Open Access Article
Open Access ArticleRetracted Article: Gm5820, an antisense RNA of FGF1, suppresses FGF1 expression at the posttranscriptional level to inactivate the ERK/STAT3 pathway and alleviates neuropathic pain in mice†
Xin Zhang‡
a,
Qing Yang‡b,
Tao Dingb,
Jingyu Xua,
Zeng Yanb,
Yanhua Menb,
Wenqi Xinb and
Haixia Xu *b
*b
aDepartment of Pain, Huaihe Hospital of Henan University, Kaifeng 475000, China
bDepartment of Anesthesiology, Huaihe Hospital of Henan University, No. 8 of Baobei Road, Kaifeng 475000, China. E-mail: haixiaxuxx@163.com; Tel: +86-371-23906620
First published on 9th September 2019
Abstract
Emerging evidence reveals that lncRNAs play important roles in various pathological processes, but precious little indicates their regulatory role in neuropathic pain. In this study, we performed unbiased whole transcriptome profiling in dorsal root ganglions (DRGs) from mice with sham operation and mice with chronic constriction injury (CCI). Gm5820 was one of the most downregulated RNA transcripts in CCI neuropathic pain model mice. Then, a pcDNA-Gm5820 expression vector was constructed and administered into CCI mice through intrathecal injection. The results showed that upregulation of Gm5820 alleviated mouse mechanical allodynia and thermal/cold hyperalgesia, and reduced the accumulation of inflammatory cytokines and ROS in the DRG tissue. Moreover, different concentrations of pcDNA-Gm5820 expression vector and Gm5820 siRNA were respectively transfected into primary DRG neurons from 4th to 6th lumbar vertebra (L4–L6). We found that Gm5820 overexpression improved cell viability and migration, and reduced the production of ROS, LDH and IL-1β. In contrast, Gm5820 knockdown had the opposite effects. Furthermore, RNA pull-down assays with FGF1 and Gm5820 cDNA probes both demonstrated that FGF1 mRNA and Gm5820 directly bound to each other. Moreover, Gm5820 negatively regulated the stability of FGF1 mRNA. Gm5820 suppressed the expression of FGF1 at the post-transcriptional level and negatively regulated the activation of ERK1/2-mediated STAT3, a critical contributor in neuropathic pain. In conclusion, Gm5820 directly binds to FGF1 mRNA and suppresses FGF1 expression at the posttranscriptional level, it functions as a negative regulator in the activation of the ERK/STAT3 pathway, and upregulation of Gm5820 alleviates neuropathic pain in CCI mice.
1. Introduction
Neuropathic pain is a most common neurological symptom, but it is still a relatively great challenge in clinical study owing to its complex mechanism. Emerging evidence revealed that inflammatory events contribute to the incidence and development of neuropathic pain.1,2 When the nervous system is damaged, multiple inflammatory cytokines are released causing local or large range of inflammation.3 Signal transducers and activators of transcription (STATs) are key regulators in the mediation of inflammation and immune response.4,5 STATs have been shown to regulate functions of the central nervous system, including synaptic plasticity, neural degeneration and memory formation.6 Recently, STATs were found to be activated by inflammatory cytokines during nerve damage.7 Among the STATs, STAT3 displays a major regulatory role in activation of macrophages and it is revealed to be centrally involved in the incidence and development of chronic pain, including chronic neuropathic pain.8–12 Stimulated by inflammatory cytokines in response to nerve damage, STAT3 protein was phosphorylated by tyrosine or serine/threonine kinases to activate, such as Janus kinase 2 (JAK2) and extracellular signal-regulated kinase 1/2 (ERK1/2).13,14 Activated STAT3 in turn transcriptionally promoted expression of inflammatory cytokines and increased production of neuronal deleterious substances, such as interleukin-1β (IL-1β), tumor necrosis factor-α (TNF-α), reactive oxygen species (ROS), and lactate dehydrogenase (LDH), etc.15 How STAT3 expression and activity are regulated and the mechanism underlying the regulatory network of STAT3-dependent pathways during the incidence and development of neuropathic pain remain largely unknown.Accumulating evidence suggests that lncRNAs are functional RNA regulators involved in diverse biological processes. Up to date, the functions of more than 1000 human lncRNAs have been annotated, most of which are involved in the regulation of carcinogenesis, development of degenerative diseases, and incidence of genetic diseases.16 LncRNAs in the development of neurological disorders are also attracting warm attentions. Many lncRNAs are highly expressed in impaired nervous tissues, displaying particularly temporal and spatial expression patterns.17,18 However, only a couple of lncRNAs were reported to regulate neuropathic pain behaviors. For example, downregulation of lncRNA uc.48+ by siRNA in DRGs decreased P2X3 expression, inactivated ERK1/2, and also alleviated neuropathic pain in diabetic rats.19
In this study, to identify potential lncRNAs relevant to neuropathic pain caused by chronic constriction injury (CCI), unbiased whole transcriptome profiling was performed in DRGs from sham mice and CCI mice. Gm5820 was one of the most downregulated RNA transcripts and was identified to be involved in the development of neuropathic pain. We also studied the role of Gm5820 in FGF1-mediated activation of ERK1/2 and STAT3 in primary neurons from the DRG.
