 41, 320 CrossRef CAS PubMed.
- I. De Pauw, F. Lardon, J. Van den Bossche, H. Baysal, E. Fransen, V. Deschoolmeester, P. Pauwels, M. Peeters, J. B. Vermorken and A. Wouters, Mol. Oncol., 2018, 12, 830 CrossRef CAS PubMed.
- S. Chikara, L. D. Nagaprashantha, J. Singhal, D. Horne, S. Awasthi and S. S. Singhal, Cancer Lett., 2018, 413, 122 CrossRef CAS PubMed.
- M. Shahzad, A. Shabbir, K. Wojcikowski, H. Wohlmuth and G. C. Gobe, Curr. Drug Targets, 2016, 17, 1331 CrossRef CAS PubMed.
- M. I. Yousef and H. M. Hussien, Food Chem. Toxicol., 2015, 78, 17 CrossRef CAS PubMed.
- N. Ridzuan, N. A. Rashid, F. Othman, S. B. Budin, F. Hussan and S. L. Teoh, Mini-Rev. Med. Chem., 2019, 19, 1134 CrossRef PubMed.
- H. Zhu, H. Luo, W. Zhang, Z. Shen, X. Hu and X. Zhu, Drug Des., Dev. Ther., 2016, 10, 1885 CrossRef CAS PubMed.
- D. Shin, E. H. Kim, J. Lee and J. L. Roh, Redox Biol., 2017, 13, 219 CrossRef CAS PubMed.
- M. W. Baek, H. S. Cho, S. H. Kim, W. J. Kim and J. Y. Jung, J. Cell. Physiol., 2017, 232, 417 CrossRef CAS PubMed.
- A. Beckmann, N. Hainz, T. Tschernig and C. Meier, Cancers, 2019, 11 Search PubMed.
- J. M. Roodhart, L. G. Daenen, E. C. Stigter, H. J. Prins, J. Gerrits, J. M. Houthuijzen, M. G. Gerritsen, H. S. Schipper, M. J. Backer, M. van Amersfoort, J. S. Vermaat, P. Moerer, K. Ishihara, E. Kalkhoven, J. H. Beijnen, P. W. Derksen, R. H. Medema, A. C. Martens, A. B. Brenkman and E. E. Voest, Cancer Cell, 2011, 20, 370 CrossRef CAS PubMed.
- L. Galluzzi, I. Vitale, J. Michels, C. Brenner, G. Szabadkai, A. Harel-Bellan, M. Castedo and G. Kroemer, Cell Death Dis., 2014, 5, e1257 CrossRef CAS PubMed.
- A. Wilmes, C. Bielow, C. Ranninger, P. Bellwon, L. Aschauer, A. Limonciel, H. Chassaigne, T. Kristl, S. Aiche, C. G. Huber, C. Guillou, P. Hewitt, M. O. Leonard, W. Dekant, F. Bois and P. Jennings, Toxicol. In Vitro, 2015, 30, 117 CrossRef CAS PubMed.
- P. Zhang, J. Q. Chen, W. Q. Huang, W. Li, Y. Huang, Z. J. Zhang and F. G. Xu, Sci. Rep., 2017, 7, 44804 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra04701h |
| ‡ These authors contributed equally to this work. |
| § http://sea.bkslab.org/ |
| ¶ http://SwissTargetPrediction.ch/ |
| || http://lsp.nwu.edu.cn/tcmsp.php |
| ** http://www.disgenet.org/ |
| †† http://string-db.org/ |
| ‡‡ http://systemsdock.unit.oist.jp/ |
| §§ http://www.rcsb.org/pdb/ |
| ¶¶ https://david.ncifcrf.gov/home.jsp |
| |||| https://portal.gdc.cancer.gov/ |
| This journal is © The Royal Society of Chemistry 2019 |