Open Access Article
Open Access ArticleStudy on enhancing sludge methanogenesis by adding acetylene black and effect on the characteristics & microbial community of anaerobic granular sludge
Haitong Ma ab,
Ming Wuab,
Hui Liuab,
Zhiwei Wang*abc,
Chenyan Guoab and
Shuangfei Wang*ab
ab,
Ming Wuab,
Hui Liuab,
Zhiwei Wang*abc,
Chenyan Guoab and
Shuangfei Wang*ab
aCollege of Light Industry and Food Engineering, Guangxi University, Nanning 530004, PR China. E-mail: wangzhiwei@gxu.edu.cn; wangsf@gxu.edu.cn; Fax: +86 18648908256; Tel: +86 18648908256
bKey Laboratory of Clean Pulp and Paper and Pollution Control, Nanning 530004, PR China
cCollege of Environmental Science and Forestry, State University of New York, Syracuse, NY 13210, USA
First published on 26th July 2019
Abstract
The effect of acetylene black (ACET) as additives on methane production, extracellular polymeric substances (EPS), microbial community structure and methanogenesis pathway during sludge anaerobic digestion (AD) was investigated in this study. The results indicated that the addition of 2 g L−1 ACET resulted in a 44.36% increase in methane accumulation. ACET, which resulted in the increase of EPS and VSS/TSS by 4.71–50.64%, effectively improved the physicochemical properties of anaerobic granular sludge (AnGS). During anaerobic digestion, the high throughput sequencing presented direct evidence that the ACET increased microbial diversity and enriched functional microorganisms such as norank_f__Synergistaceae, norank_f__Anaerolineaceae, and unclassified_f__Clostridiaceae_3, which can improve the hydrolysis acidification process and the acetotrophic pathway. These results were reaffirmed by applying metagenome inference and gene content inference (16S function prediction). Microscopically, significant enhancement in the AD efficiency can be due to the methanogenesis promoted by the ACET that can construct direct interspecies electron transfer (DIET) between the unclassified_f__Clostridiaceae_3, norank_f__Anaerolineaceae, and Methanosaeta. These results were expected to provide primary research data for improving the performance of anaerobic reactors and the development of microbial fuel cells.
1 Introduction
With the depletion of energy resources and the growing environmental problems, anaerobic digestion (AD) is considered to be one of the most promising biological treatment technologies.1,2 Compared with aerobic treatment, anaerobic digestion has advantages such as low sludge production and energy requirements, and energy recovery from methane production.2 However, in the actual operation process, COD removal efficiency decreases and methane production reduction often occurs in anaerobic bioreactors due to the complex composition of wastewater, high organic load, accumulation of toxic substances and changes in the operating environment, resulting in effluent not meeting the emissions and reuse standards.3,4 The bio-active component of anaerobic granular sludge (AnGS) in the reactor determines the anaerobic digestion efficiency and effluent quality.2,5 Consequently, researchers are concerned about how to strengthen the bio-active component of granular sludge in the wastewater treatment process.Reviewed literature indicated that the use of additives to improve anaerobic digestion efficiency and effluent quality is currently a hot topic. Additives are substances added from outside the anaerobic digestion system to increase biogas yields, such as microorganisms,6 enzymes,7 nutrients,8 and conductive materials.9 Compared with other additives, conductive materials are not only simple in operation, low in cost, and have high application value, but also can enhance the anaerobic digestion performance via direct interspecies electron transfer (DIET). Therefore, a large number of studies have reported the influence of conductive materials on anaerobic digestion performance recently.10,11
Conductive materials can be roughly divided into two categories: a class of conductive mineral materials such as magnetite,12 hematite,13 and akaganeite.14 Hematite (red mud) can ameliorate the methanogenesis by enriching Fe(III)-reducing bacteria and conductive bacteria (Geobacter) in the process of anaerobic digestion and accelerating electron transfer between the syntrophic bacteria (Geobacteraceae) and methanogens (Methanosaeta and Methanosarcina).15 Furthermore, Feng et al. also found that the addition of magnetite, hematite, and goethite was observed to enhance the methanogenesis of AGS compared with the control reactor, and the release of methane carbon increased by 8.92%, 7.09%, and 2.91%.16 Further investigation demonstrated that conductive mineral materials increased methane production by improving direct interspecies electron transfer between methanogens and syntrophic bacteria because the DIET performs distinctively faster than methanogenesis via hydrogen or formate while considering the diffusion limitation of the substrate.17
The other is a conductive carbon-based material, such as graphite,18 granular activated carbon (GAC),19 biochar,20 and carbon cloth21 have been reported to increase methane production during anaerobic digestion. The seven bio-based carbon materials prepared by Yun et al., which can significantly improve CODt removal efficiency (51.39–67.81%) and biogas yield (380–502 mL g−1 TS) compared with the control reactor (CK, control check, 294 mL g−1 TS and 29.55%). Bio-based carbon materials with the higher specific surface area are responsible for improving the AD efficiency by providing sites where substrate accumulate and thereby promote high localized substrate concentrations, which also can facilitate direct interspecies electron transfer between fermenting bacteria and methanogens, accelerating syntrophic acetate metabolism.9 Interestingly, Zhang et al. reported that the GAC can stimulate methanogenesis in anaerobic digesters by facilitating DIET between fermenting bacteria and methanogens, but the GAC does not improve methane production by increasing the number of microorganisms.19 Although conductive materials can enhance the methanogenic capacity of sludge in the anaerobic system, relevant researches are still in the initial stage, especially the key factors (or promoting factors) of DIET production and the mechanism of methanogenic enhancement still need to be further studied.22 Some researchers have tried adding different conductive materials to the same anaerobic system, and found little difference in methane-producing efficiency.23,24 Interestingly, under the same conditions, the addition of nanomagnetite and magnetite may have different promoting mechanisms. For example, nanomagnetite can complete DIET in the absence of cytochrome c, while magnetite will only promote the enrichment of methanogens.24,25 Previous study suggested that the size of the conductive material may affect DIET formation and lead to different methanogenesis enhancement mechanisms.
Currently, there are few studies on the effect of nanocarbon materials on the performance of AnGS and the structure of the microbial community, especially on the mechanism. Additionally, microorganisms tend to adhere to the surface of the carbon-based material and form a thick and dense biofilm during long-term operation of the anaerobic reactor, which limits the mass transfer of electrons and substrates between microorganisms.26 In comparison, nanocarbon materials do not have this problem. Consequently, the effect and mechanism of nanocarbon materials on anaerobic digestion are worth exploring, and acetylene black (ACET), as a common conductive material, is a good choice. In this study, we investigate the effects of ACET on the physical–chemical properties, methanogenic capacity and microbial community structure of AnGS, and the mechanism of ACET on the methanogenic activity of granular sludge. This work is expected to provide basic research data for improving the performance of anaerobic reactors and the development of microbial fuel cells.
2 Materials and methods
2.1 Materials
Acetylene black (the size of ACET 30–45 nm) was provided by Zhejiang Huimei Industry Corporation (Zhejiang, China). Anaerobic granular sludge (AnGS) was collected from a local paper mill (Nanning, China). Fresh AnGS was stored at 4 °C to maintain its freshness. In order to reduce the experimental error, AnGS with average particle size of 0.3–0.6 mm was selected for seed sludge. AnGS was pre-acclimated using glucose as a substrate before the experiment, which resuscitated microbial activity. The characteristics of the synthetic wastewater using glucose as carbon source: glucose of 25.50 g L−1; NaHCO3 of 16.25 g L−1; NH4Cl of 3.00 g L−1; KH2PO4 of 0.56 g L−1; FeSO4·4H2O of 0.032 g L−1; CaCl2 of 0.038 g L−1; MgSO4·7H2O of 0.042 g L−1, and pH of 7.20 ± 0.20.2.2 Anaerobic digestion experiment
Batch experiments were conducted in 250 mL anaerobic serum bottles. A mass of 0.15 and 0.3 g of ACET was added into bottles to construct the ACET supplemented reactors (S2 and S3) with 25 mL of AnGS and 150 mL of the substrate. The bottle without the addition of ACET was designated the control reactor (S1). The bottles were emptied of air by the flush of nitrogen for 15 min and sealed with stoppers and tapes. Then, the reactors were maintained at 37 ± 1 °C with a constant temperature incubator for 28 days. During the research, a simple drainage gas-collecting method was adopted to measure the daily biogas yield over 28 days. Before the biogas volume was measured, each reactor was shaken for 2 min to ensure the mixture was well-distributed, and the gas produced by reactors was passed through a 1.5–5% NaOH solution to remove acid gases. The samples for EPS, volatile suspended solid/total suspended solid (VSS/TSS) and average particle size analysis was obtained from anaerobic reactors every seven days. At the end of the experiment, the microbial community and function of AnGS were discussed by the high-throughput sequencing technology.2.3 Analytical method
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min, and eventually filtered through a 0.22 μm membrane to obtain the filtrate. The protein, humic acid, and polysaccharide concentrations were determined by a modified Lowry method, and the anthrone–sulfuric acid spectrophotometric method using bovine serum albumin, humic acid salt, and glucose as the respective standards.28,29 Ultraviolet/visible (UV/Vis) spectroscopy in a diffuse transmission mode was used to measure the electronic absorption spectra of the multi-heme cytochromes c (c-Cyts) in the EPS.30 EEM fluorescence spectroscopy was used to characterize EPS in this research, and excitation and emission spectra were scanned from 220 to 550 nm in 5 nm increments and from 250 to 550 nm in 5 nm increments, respectively.
000 × g for 10 min, and eventually filtered through a 0.22 μm membrane to obtain the filtrate. The protein, humic acid, and polysaccharide concentrations were determined by a modified Lowry method, and the anthrone–sulfuric acid spectrophotometric method using bovine serum albumin, humic acid salt, and glucose as the respective standards.28,29 Ultraviolet/visible (UV/Vis) spectroscopy in a diffuse transmission mode was used to measure the electronic absorption spectra of the multi-heme cytochromes c (c-Cyts) in the EPS.30 EEM fluorescence spectroscopy was used to characterize EPS in this research, and excitation and emission spectra were scanned from 220 to 550 nm in 5 nm increments and from 250 to 550 nm in 5 nm increments, respectively.For high-throughput sequencing, the V3–V4 hypervariable region of the 16S rRNA gene of the genomic DNA was amplified using a universal primer set of 341F/805R under the following conditions: 95 °C for 3 min, followed by 29 cycles at 94 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s and a final extension step at 72 °C for 5 min. The same volume of 1× loading buffer was mixed with PCR products and operate electrophoresis on 2% agarose gel for detection. Samples with a bright main strip between 300–350 bp were chosen for further experiments. Referring to the preliminary quantitative results of electrophoresis, the PCR products were quantified by QuantiFluor™-ST Blue Fluorescence Quantification System (Promega) and then mixed according to the sequencing amount of each sample. At last, the library was sequenced on an Illumina MiSeq platform. The paired-end reads obtained by MiSeq sequencing are first spliced according to the overlapping relationship. At the same time, the quality of the sequence was quality-controlled and filtered. Sequences analysis was performed by QIIME software package. The uclust method was used to pick OTU and pick a representative sequence for each OTU. Results were classified to the phylum, class, order, family and genus levels. To further estimate the function and metabolic pathway of genes, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used via the PICRUS program.
3 Results and discussion
3.1 ACET promoted methane production
The influence of acetylene black (ACET) on methane production during anaerobic digestion was explored, and it could be clearly found that methane production increased with the increase of ACET dosage (Fig. 1A). In the first 5 days of anaerobic digestion, the methane production in the ACET reactor was similar or less than that of the control reactor. This may be because AnGS needed time to adapt environmental change by external substances.31 This phenomenon also occurs in the effects of GAC (granular activated carbon) and Zeo (zeolite) on anaerobic digestion.19 After 24 days of anaerobic digestion, the maximum methane production was 25.63 mmol in the ACET reactor with a dosage of 2 g L−1, and the methane production increased by 44.36% compared to the control reactor (14.26 mmol) (Fig. 1A).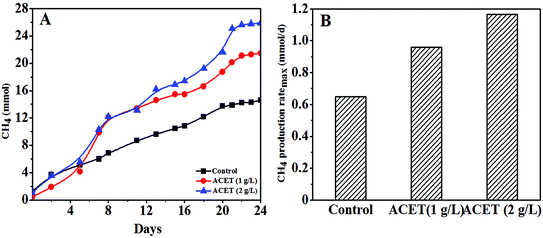 | ||
| Fig. 1 The change of methane accumulation in the control reactor and the reactor with the ACET (A); the maximum methane production rate in the control reactor and the reactor with the ACET (B). | ||
As shown in Fig. 1B, the methane production ratemax in the ACET reactor was 0.96 and 1.17 mmol d−1, respectively, and the methane production ratemax increased by 47.69% and 80.00% compared with the control reactor (0.65 mmol d−1), indicating that the addition of ACET can improve the methanogenic capacity of AnGS. The higher methane production in the ACET reactor could be owing to the improved hydrolysis-acidification process and the electron transfer between methanogenic and syntrophic bacteria by ACET. The result was similar to the result reported by Ye et al., in which the hydrolysis-acidification process in the red mud anaerobic reactor was accelerated, and the methane accumulation was increased by 35.52 ± 2.64% compared with the control reactor.15 Yun et al. also found that bio-based carbon materials can accelerate the electron transfer efficiency between methanogenic and syntrophic bacteria, thereby increasing biogas production.9
3.2 Effect of ACET on the characteristics of AnGS
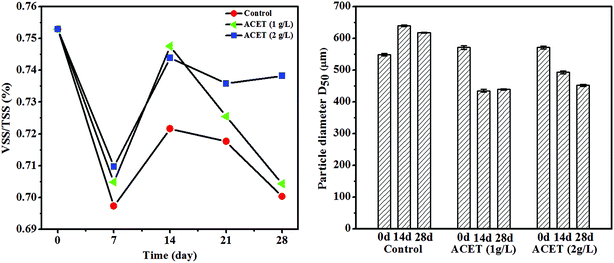
During the stable operation of anaerobic digestion, the VSS/TSS of anaerobic granular sludge in S2 (1 g L−1ACET) and S3 (2 g L−1 ACET) reactors was significantly higher than that of the control reactor, and the ratio of VSS/TSS increased as the dosage of ACET increased, which indicated that the granular sludge had a higher organic component in the S2 and S3 reactors (Fig. 2A). Ding et al. also reported that the VSS/TSS of anaerobic granular sludge in EGSB1 (with calcium) higher than that of EGSB2 (without calcium), indicating that ACET and calcium could increase sludge organic components.32 It was probably because that additives (ACET, red mud, and activated carbon, etc.) promoted microbial aggregation, and secretion of extracellular polymeric substances, which changed the properties of AnGS.In addition, the average particle size of AnGS in the ACET reactor was gradually reduced with the anaerobic digestion operation compared to the control reactor. Day 14 of anaerobic digestion, the average particle size (D50) of AnGS decreased from 580 ± 2.31 and 582 ± 2.17 μm to 440 ± 2.08 and 460 ± 1.67 μm in the ACET reactors, which decreased by 31.82 ± 0.85 and 26.52 ± 0.74% (Fig. 2B). Interestingly, the average particle size of AnGS was reduced but the bio-active was enhanced in the ACET reactor. In contrast, Wu et al. found that large granular sludge (3–3.5 mm) was highly bio-active due to their structure, i.e. big pore size, high porosity and short diffusion distance as compared to small (0.5–1 mm) and medium granular (1.5–2 mm).33 This may be because that ACET reduced the average particle size of AnGS, but it still belonged to small granular (0.5–1 mm), which was not enough to affect mass transfer efficiency and internal structure of AnGS. Furthermore, ACET as a conductive medium can accelerate the electron transfer efficiency of microorganisms and improve the bio-active of sludge.
3.3 Effect of ACET on the properties of EPS
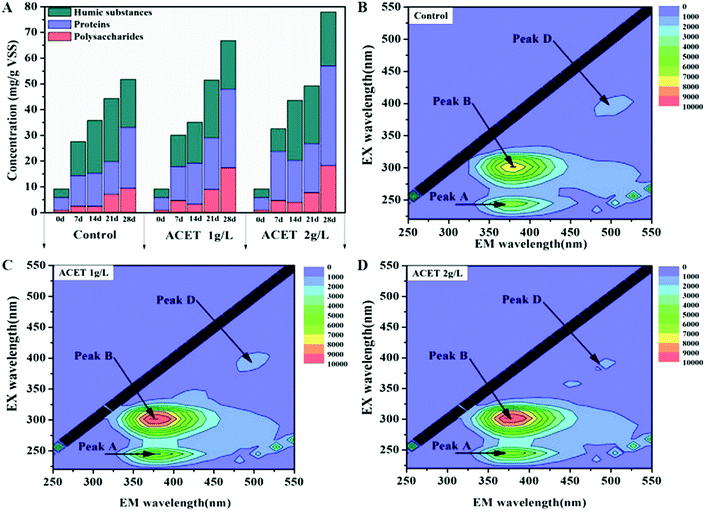
Fig. 3A–D demonstrated that the addition of ACET (2 g L−1) significantly promote the excretion of extracellular polymeric substances (EPS) during anaerobic digestion. The maximum concentrations of protein, polysaccharide, and humic substances in the 2 g L−1 ACET reactor reached 38.81, 18.13 and 20.97 mg g−1 VSS, respectively, and were higher than that in the S1 reactor (23.72, 9.35 and 18.65 mg g−1 VSS). It was possibly because that ACET promoted the growth and reproduction of microorganisms, thereby enhancing the excretion of EPS. Similarly, Jia et al. also found that the concentration of EPS gradually increased during the growth and reproduction of bacteria. In addition, the EPS concentrations increased as the anaerobic digestion proceeded in both reactors (Fig. 3A).34 The result was different from those reported by Ye et al. (2017), in which the EPS content first increase and then decrease when the red mud was added into an anaerobic reactor. This may be because that the EPS was not used as the carbon and energy source by cells during the period of sufficient nutrients.15Three-dimensional EEM spectroscopy revealed three peaks located at Ex/Em of 270–280/345–360 nm (protein-like substances), 225–250/340–360 nm (soluble microbial byproduct-like substances), and 390/470–490 nm (fulvic acid-like substances) in the EPS samples extracted from all reactors. The review literature further determined that Peak B, Peak A, and Peak D were protein-like substances (Peak B: Ex/Em = (270, 280)/345–360),35,36 the tryptophan-like and soluble microbial by-product-like component (Peak A: Ex/Em = (220, 280)/350),37,38 and the humic-like component, respectively (Peak D: Ex/Em = 390/(450–470)).36 Compared to the control reactor, the intensities of Peak A and Peak B increased as the anaerobic digestion proceeded, which was consistent with the conclusion of Fig. 3A. Interestingly, the intensities of Peak D was approximately the same in all reactors, which suggested that ACET may not affect the concentration of the humic-like component. As a conductive medium, the constant concentration of the humic-like component indicated that ACET cannot change the redox activity of EPS.
Extracellular polymeric substances was important for the stability and biochemical properties of granular sludge.39 After the addition of ACET (2 g L−1), the zeta potential of EPS changed from 20.30 ± 0.35 mV to 15.20 ± 0.56 mV, which weaken the electrical repulsion, compress the double electrode layer and then promote the cell-to-cell interaction and granulation. This may be one of the reasons for the average particle size reduction of granular sludge in the ACET reactor. Cytochromes c (c-Cyts) can play an important role in the electron transfer process between syntrophic bacteria and methanogenic.40 Fig. 4 demonstrated that the absorbance of c-Cyts (419 nm) gradually increases during anaerobic digestion in all reactors. Interestingly, the ACET did not change the absorbance of c-Cyts compared to the control reactor. The result was different from the discovery by Ye et al., in which red mud can increase the absorbance of c-Cyts.15 Fig. 3 and 4 indicated that ACET does not change c-Cyts and the concentration of the humic-like component. Therefore, we speculated that ACET may become a substitute for c-Cyts, playing the role of transferring electrons. The result was consistent with those reported by Zhang et al., in which the c-Cyts response to the supplement of GAC, it was speculated that GAC may play the role of c-Cyts' substitution.
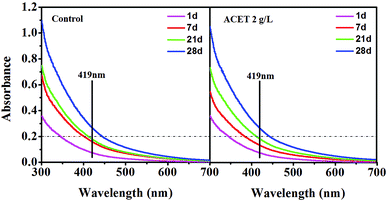 | ||
| Fig. 4 UV/Vis spectra of EPS on days 1, 7, 21, and 28 in the control reactor and the ACET reactor with a dosage of 2 g L−1. | ||
3.4 Effect of ACET on microbial community
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 635 high-quality sequences were obtained from a total of 155
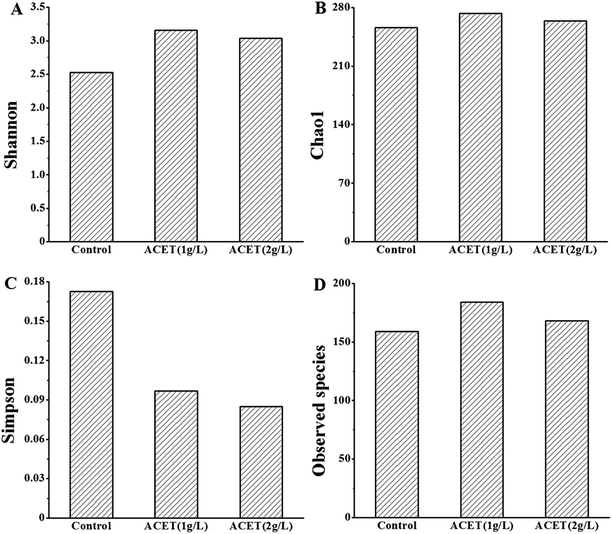
635 high-quality sequences were obtained from a total of 155![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 607 reads to form 722 representative OUTs using the Qillme platform. Fig. 5A indicated that the Shannon index of the three reactors varied between 2.53 and 3.16. The species diversity in the ACET reactor changed significantly compared with the control reactor under the same external conditions. Taking the Shannon index as an example, the maximum increase was 24.83% (Fig. 5A–C). Importantly, the number of observed species in the ACET reactor was higher than the control (Fig. 5D). The result was consistent with the results reported by Qin et al., in which the higher the diversity and the longstanding core species, the stronger the digestion performance of the sludge.2
607 reads to form 722 representative OUTs using the Qillme platform. Fig. 5A indicated that the Shannon index of the three reactors varied between 2.53 and 3.16. The species diversity in the ACET reactor changed significantly compared with the control reactor under the same external conditions. Taking the Shannon index as an example, the maximum increase was 24.83% (Fig. 5A–C). Importantly, the number of observed species in the ACET reactor was higher than the control (Fig. 5D). The result was consistent with the results reported by Qin et al., in which the higher the diversity and the longstanding core species, the stronger the digestion performance of the sludge.2
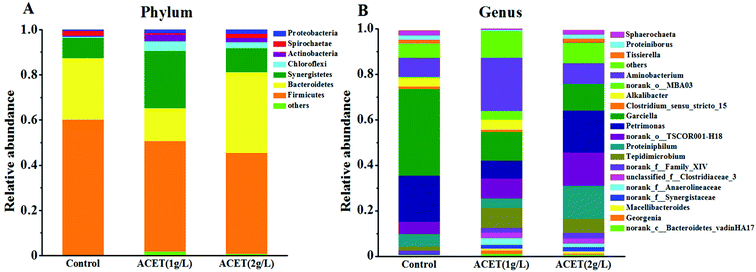 | ||
| Fig. 6 Changes in the predominant microbial community of bacteria. The distribution of dominant taxa at the phylum (A) and genus (B) level. | ||
At the phylum level, Firmicutes, Bacteroidetes, and Synergistetes were the most dominant functional species in all reactors. During the anaerobic digestion process, the ACET cannot change the predominant phylum of the bacteria as a whole. Fig. 6A demonstrated that the abundance of Firmicutes was reduced by 25.33% in the 2 g L−1 ACET reactor compared to the control reactor. Interestingly, the relative abundance of Chloroflexi, Synergistetes, and Proteobacteria, as well-known anaerobic hydrolytic and acidifying phyla, increased the most significantly, indicating that ACET can enrich functional microorganisms.2,41 At the genus level, the microbial community structure of AnGS had obvious differences (Fig. 6B). For example, the abundance of Proteiniphilum (14.60%), Tepidimicrobium (6.17%), norank_f__Family_XIV (2.40%), and norank_o__TSCOR001-H18 (14.71%) was enriched in the 2 g L−1 ACET reactor, which can metabolize organic acids to acetate.42 More importantly, the addition of ACET resulted in the abundance of unclassified_f__Clostridiaceae_3 from 0 to 2.25%. The genus (unclassified_f__Clostridiaceae_3) was close to Clostridium species, which belong to the Fe(III)-reducing genes and had the type IV pili for extracellular electron transfer.43 Meantime, the abundance of norank_f__Synergistaceae (1.63%) and norank_f__Anaerolineaceae (within the phylum of Chloroflexi) was significantly increased in the ACET reactor, which can decompose organic acids into acetate with the production of H2 in co-culture with H2-consuming methanogens.44 It was in step with the results reported by Zhao et al., in which Synergistaceae was enriched with the addition of GAC and was speculated to be able to transfer electrons to Fe(III) oxides.19 These data suggested that ACET not only accelerated the hydrolysis and acidification process, but also improved the uniformity of bacterial species distribution and promoted the expression of bacterial function.
Syntrophic organisms can play a key role in the methanogenic process. In this study, the relative abundance of norank_f__Synergistaceae, unclassified_f__Clostridiaceae_3, norank_f__Anaerolineaceae was higher in the ACET reactor than that in the control reactor, which may trigger the electron transfer between syntrophic bacteria and methanogens and the CO2 reduction to CH4 by methanogenesis. This result was in line with the conclusion of Fig. 1.
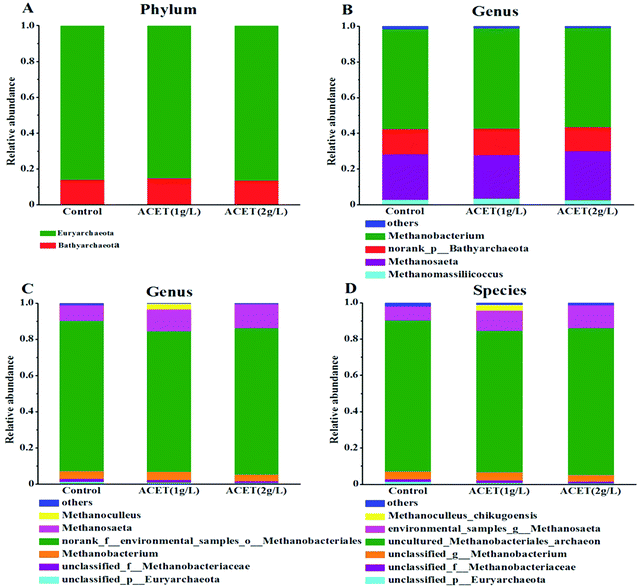
Moreover, electron-accepting methanogenic archaea are also necessary during the electron transfer process.45 To clarify the possible electronic transfer mechanism in this study. The community structure of archaea (methanogens) was further analyzed at the phylum, genus and species levels. Fig. 7A revealed that Euryarchaeota and Bathyarchaeota dominated at the phylum level in all reactors. Euryarchaeota, the famous methanogen, was an irreplaceable functional microorganism in the anaerobic reactor. The relative abundance of Euryarchaeota (86.50%) was higher compared to the control reactor (85.19%) in the 2 g L−1 ACET reactor (Fig. 7A). This result suggested that ACET can enrich Euryarchaeota in the anaerobic digestion process. As shown in Fig. 7B, at the genus level, the abundances of norank_p__Bathyarchaeota, Methanomassiliicoccus, Methanobacterium and Methanosaeta in the S1, S2 and S3 reactors was 14.06%, 14.74%, 13.44%; 2.84%, 3.44%, 2.51%; 56.00%, 56.17%, 55.87%; 25.58%, 24.56%, 27.66%; respectively. Particularly, Methanobacterium (4H2 + HCO3− + H+ → CH4 + 3H2O) was the most abundant methanogen in all reactors, which showed that the hydrogenotrophic pathway was the main methanogenic pathway. Interestingly, the abundance of Methanosaeta in the 2 g L−1 ACET reactor was higher than that of the control reactor, which can use acetic acid as a methanogenic substrate.46 Additionally, Fig. 7C and D showed that the abundance of environmental_samples_g__Methanosaeta (12.39%) was also higher than that of the control reactor (7.75%), indicating that ACET can alter the community structure of methanogens and enhance acetotrophic pathway.
In summary, ACET can increase the relative abundance of Methanosaeta (environmental_samples_g__Methanosaeta) and syntrophic bacteria (Tepidimicrobium, norank_f__Synergistaceae, norank_f__Anaerolineaceae, and unclassified_f__Clostridiaceae_3), and their enrichment enhanced acetotrophic pathway. More importantly, Methanosaeta, norank_f__Synergistaceae, norank_f__Anaerolineaceae, and unclassified_f__Clostridiaceae_3 had been proven to deliver electrons.47 For example, Ye et al., found that magnetite as an electron conductor can stimulate direct electron transfer between Clostridiaceae, Anaerolineaceae, and Methanosarcinaceae, and promote the methanation process.15 So we speculated that promoting methanogenesis with ACET probably was related with DIET between unclassified_f__Clostridiaceae_3, norank_f__Anaerolineaceae, and Methanosaeta (environmental_samples_g__Methanosaeta). Interestingly, this result was different from the results reported by Yang et al., in which Geobacter was enriched in anaerobic digestion and was proved to be able to transfer electrons to Methanosarcina.48
3.5 Analysis of the effect of ACET on the methanogenesis pathway
To further analyze the impact mechanism of ACET, the genes (treated) of the S1, S2, and S3 samples were compared with the KEGG database by using the PICRUS software, and the results were shown in Fig. 8. Each ko represents a pathway, and the three pathways with the highest relative abundance in all the reactors (S1, S2, and S3) were ABC transporters (ko02010), ribosome (ko03010), and purine metabolism (ko00230). The relative abundances were 5.60%, 6.35%, 4.97%; 4.14%, 4.03%, 3.98%; 3.69%, 3.64%, 3.62%, respectively. Ko00680 was a methane metabolic pathway that included methane production and degradation, with a relative abundance of 2.28% (S1), 2.56% (S2), and 2.33% (S3), respectively, which suggested that ACET can enhance the abundance of the methane metabolic pathway.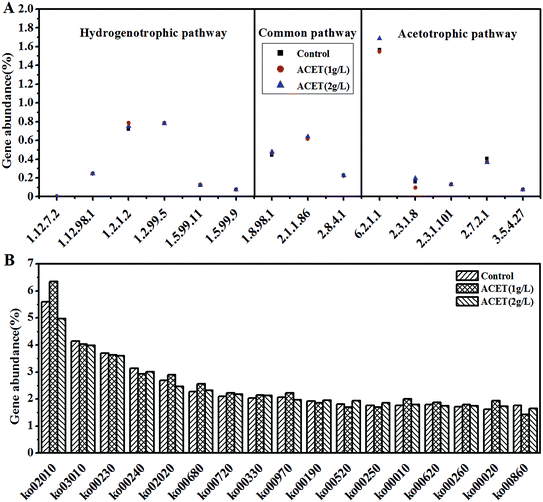 | ||
| Fig. 8 Gene abundance of methane-related enzymes in the three reactors (A). Gene abundance of KEGG pathway in the three reactors (B). | ||
The methane production process of methanogens can be divided into three pathways depending on the metabolic substrate, namely the hydrogenotrophic pathway, acetotrophic pathway, and methylotrophic pathway.48 The implementation of three methanogenic pathways must involve the participation of related coenzymes such as coenzyme M, formate dehydrogenase (EC: 1.2.1.2), acetyl-CoA synthetase (EC: 6.2.1.1), coenzyme 420 dehydrogenase (EC: 1.12.98.1), furan dehydrogenase (EC: 1.2.99.5) and the like.48,49 Fig. 8A demonstrated that the gene abundance of methanogenic enzymes in the KEGG database. In addition to the common pathway gene abundance, in the S1, S2 and S3 reactor, the abundances of the genes involved hydrogenotrophic pathways was 1.95%, 2.02%, and 1.99%, respectively. Compared to the S1 (2.33%), the abundances of the genes involved acetotrophic pathway increased by 5.58% in the S3 reactor. These data indicated that the acetotrophic pathway may be enhanced in the ACET reactor, which was the line in with the results of microbial community structure changes. Simultaneously, it was further suggested that ACET can improve the anaerobic digestion efficiency of granular sludge.
4 Conclusion
In this study, ACET can significantly increase methane production, which was increased by 44.36% in the 2 g L−1 ACET reactor compared with the control reactor. According to the high-throughput sequencing and other data, the main mechanism of promoting methanogenesis by ACET can be obtained. Microscopically, ACET changed the original microbial community structure by enriching functional microorganisms such as norank_f__Synergistaceae, norank_f__Anaerolineaceae, and unclassified_f__Clostridiaceae_3, which affects the microbial methanogenic pathway, and predictive analysis indicated that ACET can enhance the acetotrophic pathway of AnGS. More importantly, ACET can construct direct interspecies electron transfer between the unclassified_f__Clostridiaceae_3, norank_f__Anaerolineaceae, and Methanosaeta, improve the electron transfer efficiency and enhance the methanogenic activity of the granular sludge. Macroscopically, ACET can also improve the physical–chemical properties of AGS (VSS/TSS, EPS and particle size), which provided a material basis and structural basis for anaerobic digestion. For example, increased EPS concentration may enhance the self-protection ability of granular sludge and the utilization efficiency of the substrate. The direct evidence was that the VSS/TSS ratio in ACET reactor was higher than that in the control reactor. Consequently, this study provided references and ideas for the improvement of anaerobic reactor performance and the development of microbial fuel cells.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful for grants from the National Natural Science Foundation of China (31660182, 21868004), and the project of Guangxi Autonomous Region Department of Science and Technology, China (2017GXNSFAA198200) that supported this research.References
- R. Kleerebezem and H. Macarie, Chem. Eng., 2003, 4, 56–62 Search PubMed.
- Q. Xianchao, J. Mengmeng, W. Xiaogang, L. Chunjie, G. Yueshu, L. Ji, W. Qiaoyu, Z. Xiaojun and Z. Zhenjia, Sci. Total Environ., 2019, 646, 229–237 CrossRef PubMed.
- A. Patoine and J. Hawari, Water Res., 1997, 31, 825–831 CrossRef CAS.
- M. Kawai, M. Kishi, M. R. Hamersley, N. Nagao, J. Hermana and T. Toda, Int. Biodeterior. Biodegrad., 2012, 72, 46–51 CrossRef CAS.
- A. K. Friedman, J. J. Walsh, B. Juarez, S. M. Ku, D. Chaudhury, J. Wang, X. Li, D. M. Dietz, N. Pan and V. F. Vialou, Science, 2014, 344, 313–319 CrossRef CAS PubMed.
- I. Angelidaki and B. K. Ahring, Biodegradation, 1992, 3, 409–414 Search PubMed.
- M. Dohányos, J. Zábranská and P. Jenícek, Water Sci. Technol., 1997, 36, 145–153 CrossRef.
- K. Hopfnersixt, T. Amon, V. Bodiroza, V. Kruvorucko, D. Milovanovic and J. Boxberger, Landtechnik, 2006, 61, 30–31 Search PubMed.
- S. Yun, W. Fang, T. Du, X. Hu, X. Huang, X. Li, C. Zhang and P. D. Lund, Energy, 2018, 164, 898–909 CrossRef CAS.
- S. Yun, A. Hagfeldt and T. Ma, ChemInform, 2014, 26, 6210–6237 CAS.
- S. Yun, H. Zhang, H. Pu, J. Chen, A. Hagfeldt and T. Ma, Adv. Energy Mater., 2013, 3, 1407–1412 CrossRef CAS.
- Y. F. Zhang and I. Angelidaki, Biotechnol. Bioeng., 2015, 112, 1478–1482 CrossRef CAS PubMed.
- S. Kato, K. Hashimoto and K. Watanabe, Environ. Microbiol., 2012, 14, 1646–1654 CrossRef CAS PubMed.
- S. Jiang, S. Park, Y. Yoon, J. H. Lee, W. Wu, N. P. Dan, M. J. Sadowsky and H. G. Hur, Environ. Sci. Technol., 2013, 47, 10078–10084 CrossRef CAS PubMed.
- J. Ye, A. Hu, G. Ren, T. Zhou, G. Zhang and S. Zhou, Bioresour. Technol., 2017, 247, 131–137 CrossRef PubMed.
- X. M. Feng, Master thesis, HeFei University of Technology, 2015.
- V. C. Cruz, S. Rossetti, S. Fazi, P. Paiano, M. Majone and F. Aulenta, Environ. Sci. Technol., 2014, 48, 7536–7543 CrossRef PubMed.
- Z. Zhao, Y. Zhang, T. L. Woodard, K. P. Nevin and D. R. Lovley, Bioresour. Technol., 2015, 191, 140–145 CrossRef CAS PubMed.
- S. Zhang, J. Chang, C. Lin, Y. Pan, K. Cui, X. Zhang, P. Liang and X. Huang, Bioresour. Technol., 2017, 245, 132–137 CrossRef CAS PubMed.
- L. Yu, Y. Yong, T. Jia, Y. Wang and S. Zhou, Sci. Rep., 2015, 5, 16221 CrossRef CAS PubMed.
- S. Chen, A. E. Rotaru, F. Liu, J. Philips, T. L. Woodard, K. P. Nevin and D. R. Lovley, Bioresour. Technol., 2014, 173, 82–86 CrossRef CAS PubMed.
- Z. Guo, A. Zhou, C. Yang, B. Liang, T. Sangeetha, Z. He, L. Wang, W. Cai, A. Wang and W. Liu, Biotechnol. Biofuels, 2015, 8, 192 CrossRef PubMed.
- Z. Zhao, Y. Zhang, T. L. Woodard, K. P. Nevin and D. R. Lovley, Bioresour. Technol., 2015, 191, 140–145 CrossRef CAS PubMed.
- L. Fanghua, R. Amelia-Elena, P. M. Shrestha, N. S. Malvankar, K. P. Nevin and D. R. Lovley, Environ. Microbiol., 2015, 17, 648–655 CrossRef PubMed.
- K. Souichiro, H. Kazuhito and W. Kazuya, Environ. Microbiol., 2012, 14, 1646–1654 CrossRef PubMed.
- F. Zhang, S. J. Yuan, W. W. Li, J. J. Chen, C. C. Ko and H. Q. Yu, Electrochim. Acta, 2015, 152, 1–5 CrossRef CAS.
- S. S. Adav and D. J. Lee, J. Hazard. Mater., 2008, 154, 1120–1126 CrossRef CAS PubMed.
- B. Frølund, T. Griebe and P. H. Nielsen, Appl. Microbiol. Biotechnol., 1995, 43, 755–761 CrossRef.
- Y. Guo, U. Sagaram, J. Kim and N. Wang, Appl. Environ. Microbiol., 2010, 76, 2234–2242 CrossRef CAS PubMed.
- R. Nakamura, K. Ishii and K. Hashimoto, Angew. Chem., Int. Ed., 2010, 48, 1606–1608 CrossRef PubMed.
- L. M. Luis, M. Samuel and B. H. Jürg, Nat. Rev. Genet., 2008, 9, 583–593 Search PubMed.
- A. Ding, W. Pronk, F. Qu, J. Ma, G. Li, K. Li and H. Liang, J. Membr. Sci., 2015, 489, 55–63 CrossRef CAS.
- W. Jing, Z. U. Afridi, Z. P. Cao, Z. L. Zhang, S. Poncin, H. Z. Li, J. Zuo and K. J. Wang, Bioresour. Technol., 2016, 202, 165–171 CrossRef PubMed.
- X. S. Jia, H. Furumai and H. H. P. Fang, Water Res., 1996, 30, 1439–1444 CrossRef CAS.
- L. Li, M. Liu, Y. Li, X. Ma, X. Tang and Z. Li, Environ. Sci. Pollut. Res. Int., 2016, 23, 13498–13507 CrossRef CAS PubMed.
- E. Ouyang, W. Wei, N. Long and H. Li, Spectrosc. Spectral Anal., 2009, 29, 1313–1318 Search PubMed.
- W. Chen, P. Westerhoff, J. A. Leenheer and K. Booksh, Environ. Sci. Technol., 2003, 37, 5701–5710 CrossRef CAS PubMed.
- G. H. Yu, P. J. He and L. M. Shao, Water Res., 2010, 44, 797–806 CrossRef CAS PubMed.
- X. M. Liu, G. P. Sheng, H. W. Luo, Z. Feng, S. J. Yuan, J. Xu, R. J. Zeng, J. G. Wu and H. Q. Yu, Environ. Sci. Technol., 2010, 44, 4355–4360 CrossRef CAS PubMed.
- A. Okamoto, K. Hashimoto and K. H. Nealson, Angew. Chem., Int. Ed., 2015, 53, 10988–10991 CrossRef PubMed.
- S. Ahlert, R. Zimmermann, J. Ebling and H. König, MicrobiologyOpen, 2016, 5, 1027–1037 CrossRef CAS PubMed.
- S. Hahnke, T. Langer, D. E. Koeck and M. Klocke, Int. J. Syst. Evol. Microbiol., 2016, 66, 1466–1475 CrossRef CAS PubMed.
- E. Bordeleau, E. B. Purcell, D. A. Lafontaine, L. C. Fortier, R. Tamayo and V. Burrus, J. Bacteriol., 2015, 197, 819–832 CrossRef PubMed.
- D. Z. Sousa, H. Smidt, M. M. Alves and A. J. Stams, Int. J. Syst. Evol. Microbiol., 2007, 57, 609–615 CrossRef CAS PubMed.
- H. Li, J. Chang, P. Liu, L. Fu, D. Ding and Y. Lu, Environ. Microbiol., 2015, 17, 1533–1547 CrossRef PubMed.
- K. Ma, X. Liu and X. Dong, Int. J. Syst. Evol. Microbiol., 2006, 56, 127–131 CrossRef CAS PubMed.
- J. Zhang and Y. H. Lu, Microbiology, 2015, 42, 920–927 CAS.
- L. Xing, S. Yang, Q. Yin, S. Xie, P. J. Strong and G. Wu, Bioresour. Technol., 2017, 244, 982 CrossRef CAS PubMed.
- Z. Wang, Q. Yin, M. Gu, K. He and G. Wu, J. Hazard. Mater., 2018, 357, 226–234 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |