Open Access Article
Open Access ArticleIsolation and identification of an antioxidant collagen peptide from skipjack tuna (Katsuwonus pelamis) bone†
Ding Ding‡
,
Bowei Du‡,
Chao Zhang,
Fakhar Zaman and
Yaqin Huang *
*
Beijing Laboratory of Biomedical Materials, Beijing Key Laboratory of Electrochemical Process and Technology for Materials, Beijing University of Chemical Technology, Beijing 100029, People's Republic of China. E-mail: huangyq@mail.buct.edu.cn; Fax: +86-10-64438266; Tel: +86-10-64438266
First published on 28th August 2019
Abstract
To date, many researchers have developed active components that are derived from seafood processing for the purposes of healthcare. Here, an antioxidant collagen peptide was obtained from skipjack tuna (Katsuwonus pelamis) bone by using a combination of trypsin and chymotrypsin as the catalyst. The amino acid sequence of the peptide was identified as Ser–Ser–Gly–Pro–Pro–Val–Pro–Gly–Pro–Met–Gly–Pro–Met–Gly–Pro–Arg (SSGPPVPGPMGPMGPR) by liquid chromatography-electrospray ionization quadrupole time-of-flight mass spectrometry (LC-ESI-QTOF-MS) analysis. We found that the as-prepared collagen peptide can efficiently scavenge DPPH radical (IC50 3.149 mM), superoxide anion radical (IC50 3.803 mM) and ABTS radical (IC50 9.489 mM). In addition, it has been found that the methionine (Met) residue in the collagen peptide could provide a precise active site during the scavenging of DPPH radicals by Fourier transform infrared spectroscopy (FTIR) analysis and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry analysis. These results suggest that the peptide can find wide uses in the food, cosmetic and pharmaceutical industries.
Introduction
Oxidative stress, a vast threat to human health1–3 results from an imbalance between the generation and scavenging of free radicals.4 Excessive free radicals attack membrane lipids, proteins and DNA and cause many health disorders, including diabetes mellitus, neurodegenerative disorders, cancer and inflammatory diseases.5–7 Currently, many researchers focus on developing functional foods to scavenge free radicals.8–10 As ingredients to such functional foods, antioxidant peptides have received much attention.11–14Researchers have tried to find antioxidant peptides from natural sources. Shazly et al. discovered that antioxidant peptides obtained from buffalo casein hydrolysates contain a number of antioxidant amino acids (Ser, Arg, Ala and Leu), and considered these to be key to the high radical scavenging potential of the hydrolysates.15 Blanca et al. identified an antioxidant peptide in fermented milk and discovered that the Met residue of the peptide is primarily responsible for the antioxidant activities of the peptide.16
Fish collagen obtained from the byproducts of seafood processing usually contains a large number of antioxidant amino acids residues, such as Val, Met, Trp, Tyr, and Phe.17 Thus, people have been attempting to obtain antioxidant peptides from fish collagen. Wu et al. isolated an antioxidant peptide from the salmon skin collagen by using extracellular proteases from Vibrio sp. SQS2-3.18 Alemán et al. hydrolysed a squid collagen with alcalase to isolate a number of antioxidant peptides containing Leu residue which the authors speculated as playing an important role in the antioxidant activities of the peptides.19 Ngo obtained antioxidant collagen peptides from Nile tilapia and attributed the antioxidant activity of the peptides to the existence of non-aromatic amino acids such as Ala, Pro, and Val.20
Tuna (Thunnus spp.) and tuna-like species have long been significant sources of food for their high contents of bioactive proteins and unsaturated fatty acids.21 Currently, tuna generally is processed to produce raw flesh or canned food. Only about one-third of the whole tuna fish is used in the canning process, and the canning industry generates as much as 70% solid wastes from original fish materials. Waste includes viscera, gills, head, bone, and skin. In general, protein-rich by-products from the production of these tuna are processed into products with low market value, such as fishmeal and fertilizer.21 However, these by-products could be used as functional food ingredients. Tuna protein hydrolysates (TPH), which are obtained through hydrolysis of the by-products of tuna processing, can be used as an ingredient in food industries to provide whipping, gelling, and texturing properties. Many researchers have attempted to derive antioxidant peptides from TPH.22–24
Trypsin (T) and chymotrypsin (C) are enzymes of the digestive system. Each enzyme is responsible for the hydrolysis of different peptide bonds. Trypsin hydrolyses the peptide and ester bonds formed by the carboxyl group of the basic amino acids, such as Arg and Lys.25 Chymotrypsin hydrolyses the peptide bonds formed by the carboxyl group of the aromatic or large hydrophobic amino acids, such as Phe, Tyr, Trp and Met.26 Here, antioxidant collagen hydrolysates from skipjack tuna (Katsuwonus pelamis) bone were prepared by using a combination of trypsin and chymotrypsin as the catalyst. We further purified the antioxidant peptides, determined their sequences and evaluated their antioxidant activities. We also investigated the mechanism of scavenging of free radicals by these peptides via experiments and molecular dynamics (MD) simulations.
Experimental
Materials
The frozen skipjack tuna bones were supplied by Xinfa Aquatic Food Co., Ltd. (Rongcheng, China). The frozen tuna bones were packed in zip-lock bags that were contained in a polystyrene box. The samples were transported to the Beijing University of Chemical Technology within 72 h, and stored at −20 °C. Trypsin (2.50 × 104 U g−1), chymotrypsin (1.70 × 105 U g−1), reduced glutathione (GSH), 2,2-diphenyl-1-picrylhydrazyl (DPPH), 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS), diethylaminoethyl cellulose-52 (DEAE-52) and trifluoroacetic acid (TFA) were purchased from Aladdin Trading Co., Ltd. (Shanghai, China). Acetonitrile (ACN) was purchased from Thermo Fisher Scientific Co., Ltd. (Beijing, China). Synthetic peptides (SSGPPVPGPMGPMGPR, GEQGSTGPAGF, GFPGER, SSGPPVPGPGGPMGPR, purity > 99%) were obtained from Botai Co., Ltd. (Shanghai, China). Human hepatocellular carcinoma (HepG2) cell line was obtained from the BeNa Culture Collection (BNCC, Beijing, China).Preparation of skipjack tuna bone collagen hydrolysates (STCHs)
The bones were first demineralised using 1.0 M HCl solution at ambient temperature (∼27 °C) with a bone/solution ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (g mL−1) for 12 h, and the HCl solution was changed every 1.5 h. The acid treated bones were washed with tap water and soaked in 4% NaCl solution for 12 h to remove non-collagenous proteins. The pretreated bones were washed thoroughly with tap water and then subjected to a final wash with distilled water to remove any residual matter. Then tuna bone collagen was obtained.
2 (g mL−1) for 12 h, and the HCl solution was changed every 1.5 h. The acid treated bones were washed with tap water and soaked in 4% NaCl solution for 12 h to remove non-collagenous proteins. The pretreated bones were washed thoroughly with tap water and then subjected to a final wash with distilled water to remove any residual matter. Then tuna bone collagen was obtained.
We designed three sets of enzymatic hydrolysis experiments. One set involved a simultaneous addition of two enzymes at the trypsin and chymotrypsin (TC), while the other two sets involved the sequential addition of trypsin + chymotrypsin (T + C) or chymotrypsin + trypsin (C + T). The enzymolysis conditions were shown in Table S1.† The enzymes were inactivated by heating at 90–95 °C for approximately 15 min. The degree of hydrolysis (DH) was determined by the pH-state method.27 After filtering and drying, powders containing hydrolysates were obtained and used in subsequent investigations.
Determination of amino acid composition of skipjack tuna bone collagen
Amino acid analysis of collagen from skipjack tuna bone was performed using Hitachi L-8900 High Speed Amino Acid Analyzer (Tokyo, Japan) according to Liu and Sun.28Purification and identification of antioxidant peptides from STCH-TC
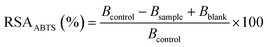
Determination of antioxidant activity
 | (1) |
 | (2) |
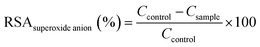 | (3) |
The cell culture and treatment were investigated using a previously described method.33 In detail, cell cytotoxicity was measured by using a tetrazolium salt WST-8 (Cell Counting Kit 8, CCK-8). HepG2 cells (6 × 103 per well) were pretreated with the synthetic peptide (SSGPPVPGPMGPMGPR, GEQGSTGPAGF and GFPGER) at different concentrations (5 and 15 μM) in a 96-well plate for 24 h. After treatment with 500 μM of H2O2 for 6 h, 10 μL of WST-8 was added and the mixture was incubated at 37 °C for 2 h. Cell viability was calculated in the light of the absorbance at 450 nm by SpectraMax® M2/M2e (CA, USA).
Determination of peptide structure
Determination of the molecular weights and sequences of the synthetic peptides
The molecular weight distribution of the samples was analyzed using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. For the identification of the sequences, the synthetic peptides were applied to MALDI-TOF/TOF mass spectrometry analysis (Ultraflextreme, Bruker, Germany).Modeling platform and simulations of peptide–DPPH radical complexes
Molecular dynamics (MD) simulation was performed using Material Studio 6.0 software with a canonical (NVT) ensemble. A cubic box with a size of 47.43 × 47.43 × 47.43 Å (x, y, and z dimensions) containing one peptide molecule and one DPPH radical molecule was built. Periodic boundary conditions were employed for all three dimensions of the box, and a PVFF force field was chosen to model the interactions among all the particles. A velocity scale thermostat was used to fix the temperature at 298 K with a 0.1 ps decay constant. Energy minimization with integration (5000 steps) was first used to relax the molecules prior to the beginning of the simulation. The system was then equilibrated using a 5 ns MD simulation with a time step of 1 fs.Statistical analysis
All experiments were conducted in triplicate (n = 3) and the values were expressed as the mean ± the standard deviation (SD). An ANOVA test was used to analyze the experiment data in SPSS 22.0 software (Statistical Program for Social Sciences, SPSS Corporation, Chicago, IL, USA).Results and discussion
Antioxidant peptides obtained from skipjack tuna bone
For skipjack tuna bone collagen, the relative content of each amino acid residue in the bone collagen was determined (Table 1). We found that the antioxidant amino acids, such as Arg, Lys, Phe and Met, were present. By comparison, skipjack tuna bone collagen contains a higher ratio of Met residues than tilapia skin collagen.34| Amino acid | Number of residues/1000 residues | |
|---|---|---|
| Skipjack tuna bone | Tilapia skin | |
| Asp | 46 | 48 |
| Thr | 30 | 24 |
| Ser | 37 | 35 |
| Glu | 77 | 69 |
| Gly | 337 | 347 |
| Ala | 125 | 123 |
| Val | 25 | 15 |
| Met | 15 | 9 |
| Ile | 13 | 8 |
| Leu | 27 | 23 |
| Tyr | 7 | 2 |
| Phe | 18 | 13 |
| Lys | 23 | 25 |
| His | 13 | 6 |
| Arg | 43 | 47 |
| Hyp | 70 | 79 |
| Pro | 94 | 119 |
Researchers have previously demonstrated that the presence of Trp, Tyr, and Phe residues within the C-terminus of peptides appeared to be essential for radical scavenging,35 and those antioxidant peptides which contain Lys or Arg residues in the C-terminus have higher antioxidant activity.36 Therefore, based on the structure and amino acid composition of skipjack tuna bone collagen, we used a combination of trypsin and chymotrypsin as the catalyst to prepare collagen hydrolysate (STCH-TC) with high antioxidant activity. Actually, chymotrypsin cleaves peptide bonds on the carboxyl side of Phe, Tyr, and Try residues, and trypsin specifically cleaves peptides on the carboxyl side of Arg and Lys residues. Here, we studied the optimal conditions for the preparation of STCH-TC by implementing Box–Behnken Design (BBD) in combination with response surface methodology (RSM). At the same time, we prepared STCH-T + C and STCH-C + T as control samples to investigate the interaction between trypsin and chymotrypsin. The results are shown in Table S1.† STCH-TC exhibited much higher antioxidant activity than the control samples, indicating that there are synergistic effect between trypsin and chymotrypsin for the hydrolysis of skipjack tuna bone collagen. We believe that peptides with high antioxidant activity could be isolated from the STCH-TC.
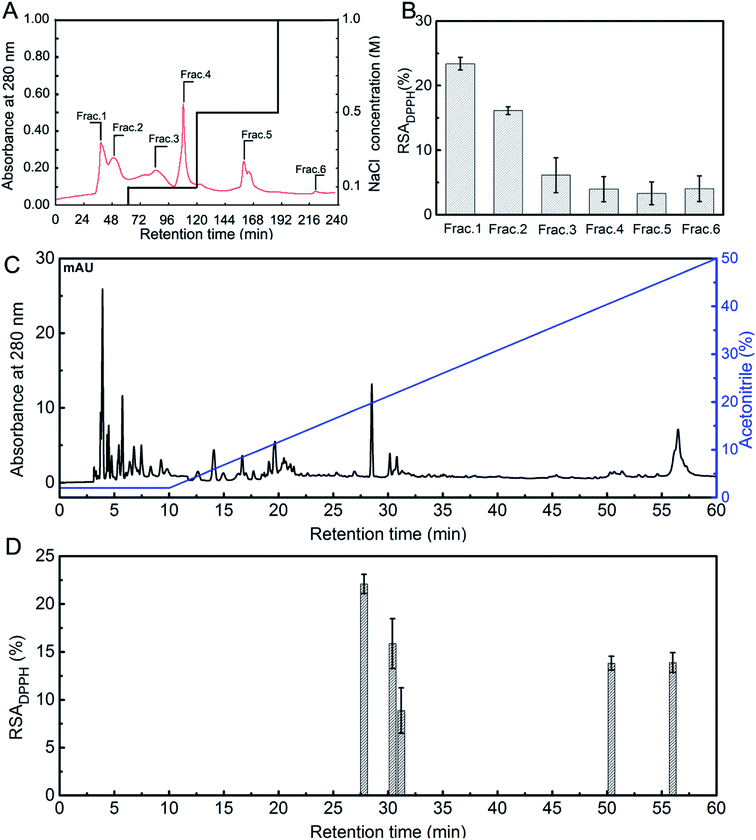
As shown in Fig. 1A, six fractions (Frac. 1 to Frac. 6) were separated from STCH-TC using a DEAE-52 cellulose column. Frac. 1 exhibited the highest DPPH radical scavenging ratio (23.41%) at 1 mg mL−1 (Fig. 1B). Thus, it was subsequently purified using RP-HPLC (Fig. 1C). According to the elution profile obtained using RP-HPLC, one major fraction with the highest DPPH activity (22.09%) was identified (27.6 min) (Fig. 1D).
The peptides with the highest antioxidant activity were determined to reside in the main peak (27.6 min) obtained from Frac. 1 and were evaluated using LC-ESI-QTOF-MS analysis to identify antioxidant peptides. As shown in Table 2, the peptides with antioxidant activity were identified. We did not find any information on these peptides in the BIOPEP database. Previous studies have shown that there is at least one antioxidant amino acid residue (Val, Try, Tyr, Phe, Met, or His) within antioxidant peptide chains.37 Therefore, we believe the peptide (SSGPPVPGPMGPMGPR) which contains a Gly–Pro–Met (GPM) fragment from STCH-TC could be an antioxidant peptide. In addition, the Arg residue in its C-terminus may contribute to the antioxidant activity of the peptide.
| Theo. MH+ [Da] | Sequence | Ions score mascot | Location |
|---|---|---|---|
| 1520.7348 | SSGPPVPGPMGPMGPR | 42 | f 159–175 |
| 702.3570 | APDPFR | 35 | f 1195–1200 |
| 1327.6641 | GFPGLPGQSGEPGK | 33 | f 959–972 |
| 976.4371 | GETGPSGPAGF | 32 | f 791–801 |
| 830.4268 | GHRGFSGL | 29 | f 253–261 |
| 662.3257 | GFPGER | 27 | f 668–673 |
| 1007.4429 | GEQGSTGPAGF | 18 | f 620–630 |
In vitro evaluation of antioxidant peptides
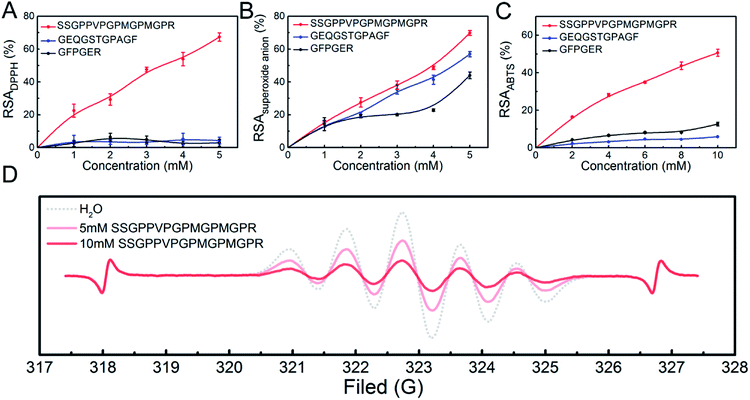
The three peptides (SSGPPVPGPMGPMGPR, GEQGSTGPAGF and GFPGER) identified in the highly-antioxidant fractions from STCH-TC were synthesized (Fig. S1†). These peptides were selected to meet certain requirements, such as the presence of Phe residues at the C-terminus or the presence of Val, Met and Phe residues within the peptide chain. To evaluate the antioxidant activities of these three peptides, DPPH, superoxide anion and ABTS radical scavenging assays were performed, and their activities were compared with those of the GSH positive controls (0–1.0 mM). The IC50 values of GSH (as standards) during the inhibition of DPPH, superoxide anion and ABTS are 0.329 mM, 0.335 mM and 0.105 mM, respectively (Fig. S2†).One peptide (SSGPPVPGPMGPMGPR) exhibited higher DPPH radical scavenging activity (IC50 3.149 mM) than the other two peptides (no more than 25%; Fig. 2A). The IC50 of the SSGPPVPGPMGPMGPR peptide was also lower than that of GPP (IC50 7.16 mM) and WDR (IC50 7.63 mM) derived from hydrolysates from bluefin leatherjacket and S. lewini proteins, respectively.38,39 DPPH is a radical that contains an unpaired electron, which could capture a proton when it encounters antioxidants. Recently, it has been reported that the radical scavenging mechanism utilized by antioxidant peptides containing Met residues is single electron transfer (SET),40 which indicates that peptides containing Met residues may have high DPPH radical scavenging activity. In addition, the position of the antioxidant amino acid residues within peptide sequences also affects the antioxidant activity of the peptide, and antioxidant peptides containing Lys and Arg residues at the C-terminus have higher antioxidant activity.36
From the Fig. 2B and C, we observed that the SSGPPVPGPMGPMGPR peptide also exhibited high scavenging activity in the presence of superoxide anion radicals (IC50 3.803 mM) and ABTS radicals (IC50 9.489 mM). Thus, we inferred that the presence of Val, Arg and Met residues within a peptide sequence may be beneficial to its antioxidant activity.16
The above results indicate that the SSGPPVPGPMGPMGPR peptide has high antioxidant activity. Using ESR measurements, the DPPH radical scavenging activity of the SSGPPVPGPMGPMGPR peptide was obtained. The changes in the ESR signal intensity of the SSGPPVPGPMGPMGPR at different concentrations are shown in Fig. 2D. The ESR signal intensities can be determined based on the peak height, which is the sum of the up-peak value and the down-peak value.41,42 For the SSGPPVPGPMGPMGPR peptide, the comparison between the signal heights of the blank and the samples in each ESR plot indicated that the decrease in the ESR signal intensity is 43.10% at a 5 mM concentration and 79.7% at the 10 mM concentration. In addition, upon searching the BLAST protein database, the sequence of this antioxidant peptide was only found in a few species of fish, almost all of which are uncommon.
In summary, the SSGPPVPGPMGPMGPR peptide not only has high bioactivity in terms of scavenging DPPH radicals, but could also be used to distinguish tuna collagen from other common collagens on the market. Some peptides with 3–9 amino acid residues have been found to have strong antioxidant activities. In fact, many factors affect the antioxidant activity of peptides, including the amino acid composition, amino acid sequence and spatial conformation.43 Nowadays, some antioxidant peptides with high molecular weight, such as NTDGSTDYGILQINSR and TVTSLDLPVLRW, have also been reported.44,45 The radical scavenging mechanism of the peptide SSGPPVPGPMGPMGPR is worth further exploring.
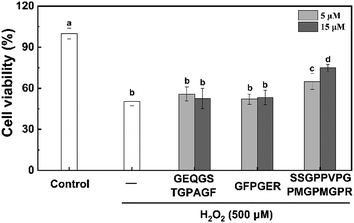
The HepG2 cell line is considered a good model with which to research in vitro toxicity of the liver, since it retains many of the specific functions that are characteristic of normal human hepatocytes.46 Prior to the evaluating the effect of the three peptides on scavenging free radicals, the cytotoxicity of the synthetic peptides in HepG2 cells was also assessed (Fig. 3). H2O2 at 500 μM significantly decreased cell viability by 50.0%. We found that pre-treatment of SSGPPVPGPMGPMGPR at concentration of 5 and 15 μM significantly enhanced cell viability to 64.92 ± 5.73% and 74.93 ± 2.55%, respectively. In contrast, the other two peptide can not reduce cytotoxicity induced by H2O2. In addition, we found that the three peptides have no toxic side effects on HepG2 cells (Fig. S4†). These results demonstrate that the peptide SSGPPVPGPMGPMGPR possess protective effect on H2O2-induced oxidative stress in HepG2 cell.
DPPH radical scavenging mechanism utilized by antioxidant peptides
The measurement of DPPH scavenging is a quick, convenient, and efficient method to determine antioxidant activity. Here, we are interested to understand the mechanism utilized by the SSGPPVPGPMGPMGPR peptide during scavenging of DPPH radicals. Due to the complexity of the peptides and the lack of effective experimental methods, there are some challenges for exploring their radical scavenging mechanism. The secondary structure of peptides affects its biological activity, and it could be investigated by CD. The CD spectra (Fig. 4A) of the SSGPPVPGPMGPMGPR peptide and the SSGPPVPGPMGPMGPR peptide in the presence of DPPH radicals exhibited one negative minimum at approximately 200 nm, which is due to a random coil conformation. Although there are only slight differences between the curves generated for the SSGPPVPGPMGPMGPR and the SSGPPVPGPMGPMGPR peptides in the presence of DPPH radicals, the conformation of the SSGPPVPGPMGPMGPR peptide was altered after the reaction. After scavenging of DPPH radicals, the proportion of β-turns increases from 34.3% to 35.1%, and the proportion of random coil increases from 46.5% to 47.6%, while the proportion of β-sheets decreases from 15.1% to 13.9%. After reacting with the DPPH radicals, the secondary structures of the peptides assumed a more relaxed conformation. Their FTIR spectra displayed similar curves between 4000 and 500 cm−1 (Fig. 4B). The CH3 stretching vibrations were blue-shifted from 2972.78 cm−1 to 2925.04 cm−1. In addition, the emergence of an S–C bond at 1056.14 cm−1 demonstrated that the structure of the Met residue was changed. These results indicate that there was a reaction between the Met residue and DPPH.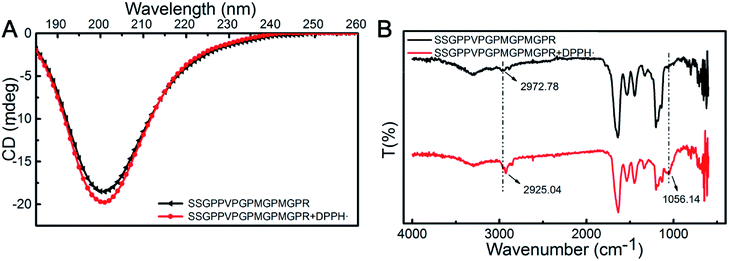 | ||
| Fig. 4 The mechanism utilized by SSGPPVPGPMGPMGPR peptide during scavenging of DPPH radicals. (A) CD spectra; (B) FTIR-ATR spectra. | ||
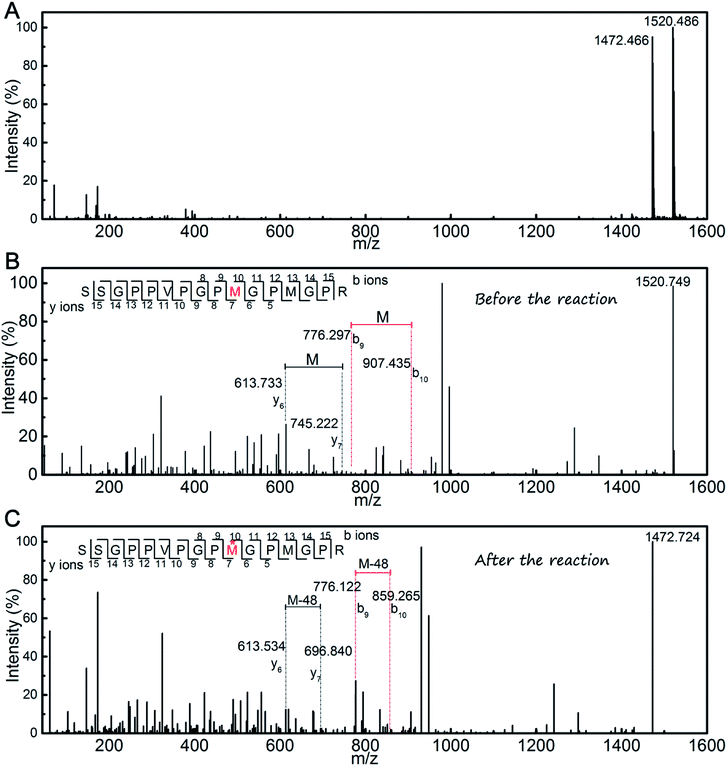
The products were measured using MALDI-TOF mass spectrometry (Fig. 5A). According to the mass spectra, the molecular weights of products were determined to be 1520.486 and 1472.466 Da, which indicated that a single novel product was present. To determine the changes in the amino acids residues that resulted in the decrease of the molecular weight, further MALDI-TOF/TOF mass spectrometry analysis was conducted (Fig. 5B and C). The results showed that the molecular weight of the Met residue was reduced by 48 Da, which is consistent with the decrease in the total molecular weight of the peptide. This means one of the Met residues in the SSGPPVPGPMGPMGPR peptide could play a key role in the scavenging of DPPH radicals. As the peptide contains two Met residues, only one serves as an active site during scavenging of DPPH radicals. It may be a result of a spatial configuration structure for the peptide.
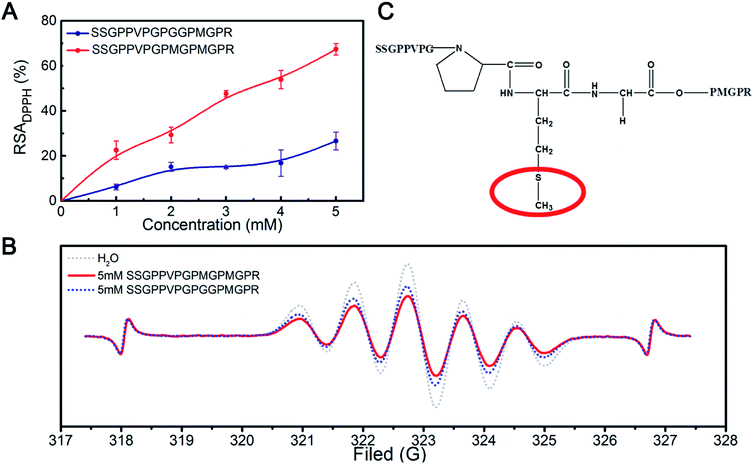
We designed a peptide that could be used to verify the above results. Gly was selected to replace the reactive Met residues, resulting in a synthetic peptide with the sequence of SSGPPVPGPGGPMGPR peptide as a control sample. The results of the scavenging DPPH radical assay (Fig. 6A) showed that the DPPH radical scavenging activity of the SSGPPVPGPGGPMGPR peptide was lower than that of the SSGPPVPGPMGPMGPR peptide. The result further demonstrates that the Met residue plays an essential role during scavenging of DPPH free radicals.
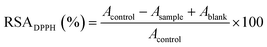
The ESR results (Fig. 6B) showed that the inhibition potency of the SSGPPVPGPMGPMGPR peptide scavenging DPPH radical is 43.10%, whereas that of the SSGPPVPGPGGMGPGR peptide scavenging DPPH radical is 29.78%, which is consistent with the above results. We deduced that the sulfur in the Met residue reacts with DPPH radicals; the structure of the SSGPPVPGPMGPMGPR peptide is shown in Fig. 6C.
In addition to the sequences of the peptides, the configuration of the peptides also strongly affects their bioactivity. This may be the key reason why only one Met residue in the SSGPPVPGPMGPMGPR peptide contributes to the scavenging of DPPH free radicals. However, because of the limitations of the experiment, the conformational structure of the peptide could not be observed directly. Therefore, a molecular dynamics (MD) simulation was used to explore the scavenging behaviour of the peptide. The conformational structure of the SSGPPVPGPMGPMGPR peptide in the presence of DPPH radicals was investigated. We placed one unfolded the SSGPPVPGPMGPMGPR peptide and one DPPH radical molecule in a box for the purposes of the simulation. The time variation of the potential energy of this system is shown in Fig. S5.† The initial potential energy was at a relatively high value and fluctuated within a large range. As the simulation progressed, the peptide folded and coalesced, and the corresponding potential energy decreased to a much lower value and was maintained at that level. This demonstrated that the curled peptide exhibited a higher stability than the unfolded peptide. In the curled peptide, the S–CH3 bond in the Met residue, which is on top of a peptide clump and has been experimentally observed to be effective, is exposed to the environment (shown inside the red circle in Fig. S5†), while another Met residue, which is inside the peptide clump, would hardly contact with the dissociated radical. This phenomenon provides further insight regarding the important role of the Met residue in the SSGPPVPGPMGPMGPR peptide during the scavenging of DPPH radicals.
Conclusions
Skipjack tuna bone has been used to obtain collagen peptides with high antioxidant activity. We found that a combination of trypsin and chymotrypsin can effectively hydrolyse the collagen of skipjack tuna bone to produce antioxidant hydrolysates. Among the antioxidant peptides isolated, the SSGPPVPGPMGPMGPR peptide exhibits particularly high antioxidant activity. In addition, a Met residue within this peptide chain was demonstrated to provide an active site for the scavenging of DPPH radicals. We believe that these findings will provide guidance for the development of peptides with high antioxidant activity from natural sources for functional foods.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support from the Fundamental Research Funds for the Central Universities (No. PYBZ1805) is gratefully acknowledged.Notes and references
- E. Hopps, D. Noto, G. Caimi and M. R. Averna, Nutr., Metab. Cardiovasc. Dis., 2010, 20, 72–77 CrossRef CAS PubMed.
- J. A. Leopold and J. Loscalzo, Free Radical Biol. Med., 2009, 47, 1673–1706 CrossRef CAS PubMed.
- T. Li, W. Xiang, F. Li and H. Xu, Biomaterials, 2017, 157, 17 CrossRef PubMed.
- H. E. Seifried, D. E. Anderson, E. I. Fisher and J. A. Milner, J. Nutr. Biochem., 2007, 18, 567–579 CrossRef CAS PubMed.
- P. Yongvanit, S. Pinlaor and H. Bartsch, Parasitol. Int., 2012, 61, 130–135 CrossRef CAS PubMed.
- Z. Yu, Y. Yin, W. Zhao, J. Liu and F. Chen, Food Chem., 2012, 135, 2078–2085 CrossRef CAS PubMed.
- F. Li, T. Li, W. Cao, L. Wang and H. Xu, Biomaterials, 2017, 133, 208–218 CrossRef CAS PubMed.
- G. F. Rocha, F. Kise, A. M. Rosso and M. G. Parisi, Food Chem., 2017, 237, 350 CrossRef CAS PubMed.
- B. M. Kimatu, L. Zhao, Y. Biao, G. Ma, W. Yang, F. Pei and Q. Hu, Food Chem., 2017, 230, 58–67 CrossRef CAS PubMed.
- H. Cao, Q. Luo, H. Wang, Z. Liu, G. Li and J. Liu, RSC Adv., 2019, 9, 9289–9300 RSC.
- L. Cai, X. Wu, Y. Zhang, X. Li, S. Ma and J. Li, J. Funct. Foods, 2015, 16, 234–242 CrossRef CAS.
- N. M. Sarbon, F. Badii and N. K. Howell, Food Hydrocolloids, 2018, 85, 311–320 CrossRef CAS.
- B. Liu, H. A. Aisa and A. Yili, Eur. Food Res. Technol., 2018, 244, 1615–1625 CrossRef CAS.
- Y. Zheng, Y. Li and G. Li, RSC Adv., 2019, 9, 5925–5936 RSC.
- A. B. Shazly, Z. He, M. A. Elaziz, M. Zeng, S. Zhang, F. Qin and J. Chen, Food Chem., 2017, 232, 753–762 CrossRef CAS PubMed.
- B. Hernández-Ledesma, B. Miralles, L. Amigo, M. Ramos and I. Recio, J. Sci. Food Agric., 2010, 85, 1041–1048 CrossRef.
- K. Kawashima, H. Itoh, M. Miyoshi and I. Chibata, Chem. Pharm. Bull., 1979, 27, 1912 CrossRef CAS PubMed.
- R. Wu, C. Wu, D. Liu, X. Yang, J. Huang, J. Zhang, B. Liao and H. He, Food Chem., 2018, 248, 346 CrossRef CAS PubMed.
- B. Giménez, E. Pérez-Santin, M. C. Gómez-Guillén, A. Alemán and P. Montero, Food Chem., 2010, 125, 334–341 Search PubMed.
- D. H. Ngo, Z. J. Qian, B. M. Ryu, J. W. Park and S. K. Kim, J. Funct. Foods, 2010, 2, 107–117 CrossRef CAS.
- N. H. Herpandi, A. Rosma and W. A. W. Nadiah, Compr. Rev. Food Sci. Food Saf., 2011, 10, 195–207 CrossRef CAS.
- X. R. Yang, Y. Q. Zhao, Y. T. Qiu, C. F. Chi and B. Wang, Mar. Drugs, 2019, 17, 78 CrossRef CAS PubMed.
- S. Nalinanon, S. Benjakul, H. Kishimura and F. Shahidi, Food Chem., 2011, 124, 1354–1362 CrossRef CAS.
- S. Saidi, M. Saoudi and R. Ben Amar, Environ. Sci. Pollut. Res. Int., 2018, 25, 17383–17392 CrossRef CAS PubMed.
- A. I. Martinez-Gonzalez, Á. G. Díaz-Sánchez, L. A. Rosa, C. L. Vargas-Requena, I. Bustos-Jaimes and E. Alvarez-Parrilla, Molecules, 2017, 22, 669 CrossRef PubMed.
- V. U. Ahmad, M. A. Lodhi, M. A. Abbasi and M. I. Choudhary, Fitoterapia, 2008, 79, 505–508 CrossRef CAS PubMed.
- J. Adler-Nissen, Enzymic hydrolysis of food proteins, Elsevier Applied Science Publishers, 1986 Search PubMed.
- L. Chao and S. Jiao, Biomacromolecules, 2014, 15, 436 CrossRef PubMed.
- L. D. Piu, A. Tassoni, D. I. Serrazanetti, M. Ferri, E. Babini, D. Tagliazucchi and A. Gianotti, Food Chem., 2014, 155, 199–206 CrossRef PubMed.
- A. J. Kirby and R. J. Schmidt, J. Ethnopharmacol., 1997, 56, 103–108 CrossRef CAS.
- Y. Ma and Y. L. Xiong, J. Agric. Food Chem., 2009, 57, 4372–4380 CrossRef CAS PubMed.
- F. Bamdad and L. Chen, Mol. Nutr. Food Res., 2013, 57, 493–503 CrossRef CAS PubMed.
- J.-R. Han, J.-N. Yan, S.-G. Sun, Y. Tang, W.-H. Shang, A.-T. Li, X.-K. Guo, Y.-N. Du, H.-T. Wu and B.-W. Zhu, Food Chem., 2018, 261, 337–347 CrossRef CAS PubMed.
- A. A. Karim and R. Bhat, Food Hydrocolloids, 2009, 23, 563–576 CrossRef CAS.
- E. Babini, D. Tagliazucchi, S. Martini, P. L. Dei and A. Gianotti, Food Chem., 2017, 228, 186 CrossRef CAS PubMed.
- H. Guo, Y. Kouzuma and M. Yonekura, Food Chem., 2009, 113, 238–245 CrossRef CAS.
- R. J. Elias, S. S. Kellerby and E. A. Decker, Crit. Rev. Food Sci. Nutr., 2008, 48, 430 CrossRef CAS PubMed.
- C. F. Chi, B. Wang, Y. M. Wang, B. Zhang and S. G. Deng, J. Funct. Foods, 2015, 12, 1–10 CrossRef CAS.
- H. Y. Luo, B. Wang, Z. R. Li, C. F. Chi, Q. H. Zhang and G. Y. He, LWT--Food Sci. Technol., 2013, 51, 281–288 CrossRef CAS.
- L. Gu, M. Zhao, W. Li, L. You, J. Wang, H. Wang and J. Ren, Food Chem. Toxicol., 2012, 50, 4085–4091 CrossRef CAS PubMed.
- Y. Yang and S. Mu, Electrochim. Acta, 2013, 109, 663–670 CrossRef CAS.
- J. Li, B. Kuttich, M. Gallei, J. Elbert, M. Rehahn and B. Stühn, Polymer, 2013, 54, 5703–5711 CrossRef CAS.
- Y. Ma, Y. Wu and L. Li, Food Chem., 2018, 264, 108–117 CrossRef CAS PubMed.
- E. Babini, D. Tagliazucchi, S. Martini, L. D. Più and A. Gianotti, Food Chem., 2017, 228, 186–196 CrossRef CAS PubMed.
- M. Memarpoor-Yazdi, A. Asoodeh and J. K. Chamani, J. Funct. Foods, 2012, 4, 278–286 CrossRef CAS.
- S. Knasmüller, W. Parzefall, R. Sanyal, S. Ecker, C. Schwab, M. Uhl, V. Mersch-Sundermann, G. Williamson, G. Hietsch and T. Langer, Mutat. Res., Fundam. Mol. Mech. Mutagen., 1998, 402, 185–202 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra04665h |
| ‡ These authors contributed equally to this work and should be considered co-first authors. |
| This journal is © The Royal Society of Chemistry 2019 |