2. Results
2.1. Transcriptomic response of the DRGs in sham and CCI stress
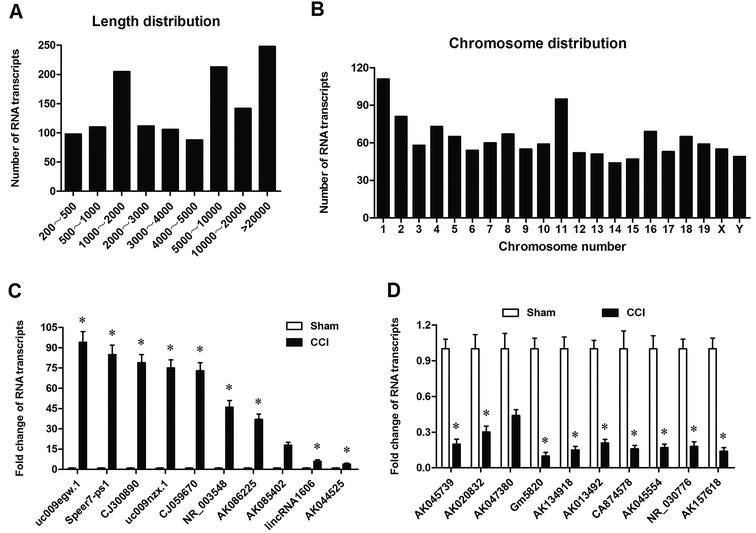
In order to assess the transcriptomic response of the DRG to CCI stress, we performed RNA sequencing on DRG tissues that were from L4–L6 of sham and CCI mice. We found a total of 6112 expressed transcripts, including lncRNAs and uncharacterized RNA transcripts that may be mRNA or lncRNAs. CCI led to differential expression of 1322 transcripts. We summarized a couple of characteristics of these transcripts, including length distribution and chromosomal distribution. Their length distribution was relatively uniform except for the transcripts with length of 1000–2000 nt (Fig. 1A). As for chromosome distribution, the largest number of these transcripts was located on Chr.1 and Chr.11, and the remaining was relatively evenly distributed over other chromosomes (Fig. 1B). The detail information of the top 10 up-regulated and down-regulated transcripts were listed in Table 1. Then, qPCR analysis was applied to confirm the sequencing results (Fig. 1C and D). The results showed that the level of RNA Gm5820 was the most downregulated transcript in CCI mice (Fig. 1D).| RNA transcript | Fold change (CCI/sham) | Adjusted P-value |
|---|---|---|
| uc009egw.1 | 36.89 | 7.50 × 10−8 |
| Speer7-ps1 | 27.23 | 4.05 × 10−10 |
| CJ300890 | 22.99 | 0.0000563 |
| uc009nzx.1 | 19.54 | 1.51 × 10−7 |
| CJ059670 | 17.01 | 7.99 × 10−5 |
| NR_003548 | 16.45 | 3.25 × 10−7 |
| AK086225 | 14.90 | 1.28 × 10−6 |
| AK085402 | 14.30 | 0.000327 |
| lincRNA1606 | 12.01 | 4.12 × 10−6 |
| AK044525 | 10.55 | 0.000423 |
| AK045739 | −16.89 | 0.0335 |
| AK020832 | −13.23 | 0.0167 |
| AK047380 | −11.99 | 0.0178 |
| Gm5820 | −11.54 | 4.51 × 10−6 |
| AK134918 | −9.01 | 3.99 × 10−6 |
| AK013492 | −8.45 | 5.16 × 10−8 |
| CA874578 | −7.99 | 5.28 × 10−7 |
| AK045554 | −7.30 | 0.000111 |
| NR_030776 | −7.01 | 0.000157 |
| AK157618 | −6.55 | 0.0000543 |
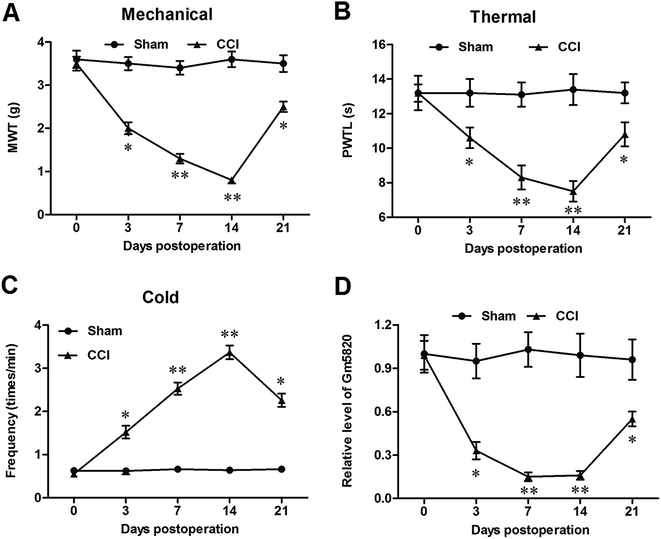
2.2. Gm5820 was significantly downregulated and its expression level was consistent with the change trend of neuropathic pain in CCI mice
The expression of Gm5820 was studied in sham and CCI mice at different time points. The time course of difference scores in the withdrawal latencies to mechanical, thermal, and cold stimuli were tested from day 0 to day 21 post CCI surgery. The results showed that, before and after surgery, withdrawal latencies displayed no significant difference (Fig. 2A–C). On day 3, CCI mice began to show hypersensitivity to stimuli. The withdrawal latencies of the CCI group decreased significantly from day 3 to day 14, compared with the sham group (Fig. 2A–C), indicating that CCI mice were more sensitive as compared to the sham group. The results from qPCR analysis revealed that Gm5820 expression in the sham group was stable from day 0 to day 21, whereas, in the CCI group, Gm5820 expression was decreased significantly from day 3 to day 14, but rebounded at day 21 (Fig. 2D), suggesting that the downregulation of Gm5820 from day 3 to day 14 might be contributed to CCI caused neuropathic pain.2.3. Gm5820 overexpression by intrathecal injection of pcDNA-Gm5820 alleviated mechanical, thermal and cold allodynia in CCI mice
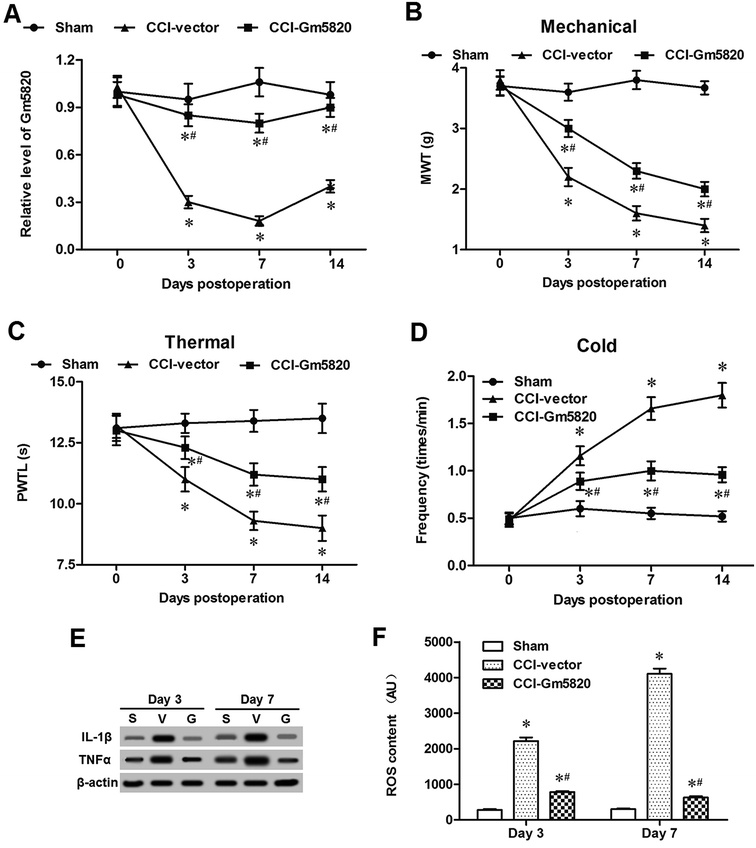
To explore the role of Gm5820 in CCI-caused neuropathic pain, a pcDNA-Gm5820 expression vector was constructed and administered into CCI mice through intrathecal injection at L4–L6. The results from qPCR analysis in L4–L6 spinal cord neurons showed that the injection of the pcDNA-Gm5820 dramatically increased the level of Gm5820 from day 3 to day 14 in mice (Fig. 3A). The data on the withdrawal latencies showed that Gm5820 overexpression caused a significantly increase in mechanical withdrawal threshold (MWT) and paw withdraw thermal latency (PWTL), and decrease in the frequency of withdrawal response to cold allodynia (Fig. 3B–D). Moreover, we found that Gm5820 overexpression give rise to a sharp decrease in the expression of inflammatory genes IL-1β and TNF-α (Fig. 3E) and accumulation of ROS in spinal cord neurons (Fig. 3F). These data indicated that upregulation of Gm5820 could alleviate mechanical, thermal and cold allodynia and production of inflammatory cytokines in CCI mice.2.4. Gm5820 improved proliferation and migration, and reduced oxidative stress and inflammation in mouse primary DRG neurons
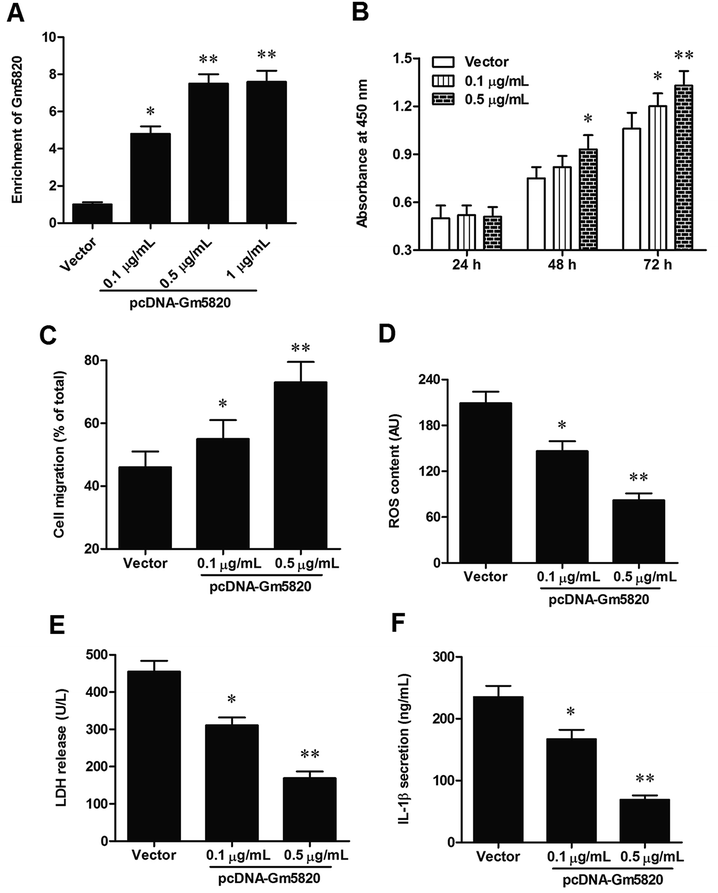
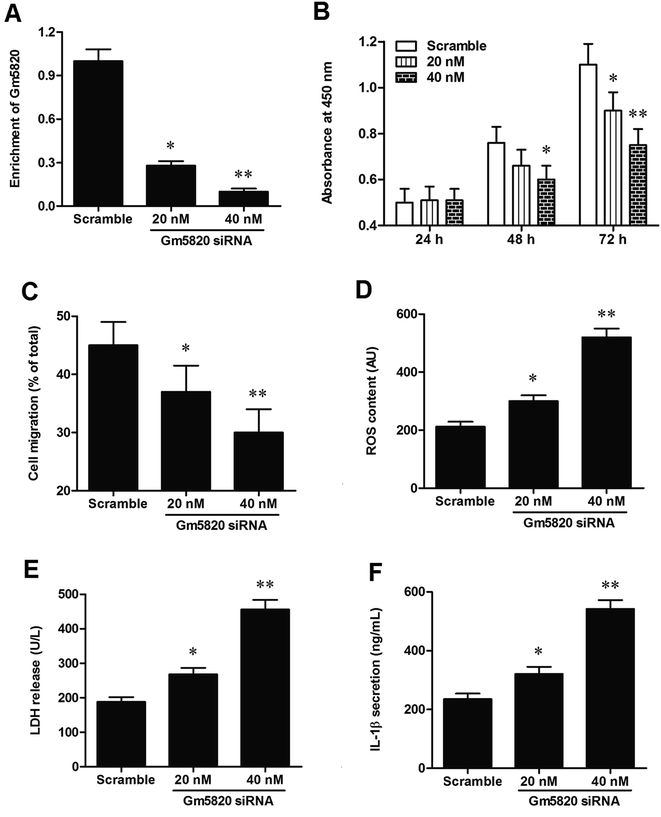
Then, to further investigate the role of Gm5820 in mechanism of neuropathic pain, primary DRG neurons were isolated from L4–L6 of mice and cultured in vitro, and different concentrations of pcDNA-Gm5820 and Gm5820 siRNA were respectively transfected into the cells. The overexpression efficiencies were detected by qPCR. The results showed that transfection with 0.1 μg mL−1 pcDNA-Gm5820 caused a 4.5-fold increase in Gm5820 level (P < 0.05), while 0.5 μg mL−1 and 1 μg mL−1 pcDNA-Gm5820 both caused an about 7-fold increase in Gm5820 level (P < 0.01, Fig. 4A). Capacities of proliferation and migration have been widely regarded to be important for nerve damage repair and regeneration. Adverse environments such as oxidative stress and inflammation are important factors affecting nerve repair and contributing to neuropathic pain. The data on CCK-8 assay showed that 0.1 μg mL−1 pcDNA-Gm5820 transfection modestly increased and 0.5 μg mL−1 pcDNA-Gm5820 transfection distinctly increased the capacity of proliferation of primary DRG neurons (Fig. 4B). The results from Transwell Cell Migration showed that 0.1 μg mL−1 pcDNA-Gm5820 transfection modestly increased and 0.5 μg mL−1 pcDNA-Gm5820 transfection distinctly increased the capacity of migration of primary DRG neurons (Fig. 4C). Moreover, the data on the detection of ROS, LDH and II-1β contents revealed that 0.1 μg mL−1 pcDNA-Gm5820 transfection modestly decreased and 0.5 μg mL−1 pcDNA-Gm5820 transfection sharply decreased the ROS accumulation, LDH release and II-1β secretion in spinal cord neurons (Fig. 4D–F).The knockdown efficiencies of different concentrations of Gm5820 siRNA were detected by qPCR, and the results showed that transfection with 20 nM Gm5820 siRNA caused a 70% decrease in Gm5820 level (P < 0.05), while 40 nM Gm5820 siRNA caused a 91% decrease in Gm5820 level (P < 0.01, Fig. 5A). In contrast to the results from Gm5820 overexpression, 20 nM Gm5820 siRNA modestly decreased and 40 nM Gm5820 siRNA sharply decreased the proliferation and migration of primary spinal cord neurons (Fig. 5B and C). Moreover, 20 nM Gm5820 siRNA modestly promoted and 40 nM Gm5820 siRNA markedly promoted the ROS accumulation, LDH release and II-1β secretion in DRG neurons (Fig. 5D–F). These data above indicated that Gm5820 had a positive effect on the proliferation and migration and a suppressive on oxidative stress and inflammation in DRG neurons.
2.5. Gm5820 directly bound to FGF1 mRNA and suppressed FGF1 expression at the posttranscriptional level to inactivate the ERK/STAT3 pathway
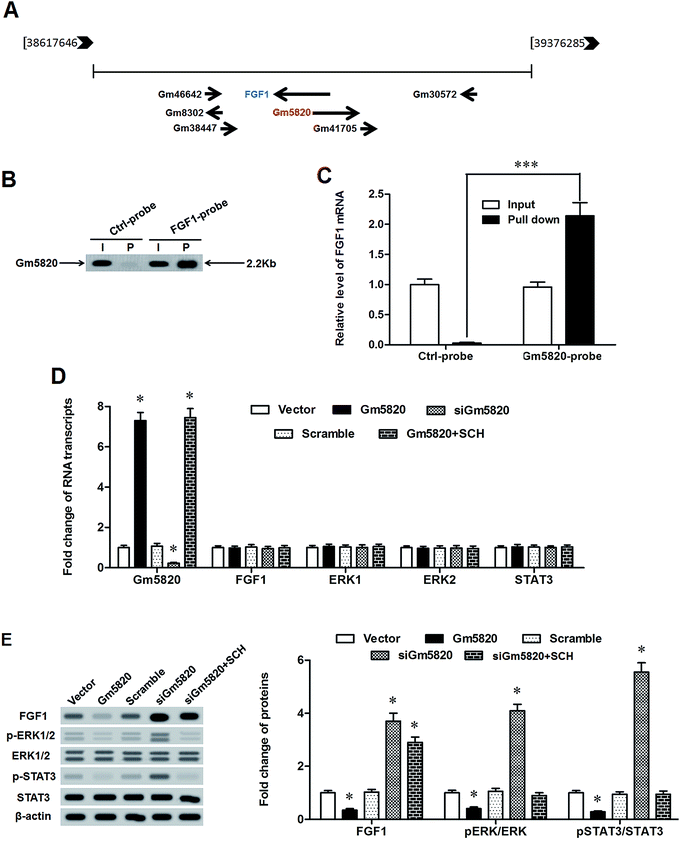
Furthermore, to explore the underlying mechanism from which Gm5820 regulate functions of DRG neurons, the landscape of Gm5820 gene in the mouse genome was analyzed. The results showed that Gm5820 gene was located at the locus of 38617646 to 39376285 on mouse chromosome 18, near which FGF1 gene and several uncharacterized genes were located, including Gm46642, Gm8302, Gm38447, Gm41705 and Gm30572 (Fig. 6A). It is worth pointing out that Gm5820 and FGF1 are located on opposite strands and they reversely overlap with each other. Alignment of Gm5820 RNA and FGF1 mRNA by the online bioinformatics tool RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) revealed that Gm5820 RNA is an antisense RNA of FGF1 that could activate STAT3 and impaired functions of primary DRG neurons (ESI Fig. 1†). To study whether they interact with each other, biotinylated cDNA probes for FGF1 mRNA and Gm5820 RNA were applied into RNA pull-down assays. Then, northern blotting and qPCR were respectively used to detect the expression of Gm5820 RNA and FGF1 mRNA. We detected high abundance of Gm5820 RNA in the RNA complexes pulled down by the FGF1 probe (Fig. 6B). High abundance of FGF1 mRNA was also detected in the RNA complexes pulled down by the Gm5820 probe (Fig. 6C). Moreover, the effect of Gm5820 on the expression of FGF1 gene was investigated. The data on qPCR and western blotting showed that neither upregulation nor downregulation of Gm5820 had no effect on the expression of FGF1 mRNA and mRNA molecules of its downstream genes ERK1/2 and STAT3 (Fig. 6D), however, upregulation of Gm5820 by pcDNA-Gm5820 transfection sharply reduced the level of FGF1 protein and activation of ERK1/2 and STAT3, and downregulation of Gm5820 by Gm5820 siRNA distinctly increased the level of FGF1 protein and activation of ERK1/2 and STAT3 (Fig. 6E). In CCI mice, upregulation of Gm5820 by pcDNA-Gm5820 injection also had no impact on expression of mRNA of FGF1 and STAT3, but increased the expression of FGF1 and phosphorylation of STAT3 (ESI Fig. 2†). As a positive control, treatment with 0.1 nM SCH772984, a specific inhibitor of ERK1/2 inhibitor, abolished the promoting effect of Gm5820 knockdown on FGF1-induced activation of ERK1/2 and STAT3 (Fig. 6E). These data indicated that Gm5820 directly bound to FGF1 mRNA and suppressed expression of FGF1 at the post-transcription level to inactivate the ERK/STAT3 pathway in DRG neurons.3. Discussion
Gm5820 was originally found in the generation of full-length cDNA libraries for human and mouse genomes in 2002.20 From then on to now, there have no documents that report the exact role in any biological processes. In this study, our RNA sequencing for unbiased whole transcriptome profiling in DRG of sham and CCI mice unearthed 1322 differentially expressed transcripts. Length distribution of these transcripts was relatively uniform except for the transcripts with length of 1000–2000 nt, and the largest number of these transcripts was located on Chr.1 and Chr.11 in which it is revealed that a considerable number of genes related to immunity, functions of lymphocytes and inflammation (an important pathogenesis of neuropathic pain). We reported that Gm5820 RNA was one of the most downregulated RNA transcripts in CCI mice and found that upregulation of Gm5820 was a relief on CCI caused neuropathic pain in vivo.Neuropathic pain is a typical external manifestation of the increased responsiveness of nociceptive neurons. The DRG is an expansive nodule of the dorsal root of the spinal cord near the medial surface of the intervertebral foramen. It consists of centripetal sensory fibers that are responsible for receiving all nerve impulses from body receptors, including general somatosensory and visceral sensations.21 It's the starting point of body feeling, the key to study the mechanism of neuropathic pain and also an important target for pain treatment.22 In current study, Gm5820 RNA was identified as a most downregulated RNA transcript in the DRG of CCI mice. Our in vitro experiments also showed that Gm5820 could improve the capacities of proliferation and migration, and reduce oxidative stress and inflammation in primary DRG neurons. Upregulation of Gm5820 displayed a diminishing effect to neuropathic pain in vivo.
RNAs are also important regulators in various biological processes. High throughput RNA sequencing has revealed that many RNA molecules, most of which are lncRNAs, potentially regulate functions of cells in the nervous system. The previous study, finding Gm5820, revealed that there was an Open Reading Frame (ORF) in Gm5820 RNA and proposed the potential protein product with length of 117 amino acids (weighing about 13 KD).20 Later, Gm5820 was annotated as an antisense gene against FGF1.23 Here, we found that Gm5820 RNA bound to FGF1 mRNA and suppressed expression of FGF1 protein, indicating that Gm5820 could functioned as a lncRNA-like regulator. Similar to our research, a quite recent study in mouse model of obliterative bronchiolitis also annotated Gm5820 as a lncRNA.24 Antisense lncRNAs are a specialized type of lncRNAs with their sequences complementarily to other transcripts. This type of lncRNA regulates gene expression at either transcriptional or post-transcriptional level through a variety of biological mechanisms including transcription interference, RNA–DNA interaction and RNA–RNA interaction, providing a theoretical basis for antisense lncRNA as a therapeutic target.25 Antisense lncRNAs may positively or negatively regulate the sense gene via RNA–RNA interaction.26,27 For example, PU.1 antisense lncRNA promotes adipogenesis through preventing PU.1 mRNA translation via binding to PU.1 mRNA to form mRNA-antisense lncRNA duplex in preadipocytes.28 Here, our pull-down assay demonstrated that Gm5820 directly bind with its sense transcript FGF1 mRNA and form sense-antisense RNA duplex. Therefore, it remains to be further studied whether Gm5820 gene encodes proteins or not, but our current study confirms that the molecular mechanism of Gm5820 RNA in the regulation of FGF1 expression is like an antisense lncRNA.
FGF1 belongs to the fibroblast growth factor family which are involved in various biological processes, including embryonic development, tissue repair, organogenesis, and carcinogenesis.29 FGF1 regulates proliferation and migration in multiple cell types, especially mesoderm- and neuroectoderm-derived cells.30,31 FGF1 has been found to induce the activation of ERK1/2 and STAT3 in some types of cells, such as prostate cancer cells and retina precursor cells.31,32 Recently, FGF1 was shown to be aberrantly expressed in spinal cord injury and regulate neuron growth and inflammation. In this study, our experiments in primary DRG neurons revealed that Gm5820 could negatively regulate the FGF1 at the post-transcriptional level, improve cell proliferation and migration, and alleviate oxidative stress and inflammation. Our results indicated that FGF1 functioned as a negative regulator of DRG neuron function and a contributor to neuropathic pain. Although a couple of previous studies revealed that FGF1 may have an improvement effect on neuron/neurite growth,33,34 more recent studies indicated that FGF-1 motivated hemichannels to promote inflammatory response in injured spinal cords.35–37
Taken together, our current study reports a novel antisense RNA of FGF1, named Gm5820, can alleviate chronic constriction injury caused neuropathic pain in mice. Gm5820 suppresses expression of FGF1 at the posttranscriptional level to inactivate the ERK/STAT3 pathway, improves cell proliferation and migration, and alleviates oxidative stress and inflammation in spinal cord neurons.
4. Materials and methods
4.1. Animals and ethics statement
Male C57BL/6 mice (aged 3 months, weighing about 26 g) were obtained from Laboratory Animal Center of Huaihe Hospital of Henan University (Kaifeng, China). They were acclimatized for 2 weeks before the CCI model was established. This study was approved by the Ethics Committee of Huaihe Hospital of Henan University (code: 18HHHU013).4.2. Establishment of CCI model
For the establishment of CCI model, 3% pentobarbital was used to anaesthetize the animals and the sciatic nerve was exposed after cutting the middle of the thigh. 4-0 nylon thread was used to ligate 4 nodes. The interval between the nodes was about 1 mm and the tightness was moderate. In the sham operation group, the sciatic nerve was exposed without ligation. The incision was washed with sterile physiological saline and then layer sutured. To confirm the success of establishment, their mechanical allodynia and thermal/cold hyperalgesia were detected at about 13:00 on days 0, 3, 7, 14 and 21 about.4.3. Mechanical allodynia
Mice were acclimatized in suspended cages. The hind paws were probed with calibrated Electronic von Frey device (IITC Life) erected to the plantar surface and kept for about 5 s. A positive response represented a sharp withdrawal of the paw. The mechanical withdrawal threshold (MWT) was indicated as the average of the measurements in each test session.4.4. Thermal hyperalgesia
Thermal hyperalgesia was detected with a Plantar Test Apparatus for mice and rats (IITC). Mice were acclimated on a glass floor above a radiant heat producer (temperature is set at 120 °C) that was aimed at the plantar surface of the hind paw. In each test session, triple measurements of latency were done for each hind paw. Each test was done with intervals greater than 3 min and the device was cutoff for 30 s in the intervals of each test to avoid tissue damage. The thermal withdrawal latency (TWL) was indicated as the average of the measurements in each test session.4.5. Cold hyperalgesia
A drop of acetone was gently applied to each hind paw at room temperature (26 ± 1 °C). A rapid hind paw withdrawal in response to the volatilization of acetone was considered as a sign of cold allodynia. The test was repeated 3 times for every hind paw, with an interval of 3 min. Times of shaking, lifting or licking the paw during 1 min were recorded. An increase in the rate of withdrawal response interpreted as increased cold sensitivity.4.6. RNA sequencing
Total RNA was isolated from the sciatic nerve tissues using the miRNeasy kit (Qiagen). RNA used for RNA sequencing had a minimum integrity index of 9.5 (checked by Agilent 2100 Bioanalyzer). Strand-specific barcoded libraries were generated using the TruSeq Stranded mRNA Library Prep Kit (Illumina) according to the manufacturer's protocol with the following modifications: PCR amplification was limited to 12 cycles to avoid bias from PCR “jackpot” mutations. RNA sequencing was performed on the HiSeq 2500 (Illumina). About 32 million filtered reads per sample uniquely mapped to the mouse genome. Transcripts were annotated using publicly available datasets from RefSeq for protein-coding transcripts and the Mouse Body Map lncRNAs annotation for long noncoding genes. The enrichment of each RNA transcript was measured in fragments per kilobase of exon per million reads mapped (FPKM) using Cufflinks 2.1.1. Differential expression (DE) between sham and CCI individuals was tested with Cuffdiff. RNA transcripts (mRNAs and lncRNAs) with a fold change of 2.0 and a false discovery rate-adjusted P value of <0.05 were considered as differentially expressed. Ingenuity Pathway Analysis software (Qiagen) was used to conduct gene ontology (GO) analysis.4.7. Grouping and administrations
To explore the function of Gm5820 in CCI mice, a total of 45 male C57BL/6 mice were enrolled in the in vivo experiments, including 15 mice with sham operation (sham), 15 CCI mice administered with 5 mg kg−1 empty vector by intrathecal injection (CCI-vector), and 15 CCI mice administered with 5 mg kg−1 pcDNA-Gm5820 by intrathecal injection (CCI-Gm5820) every 3 days.4.8. Real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from spinal cord neurons using TRIzol reagent (Invitrogen) in accordance with the manufacturers' instructions. RNA quality and quantity were checked, and then, 500 ng of total RNA from each sample was applied in the reverse transcription reaction with SuperScriptIII Reverse Transcriptase Kit (Invitrogen). RT-qPCR was carried out in triple wells with SYBR Green Realtime PCR Master Mix (Invitrogen) in Applied Biosystems Real-Time PCR Instruments (Thermo Fisher Scientific). The abundances of the RNA transcripts were analyzed by step-one System SDS Software 1.3.1 (Thermo Fisher Scientific) using the 2−ΔΔCT method. All values were normalized to that of β-actin.4.9. Western blotting
Total protein was exacted using RIPA lysis buffer (Sigma) and protease/phosphatase inhibitors. 50 μg of protein from each sample was separated by 12% SDS-PAGE and then transferred onto a PVDF membrane (Millipore, Billerica, MA, USA). The blots were blocked with TBST containing 5% non-fat milk and incubated with primary antibodies as follows: rabbit anti-TNFα (Cell Signaling Technology, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), rabbit anti-IL-1β (Cell Signaling Technology, 1
500), rabbit anti-IL-1β (Cell Signaling Technology, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), rabbit anti-FGF1 (Cell Signaling Technology, 1
500), rabbit anti-FGF1 (Cell Signaling Technology, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400), mouse anti-ERK1/2 (Cell Signaling Technology, 1
400), mouse anti-ERK1/2 (Cell Signaling Technology, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400), mouse anti-pERK1/2 (Cell Signaling Technology, 1
400), mouse anti-pERK1/2 (Cell Signaling Technology, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), mouse anti-STAT3 (Cell Signaling Technology, 1
200), mouse anti-STAT3 (Cell Signaling Technology, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), mouse anti-pSTAT3 (Cell Signaling Technology, 1
500), mouse anti-pSTAT3 (Cell Signaling Technology, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) and mouse anti-β-actin (Abcam, 1
200) and mouse anti-β-actin (Abcam, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) overnight at 4 °C. After rinsing with TBST, membranes were incubated with horseradish peroxidase-linked anti-rabbit or anti-mouse IgG (Santa Cruz Biotechnology, 1
1000) overnight at 4 °C. After rinsing with TBST, membranes were incubated with horseradish peroxidase-linked anti-rabbit or anti-mouse IgG (Santa Cruz Biotechnology, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) for 1 hour at room temperature. Membranes were processed with ECL Plus kit (Thermo Fisher Scientific) and exposed in a Gel imaging system (Thermo Fisher Scientific). Relative intensities of the proteins were normalized to β-actin.
2000) for 1 hour at room temperature. Membranes were processed with ECL Plus kit (Thermo Fisher Scientific) and exposed in a Gel imaging system (Thermo Fisher Scientific). Relative intensities of the proteins were normalized to β-actin.
4.10. Culture of primary DRGs and cell transfection
Spinal cord was isolated from L4–L6 of 3-month-old mice and digested with 0.25% trypsin for 30 min followed by 0.2% collagenase II for 2 h. The cells were washed with D-Hank's buffer (Invitrogen) for 3 times and then seeded in the plates with DMEM/F12 (containing 4.5 g L−1 glucose) supplemented with 20% FBS and 4 mM L-glutamine. Cells were cultured in a 37 °C humidified cell culture chamber with 5% CO2.On reaching 70% confluence, different concentrations of pcDNA-Gm5820 (constructed by GenScript Company, Nanjing, China) and Gm5820 siRNA (designed and synthesized by Ribobio Technology, Guangzhou, China) were respectively transfected in to the cells with Lipofectamine® 3000 reagent (Thermo Fisher Scientific) in accordance with the manufacturers' instructions. After 48 h, the cells were harvested. The transfection efficiencies were about 75%.
4.11. Cell apoptosis and proliferation
Cell apoptosis was detected with the Annexin V-FITC/PI cell apoptosis kit in accordance with the manufacturers' instructions.Cell proliferation was tested using the Cell Counting Kit-8 cell proliferation kit (Dojindo Laboratories, Kumamoto, Japan) in accordance with the manufacturers' instructions.
4.12. Cell migration
Cell migration was detected with the Transwell migration assay. 3.5 × 104 cells were grown in the top chamber with a non-coated 24-well insert membrane (8 μm; Corning) with serum-free medium. Medium plus 10% FBS was used as a chemoattractant in the lower chamber. The cells were incubated for 24 h and then the non-migrated cells in the upper chamber were removed by a cotton swab. The filters were stained with 2% crystal violet. The migrated cells adhering to the filter were examined and counted under an Olympus IX70 microscope (Olympus Corporation, Osaka, Japan).4.13. Detection of ROS content
The spinal cord neurons were split with cell lysis buffer (Sigma) and ROS content in the cells was detected with a DCFH-DA ROS Assay kit (Sigma).4.14. Detection of LDH and IL-1β contents in the supernatant
Following treatment, the cells and their medium were harvested, and then centrifugated at 1000 rpm for 5 min. The LDH content in the supernatant was detected with LDH Detection Kit (Roche), and the IL-1β content in the supernatant was detected with an IL-1β ELISA Kit (MBL, Naka-ku Nagoya, Japan).4.15. Pull-down assays with biotinylated cDNA probes
The biotinylated DNA probes complementary to Gm5820 and FGF mRNA (synthesized by Invitrogen) were dissolved in 500 μL of wash/binding buffer (containing 0.5 M NaCl, 20 mM Tris–HCl, pH 7.5, and 1 mM EDTA). The probes were incubated with streptavidin-coated magnetic beads (Sigma) at 25 °C for 2 h to generate probe-coated magnetic beads. The esophageal cancer cell lysates were incubated with probe-coated beads. After washing with the wash/binding buffer, the RNA complexes bound to the beads were eluted and extracted for northern blot analysis. Gm5820 pull-down probe: 5′-TCC CCA AGA GTC ATG GAA TAT C-3′. FGF mRNA pull-down probe: 5′-GAA ACA CTA TTC ATC TCT GAC AT-3′. Random pull-down probe: 5′-TGA TGT CTA GCG CTT GGG CTT TG-3′.4.16. Northern blot analysis
The pulled down RNA samples were collected and run on a 15% polyacrylamide–urea gel, transferred to positively charged nylon membranes (Millipore) followed by cross-linking through UV irradiation. The membranes were subjected to hybridization with 100 pmol 3′-digoxigenin (DIG)-labelled probes overnight at 43 °C. The probes were labelled with DIG using a 3′-End DIG Labelling Kit (Roche). The detection was performed using a DIG luminescent detection kit (MyLab).4.17. Statistical analysis
Data were shown as the means ± SD. Statistical analyses were performed using Student's t-test between two groups and one-way ANOVA among multiple groups by SPSS 16.0 software. Differences were statistically significant when P < 0.05.Conflicts of interest
The authors declared that they have no conflict of interest.Acknowledgements
This study was supported by the Social Development Science and Technology Project of Kaifeng City (No. 1503113) and the Scientific and Technological Project of Henan Province in China (No. 172102310689).References
- J. T. Liou, F. C. Liu, C. C. Mao, Y. S. Lai and Y. J. Day, Inflammation confers dual effects on nociceptive processing in chronic neuropathic pain model, Anesthesiology, 2011, 114, 660–672 CrossRef PubMed.
- P. L. Stemkowski and P. A. Smith, Sensory neurons, ion channels, inflammation and the onset of neuropathic pain, Can. J. Neurol. Sci., 2012, 39, 416–435 CrossRef.
- G. Tenorio, A. Kulkarni and B. J. Kerr, Resident glial cell activation in response to perispinal inflammation leads to acute changes in nociceptive sensitivity: implications for the generation of neuropathic pain, Pain, 2013, 154, 71–81 CrossRef.
- H. Siavash, N. G. Nikitakis and J. J. Sauk, Signal transducers and activators of transcription: insights into the molecular basis of oral cancer, Crit. Rev. Oral Biol. Med., 2004, 15, 298–307 CrossRef CAS.
- M. Liao, J. Xu, A. J. Clair, B. Ehrman, L. M. Graham and M. J. Eagleton, Local and Systemic Alterations in Signal Transducers and Activators of Transcription (STAT) Associated with Human Abdominal Aortic Aneurysms 1, J. Surg. Res., 2012, 176, 321–328 CrossRef CAS.
- P. Dell'Albani, R. Santangelo, L. Torrisi, V. G. Nicoletti and A. M. G. Stella, Role of the JAK/STAT Signal Transduction Pathway in the Regulation of Gene Expression in CNS, Neurochem. Res., 2003, 28, 53–64 CrossRef PubMed.
- G. V. Sherbet, Molecular Approach to Targeted Therapy for Multiple Sclerosis, CNS Neurol. Disord. - Drug Targets, 2016, 15, 20–34 CrossRef CAS.
- L. Williams, L. Bradley, A. Smith and B. Foxwell, Signal transducer and activator of transcription 3 is the dominant mediator of the anti-inflammatory effects of IL-10 in human macrophages, J. Immunol., 2004, 172, 567–576 CrossRef CAS.
- Y. Komohara, H. Horlad, K. Ohnishi, K. Ohta, K. Makino, H. Hondo, R. Yamanaka, K. Kajiwara, T. Saito and J. Kuratsu, M2 macrophage/microglial cells induce activation of Stat3 in primary central nervous system lymphoma, J. Clin. Exp. Hematop., 2011, 51, 93–99 CrossRef.
- J. Tang, Z. H. Li, S. N. Ge, W. Wang, X. P. Mei, T. Zhang, L. X. Xu and J. L. Li, The Inhibition of Spinal Astrocytic JAK2-STAT3 Pathway Activation Correlates with the Analgesic Effects of Triptolide in the Rat Neuropathic Pain Model, J. Evidence-Based Complementary Altern. Med., 2012, 2012, 185167–185180 Search PubMed.
- M. Tsuda, Y. Kohro, T. Yano, T. Tsujikawa, J. Kitano, H. Tozakisaitoh, S. Koyanagi, S. Ohdo, R. R. Ji and M. W. Salter, JAK-STAT3 pathway regulates spinal astrocyte proliferation and neuropathic pain maintenance in rats, Brain A J. Neurol., 2011, 134, 1127–1139 CrossRef.
- Z. J. Xue, L. Shen, Z. Y. Wang, S. Y. Hui, Y. G. Huang and C. Ma, STAT3 inhibitor WP1066 as a novel therapeutic agent for bCCI neuropathic pain rats, Brain Res., 2014, 1583, 79–88 CrossRef CAS PubMed.
- Z. F. Wang, Q. Li, S. B. Liu, W. L. Mi, S. Hu, J. Zhao, Y. Tian, Q. L. Mao-Ying, J. W. Jiang and H. J. Ma, Aspirin-triggered Lipoxin A4 attenuates mechanical allodynia in association with inhibiting spinal JAK2/STAT3 signaling in neuropathic pain in rats, Neuroscience, 2014, 273, 65–78 CrossRef CAS PubMed.
- J. Donnerer and I. Liebmann, A fluorescence-immunohistochemical study on phosphorylation of ERK1/2, p38, and STAT3 in rat dorsal root ganglia following noxious stimulation of hind paw sensory neurons, Tissue Cell, 2011, 43, 178–189 CrossRef CAS PubMed.
- Q. Chen, H. Wang, Y. Liu, Y. Song, L. Lai, Q. Han, X. Cao and Q. Wang, Inducible microRNA-223 down-regulation promotes TLR-triggered IL-6 and IL-1β production in macrophages by targeting STAT3, PLoS One, 2012, 7, e42971 CrossRef CAS PubMed.
- J. Li, Z. Xuan and C. Liu, Long Non-Coding RNAs and Complex Human Diseases, Int. J. Mol. Sci., 2013, 14, 18790–18808 CrossRef CAS PubMed.
- L. Han, K. Zhang, Z. Shi, J. Zhang, J. Zhu, S. Zhu, A. Zhang, Z. Jia, G. Wang and S. Yu, LncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis, Int. J. Oncol., 2012, 40, 2004–2012 CAS.
- J. Briggs, E. Wolvetang, J. Mattick, J. Rinn and G. Barry, Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution, Neuron, 2015, 88, 861–877 CrossRef CAS.
- S. Wang, X. Hong, L. Zou, J. Xie, W. Hong, W. Bing, Z. Yi, Q. Lv, Z. Xi and M. Ying, LncRNA uc.48+ is involved in diabetic neuropathic pain mediated by the P2X3 receptor in the dorsal root ganglia, Purinergic Signalling, 2016, 12, 139–148 CrossRef CAS PubMed.
- R. L. Strausberg, E. A. Feingold, L. H. Grouse, J. G. Derge, R. D. Klausner, F. S. Collins, L. Wagner, C. M. Shenmen, G. D. Schuler and S. F. Altschul, Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 16899–16930 CrossRef PubMed.
- S. Ochs, J. Erdman, R. A. Jersild Jr and V. McAdoo, Routing of transported materials in the dorsal root and nerve fiber branches of the dorsal root ganglion, J. Neurobiol., 1978, 9, 465–481 CrossRef CAS.
- D. Sapunar, S. Kostic, A. Banozic and L. Puljak, Dorsal root ganglion – a potential new therapeutic target for neuropathic pain, J. Pain Res., 2012, 5, 31–38 CrossRef.
- S. Katayama, Y. Tomaru, T. Kasukawa, K. Waki, M. Nakanishi, M. Nakamura, H. Nishida, C. C. Yap, M. Suzuki and J. Kawai, Antisense transcription in the mammalian transcriptome, Science, 2005, 309, 1564–1566 CrossRef.
- M. Dong, X. Wang, H. L. Zhao, X. L. Chen, J. H. Yuan, J. Y. Guo, K. Q. Li and G. Li, Integrated analysis of transcription factor, microRNA and LncRNA in an animal model of obliterative bronchiolitis, Int. J. Clin. Exp. Pathol., 2015, 8, 7050–7058 CAS.
- M. Magistri, M. A. Faghihi, G. St Laurent 3rd and C. Wahlestedt, Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts, Trends Genet., 2012, 28, 389–396 CrossRef CAS PubMed.
- J. Huang, J. Li, Y. Li, Z. Lu, Y. Che, S. Mao, Y. Lei, R. Zang, L. Zheng, C. Liu, X. Wang, N. Li, N. Sun and J. He, Interferon-inducible lncRNA IRF1-AS represses esophageal squamous cell carcinoma by promoting interferon response, Cancer Lett., 2019, 459, 86–99 CrossRef CAS PubMed.
- N. Wei, Y. Wang, R. X. Xu, G. Q. Wang, Y. Xiong, T. Y. Yu, G. S. Yang and W. J. Pang, PU.1 antisense lncRNA against its mRNA translation promotes adipogenesis in porcine preadipocytes, Anim. Genet., 2015, 46, 133–140 CrossRef CAS PubMed.
- W. J. Pang, L. G. Lin, Y. Xiong, N. Wei, Y. Wang, Q. W. Shen and G. S. Yang, Knockdown of PU.1 AS lncRNA inhibits adipogenesis through enhancing PU.1 mRNA translation, J. Cell. Biochem., 2013, 114, 2500–2512 CrossRef CAS PubMed.
- V. P. Eswarakumar, I. Lax and J. Schlessinger, Cellular signaling by fibroblast growth factor receptors, Cytokine Growth Factor Rev., 2005, 16, 139–149 CrossRef CAS PubMed.
- T. M. Lavallee, I. A. Prudovsky, G. A. Mcmahon, X. Hu and T. Maciag, Activation of the MAP kinase pathway by FGF-1 correlates with cell proliferation induction while activation of the Src pathway correlates with migration, J. Cell Biol., 1998, 141, 1647–1658 CrossRef CAS PubMed.
- S. S. Zhang, M. G. Liu, A. Kano, C. Zhang, X. Y. Fu and C. J. Barnstable, STAT3 activation in response to growth factors or cytokines participates in retina precursor proliferation, Exp. Eye Res., 2005, 81, 103–115 CrossRef CAS PubMed.
- T. S. Udayakumar, M. S. Stratton, R. B. Nagle and G. T. Bowden, Fibroblast growth factor-1 induced promatrilysin expression through the activation of extracellular-regulated kinases and STAT3, Neoplasia, 1900, 4, 60–67 CrossRef PubMed.
- W. Lin, C. Chen, Y. Chang, S. Chen, I. Chiu and L. Chen, SH2B1 beta enhances fibroblast growth factor 1 (FGF1)-induced neurite outgrowth through MEK-ERK1/2-STAT3-Egr1 pathway, Cell. Signalling, 2009, 21, 1060–1072 CrossRef CAS PubMed.
- H. R. Lv, lncRNA-Map2k4 sequesters miR-199a to promote FGF1 expression and spinal cord neuron growth, Biochem. Biophys. Res. Commun., 2017, 490, 948–954 CrossRef CAS PubMed.
- J. M. Garré, G. Yang, F. F. Bukauskas and M. V. Bennett, FGF-1 Triggers Pannexin-1 Hemichannel Opening in Spinal Astrocytes of Rodents and Promotes Inflammatory Responses in Acute Spinal Cord Slices, J. Neurosci., 2016, 36, 4785–4801 CrossRef PubMed.
- M. V. L. Bennett, J. M. Garré, J. A. Orellana, F. F. Bukauskas, M. Nedergaard, C. Giaume and J. C. Sáez, Connexin and pannexin hemichannels in inflammatory responses of glia and neurons, Brain Res., 2012, 1487, 3–15 CrossRef CAS PubMed.
- J. M. Garré, M. A. Retamal, P. Cassina, L. Barbeito, F. F. Bukauskas, J. C. Sáez, M. V. Bennett and V. Abudara, FGF-1 induces ATP release from spinal astrocytes in culture and opens pannexin and connexin hemichannels, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 22659–22664 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra03791h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |