Open Access Article
Open Access ArticleHighly malleable haem-binding site of the haemoprotein HasA permits stable accommodation of bulky tetraphenylporphycenes†
Erika Sakakibara a,
Yuma Shisaka
a,
Yuma Shisaka a,
Hiroki Onoda
a,
Hiroki Onoda a,
Daiki Kogab,
Ning Xub,
Toshikazu Ono
a,
Daiki Kogab,
Ning Xub,
Toshikazu Ono b,
Yoshio Hisaeda
b,
Yoshio Hisaeda b,
Hiroshi Sugimotocd,
Yoshitsugu Shiroe,
Yoshihito Watanabe
b,
Hiroshi Sugimotocd,
Yoshitsugu Shiroe,
Yoshihito Watanabe f and
Osami Shoji
f and
Osami Shoji *a
*a
aDepartment of Chemistry, Graduate School of Science, Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-0802, Japan. E-mail: shoji.osami@a.mbox.nagoya-u.ac.jp
bDepartment of Chemistry and Biochemistry, Graduate School of Engineering, Kyushu University, Fukuoka 819-0395, Japan
cCore Research for Evolutional Science and Technology (CREST), Japan Science and Technology Agency, 5 Sanban-cho, Chiyoda-ku, Tokyo, 102-0075, Japan
dRIKEN SPring-8 Center, 1-1-1 Kouto, Sayo-cho, Hyogo 679-5148, Japan
eDepartment of Life Science, Graduate School of Life Science, University of Hyogo, 3-2-1 Kouto, Kamighori, Akoh, Hyogo 678-1297, Japan
fResearch Center for Materials Science, Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-0802, Japan
First published on 13th June 2019
Abstract
Iron(III)- and cobalt(III)-9,10,19,20-tetraphenylporphycenes, which possess bulky phenyl groups at the four meso positions of porphycene, were successfully incorporated into the haem acquisition protein HasA secreted by Pseudomonas aeruginosa. Crystal structure analysis revealed that loops surrounding the haem-binding site are highly flexible, remodelling themselves to accommodate bulky metal complexes with significantly different structures from the native haem cofactor.
Introduction
The substitution of protein cofactors with non-natural analogues is a powerful means to control the function of proteins, and has been intensively investigated, procuring novel biocatalysts,1–4 biosensors,5–7 and supramolecular structures.8,9 Haemoproteins, such as myoglobin (oxygen binding),10 P450s (metabolism),11 and haem oxygenase (degradation of haem),12 which contain iron protoporphyrin IX (haem) as their cofactor, are some of the most promising targets for the reconstitution of haem with other metal complexes. The function of haemoproteins has been enhanced by substituting haem for analogues that have been decorated with functional groups and/or exchanged with metals other than iron. Unfortunately, metal complexes with structures differing from haem, such as artificial metal complexes possessing bulky substituents, can generally not be incorporated into wild-type haemoproteins, as their haem-binding cavity has evolved to only accommodate haem. Recently, we reported that the haem acquisition protein HasA secreted by Pseudomonas aeruginosa is rather promiscuous and can accommodate other metal complexes, such as insoluble iron(III)-phthalocyanine, small iron(III)-salophen, and bulky iron(III)-5,15-diphenylporphyrin.13,14HasA is a haem-binding protein that is secreted by certain pathogens, such as P. aeruginosa, when in iron deficient environments. Following acquisition of haem from its host by HasA, haem is transferred to the HasA-specific outer membrane receptor HasR, where it is taken up into the cell and used as an iron source for the pathogen's survival (Fig. 1a).15,16 HasA captures haem by using two of its loops like tweezers, where His32 and Tyr75 coordinate each side of the haem iron. Binding of haem to HasA is accompanied by significant structural changes in the His32-loop from the haem-free open-form (apo-HasA) to the haem-bound closed-form (holo-HasA). The crystal structure of holo-HasA reveals a large portion of the haem cofactor protruding out of HasA and exposed to the solvent (Fig. 1a).17,18 This unusual solvent-exposed mode of haem binding offers an enticing explanation as to why metal complexes with significantly different structures and properties from haem can be incorporation into HasA. In this study we selected the sterically demanding synthetic cofactor metallo-9,10,19,20-tetraphenylporphycene (Fig. 1b) for incorporation into HasA, as the gram-scale synthesis of 9,10,19,20-tetraphenylporphycenes (Ph4Pc) has been reported recently.19 Despite the alluring photophysical and catalytic properties of metallo-9,10,19,20-tetraphenylporphycenes, they have never been considered as potential targets for incorporation into haemoproteins.
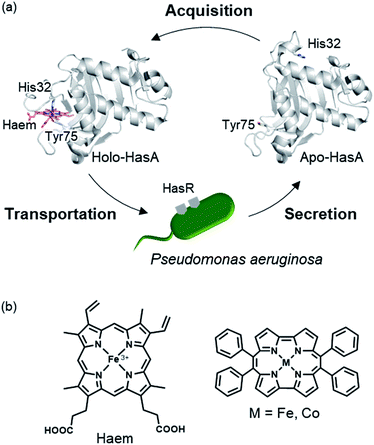 | ||
| Fig. 1 (a) Schematic of the haem acquisition system of Pseudomonas aeruginosa. Under low-iron conditions, apo-HasA (PDB ID: 3MOK) secreted by P. aeruginosa captures and transports haem (holo-HasA, PDB ID: 3ELL) to the HasA specific receptor HasR expressed on outer membrane. (b) The structure of haem (left) and metallo-9,10,19,20-tetraphenylporphycene (right). | ||
Result and discussion
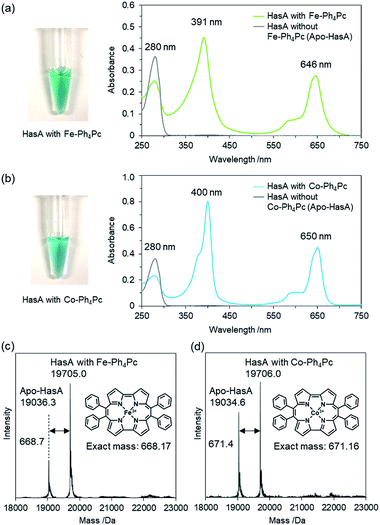
Initially, we attempted to incorporate iron(III)-9,10,19,20-tetraphenylporphycene (Fe-Ph4Pc) into HasA. A solution of Fe-Ph4Pc dissolved in DMSO was added to apo-HasA. After dialysis to remove DMSO, HasA containing Fe-Ph4Pc was purified via anion-exchange chromatography. UV/Vis spectroscopic analysis of HasA with Fe-Ph4Pc exhibited characteristic absorption at 391 and 646 nm, which was assigned to the Soret and Q band of Fe-Ph4Pc, respectively (Fig. 2a). ESI-TOF mass spectrometry gave a peak at 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 705.0 Da, which corresponded to the mass expected for HasA with Fe-Ph4Pc (Fig. 2c). Furthermore, we also succeeded in the preparation of HasA capturing cobalt(III)-9,10,19,20-tetraphenylporphycene (Co-Ph4Pc) using a similar procedure to aforementioned HasA with Fe-Ph4Pc (Fig. 2b and d).
705.0 Da, which corresponded to the mass expected for HasA with Fe-Ph4Pc (Fig. 2c). Furthermore, we also succeeded in the preparation of HasA capturing cobalt(III)-9,10,19,20-tetraphenylporphycene (Co-Ph4Pc) using a similar procedure to aforementioned HasA with Fe-Ph4Pc (Fig. 2b and d).
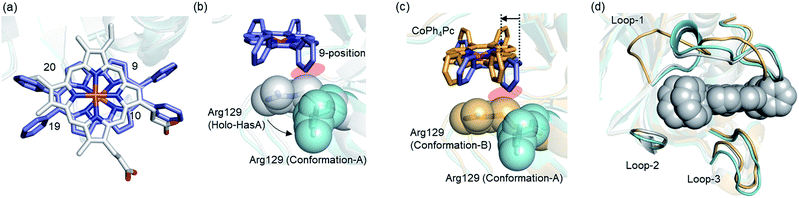
To comprehend the intricate changes HasA must undergo to enable accommodation of bulky metallo-Ph4Pcs, X-ray crystal structure analysis of HasA with Fe-Ph4Pc and Co-Ph4Pc was attempted, and the structure of HasA capturing Co-Ph4Pc was successfully determined at 2.5 Å resolution (Fig. 3a). The overall structure of HasA capturing Co-Ph4Pc resembled that of holo-HasA (root-mean-square deviation (RMSD) over 2–182 amino acid residues for Cα atoms against holo-HasA: 0.73). Electron density clearly corresponding to Co-Ph4Pc was observed in the haem-binding site of HasA (Fig. 3b). Analogous to haem, the cobalt ion of Co-Ph4Pc was ligated by a nitrogen of His32 (Nε) and oxygen of Tyr75 (Oη) (Fig. 3b). The two phenyl groups at the 9- and 10-positions of Co-Ph4Pc were accommodated in a cavity formed by His32(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), Pro34, Arg129, His134, and Tyr138, whereas the other two phenyl groups at 19- and 20-positions were accommodated in another cavity formed by Thr43, Gly44(C
O), Pro34, Arg129, His134, and Tyr138, whereas the other two phenyl groups at 19- and 20-positions were accommodated in another cavity formed by Thr43, Gly44(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), Gly45, Phe46, Pro50(C
O), Gly45, Phe46, Pro50(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), and Phe51 (Fig. 3c). The overall shape of the cavity closely resembled that of HasA with iron(III)-5,15-diphenylporphyrin, which also possesses bulky phenyl groups at a similar position.14 Superimposition of HasA with Co-Ph4Pc over haem revealed that the phenyl groups at positions 10 and 20 of Co-Ph4Pc were positioned where a propionate and vinyl group of haem are usual found, respectively (Fig. 4a). Intriguingly, two different conformations (conformation-A and -B) could be observed in the crystal structure of HasA with Co-Ph4Pc. Regarding conformation-A, when compared to the structure of holo-HasA, the side chain of Arg129 was flipped to prevent steric repulsion with the 9-phenyl of Co-Ph4Pc (Fig. 4b). Conversely, Arg129 is not flipped in conformation-B, but steric repulsion is avoided by further extrusion of Co-Ph4Pc from HasA (Fig. 4c). From these two conformations, we concluded that Arg129 actually interferes with the incorporation of bulky phenyl rings. However, owing to the high flexibility of HasA's loops (Loop-1: Asp29 to Gly44; Loop-2: Gly49 to Arg52 and Loop-3: Thr76 to Leu85), which form the haem-binding pocket, Co-Ph4Pc can be stably incorporated, following adequate changes of the cavity's shape to accommodate bulky phenyl groups (Fig. 4d).
O), and Phe51 (Fig. 3c). The overall shape of the cavity closely resembled that of HasA with iron(III)-5,15-diphenylporphyrin, which also possesses bulky phenyl groups at a similar position.14 Superimposition of HasA with Co-Ph4Pc over haem revealed that the phenyl groups at positions 10 and 20 of Co-Ph4Pc were positioned where a propionate and vinyl group of haem are usual found, respectively (Fig. 4a). Intriguingly, two different conformations (conformation-A and -B) could be observed in the crystal structure of HasA with Co-Ph4Pc. Regarding conformation-A, when compared to the structure of holo-HasA, the side chain of Arg129 was flipped to prevent steric repulsion with the 9-phenyl of Co-Ph4Pc (Fig. 4b). Conversely, Arg129 is not flipped in conformation-B, but steric repulsion is avoided by further extrusion of Co-Ph4Pc from HasA (Fig. 4c). From these two conformations, we concluded that Arg129 actually interferes with the incorporation of bulky phenyl rings. However, owing to the high flexibility of HasA's loops (Loop-1: Asp29 to Gly44; Loop-2: Gly49 to Arg52 and Loop-3: Thr76 to Leu85), which form the haem-binding pocket, Co-Ph4Pc can be stably incorporated, following adequate changes of the cavity's shape to accommodate bulky phenyl groups (Fig. 4d).
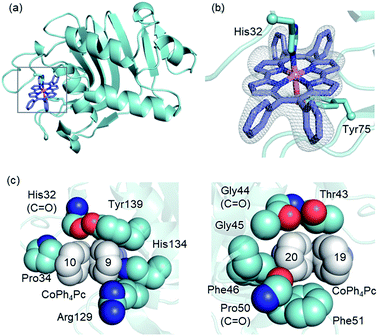 | ||
| Fig. 3 (a) Overall structure of HasA capturing Co-Ph4Pc. Co-Ph4Pc is shown as a purple stick model (PDB ID: 6JLG). (b) Enlarged view of the Co-Ph4Pc-binding site in HasA. The Fo–Fc electron density map of Co-Ph4Pc contoured at the 2.5 σ level is shown as a grey mesh. (c) Surrounding amino acids that form the cavity for accommodation of the phenyl groups of Co-Ph4Pc. Phenyl groups are shown in white. | ||
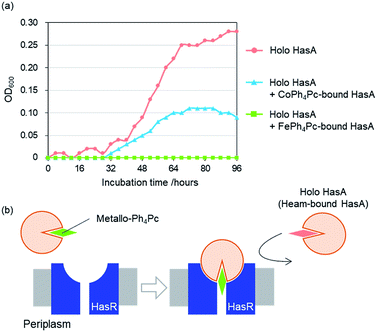
To examine whether HasA with Fe-Ph4Pc or Co-Ph4Pc has retained the ability to interact with its receptor HasR, we investigated the inhibition of P. aeruginosa growth in the presence of HasA with either Fe-Ph4Pc or Co-Ph4Pc. Growth of P. aeruginosa was monitored by tracking changes in the optical density at 600 nm (OD600) of bacterial cultures. P. aeruginosa was cultured in an M9-based medium containing EDTA as a free iron scavenger and holo-HasA as the sole iron source. Into this, HasA with either Fe-Ph4Pc or Co-Ph4Pc was also added. When compared to a control without any added metallo-Ph4Pcs, both HasA with Fe-Ph4Pc and Co-Ph4Pc were observed to significantly inhibit P. aeruginosa growth (Fig. 5a). Inhibition of growth is a strong indicator that HasA with metallo-Ph4Pcs retained the ability to interact with HasR. Moreover, these results imply that HasA containing metallo-Ph4Pcs may stay bound to HasR. As a consequence, the haem-uptake port of HasR is blocked and further acquisition of haem from holo-HasA is inhibited (Fig. 5b). We also confirmed that metallo-Ph4Pcs showed no cytotoxicity (Fig. S1†). Growth experiments revealed that HasA capturing metallo-Ph4Pcs works as an inhibitor of P. aeruginosa growth by preventing acquisition of haem from holo-HasA via HasR. This indicates that these “fake-HasAs” most probably behave similar to native holo-HasA, as the overall structure of HasA with Co-Ph4Pc resembles that of native holo-HasA. Interestingly, Ph4Pc-HasA with iron behaves like a decidedly stronger growth inhibitor than with cobalt, indicating that exchange of metals represents a potent means to regulate the efficiency of growth inhibition (Fig. 5a).
Conclusions
We have demonstrated that Fe- and Co-Ph4Pc can be incorporated into the natural haemoprotein HasA secreted by P. aeruginosa. Moreover, we succeeded in the X-ray crystal structure analysis of HasA capturing Co-Ph4Pc. The crystal structure suggests that HasA possesses an unusually adaptable haem-binding site and reveals a hidden capability of HasA to adjust its haem-binding cavity to accommodate synthetic metal complexes possessing considerably different structures from haem. HasA-loops forming the haem-binding pocket possess sufficient flexibility to accommodate the bulky phenyl groups of Co-Ph4Pc on the outside of HasA. Our findings also suggest that mutagenesis of amino acids lining the haem-binding cavity may allow for an even richer variety of metal complexes to be incorporated into HasA. Furthermore, we anticipate that HasAs containing synthetic metal complexes may also find use as novel biocatalysts unlocking access to characteristic properties unique to each metal complex. Work along this line is currently underway in our research group and will be reported in future publications.Materials and methods
Expression and purification of apo-HasA
Expression and purification of truncated HasA derived from P. aeruginosa PAO1 was performed according to similar methods reported previously.13,14 The details are given in the ESI.† The purity of the protein was checked by SDS-PAGE. Purified apo-HasA solution was flash frozen with liquid nitrogen and stored at −80 °C until use.Preparation of HasA with iron(III)-9,10,19,20-tetraphenylporphycene (Fe-Ph4Pc)
The μ-oxodimer of Fe-Ph4Pc was sufficiently dissolved in DMSO and added to a solution of apo-HasA. The mixture was dialysed into a solution of phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3) and filtrated to remove any precipitates derived from excess Fe-Ph4Pc. To remove Fe-Ph4Pc-free HasA (apo-HasA), filtered HasA solution was loaded onto an anion exchange column (HiTrap capto DEAE; GE Healthcare) equilibrated in buffer A (100 mM CHES-KOH, pH 9.5) and washed with 1 column volume of buffer A containing 10% (v/v) buffer B (100 mM CHES-KOH, 0.8 M NaCl, pH 9.5). The bound proteins were then eluted over 20 column volumes of a linear gradient from 10% to 80% buffer B, and sample fractions containing the complex of HasA with FePh4Pc were collected. This partially purified HasA solution was loaded onto the same anion exchange column equilibrated in buffer C (20 mM Tris–HCl, pH 7.5). The bound proteins were eluted over 20 column volumes of a linear gradient from 10% to 80% buffer D (20 mM Tris–HCl, 1 M NaCl, pH 7.5). Eluted fractions containing pure HasA with Fe-Ph4Pc were concentrated and purified via a desalting column (PD-10; GE Healthcare) into a PBS solution. Concentration of HasA with Fe-Ph4Pc was determined by a bicinchoninic acid (BCA) method using apo-HasA as a protein standard. The molar extinction coefficient of HasA with Fe-Ph4Pc was estimated to be 84.1 mM−1 cm−1 at 391 nm.Preparation of HasA with cobalt(III)-9,10,19,20-tetraphenylporphycene (Co-Ph4Pc)
HasA capturing Co-Ph4Pc was prepared using a similar procedure to that described for HasA with Fe-Ph4Pc. Co-Ph4Pc-free HasA (apo-HasA) was completely removed in a single-step by anion exchange chromatography using buffer C and D, as used in the 2nd step of the purification of HasA-bound Fe-Ph4Pc, and a pure solution of HasA with Co-Ph4Pc could be obtained. After exchanging buffer to PBS solution using a desalting column (PD-10; GE Healthcare), the resulting solution of HasA with Co-Ph4Pc was flash frozen with liquid nitrogen and stored at −80 °C until use. The molar extinction coefficient of HasA with Co-Ph4Pc was estimated by BCA assay to be 149.0 mM−1 cm−1 at 400 nm.Measurement
Ultraviolet-Visible spectra were recorded on a UV-2600 PC spectrophotometer (Shimadzu) and U-3310 spectrophotometer (Hitachi). ESI-TOF mass spectra were recorded on a micrOTOF II (Bruker Daltonics) using positive mode ESI-TOF method for protein solutions in 5 mM ammonium acetate buffer. FAB-MS measurements were performed on a JEOL JMS-700 instrument.Crystallisation of HasA with Co-Ph4Pc
A buffer solution of purified HasA with Co-Ph4Pc was exchanged with 100 mM KPi (pH 7.0) using desalting column (PD-10; GE Healthcare). After concentration using an Amicon Ultra (Merck Millipore; 3 kDa MWCO), the concentration of HasA capturing Co-Ph4Pc was determined by UV-Vis spectroscopy. A 1.0 μL aliquot of concentrated HasA with Co-Ph4Pc solution (30 mg mL−1 (1.6 mM) in 100 mM KPi, pH 7.0) was mixed with an equal volume of a reservoir solution composed of 1.26 M ammonium sulphate and 100 mM HEPES-HCl (pH 7.5). A drop of the protein-reservoir mixture was equilibrated with 50 μL of a reservoir solution. HasA with Co-Ph4Pc was crystallised by sitting-drop vapour-diffusion method at 20 °C for 3 weeks.Data collection and refinement
Crystals were flash-cooled by liquid nitrogen. X-ray diffraction data sets were collected at SPring-8 (Hyogo, Japan) on the BL26B1 beamline equipped with an EIGER X 4M detector at a wavelength of 1.0 Å at 100 K. The program XDS20 was used for integration of diffraction intensities and scaling. The model structure was solved by molecular replacement using MOLREP,21 with the structure of HasA with haem (3ELL)18 serving as a search model. Model building and refinement were performed on Coot,22 Phenix,23 and REFMAC5.24 The model of Co-Ph4Pc was generated by the PRODRG server25 and SKECHER.26 All protein figures were depicted by using PyMOL.27 The final refinement statistics are summarised in Table S1 (ESI).†Evaluating the inhibitory effect of HasA with metallo-Ph4Pcs on P. aeruginosa growth
Growth experiments were performed according to a previously reported procedure.14 The concentration of HasA-bound haem (holo-HasA) was estimated from the maximum absorption of the Soret band and the corresponding extinction coefficient.17Evaluating the cytotoxicity of HasA with metallo-Ph4Pcs
Using M9-based medium without EDTA, the evaluating of cytotoxicity was performed by the same procedure described in the previous section.Synthesis of metallo-Ph4Pcs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 to collect target product) to yield (Fe-Ph4Pc) μ-oxodimer as a black-purple solid (40 mg, 71.4%). HRMS (FAB, m/z): calcd (%) for C90H64Fe2N8O [M]+ 1348.3902; found for [M/2-O]+ mono Fe-Ph4Pc 668.1662. The μ-oxodimer was characterized by single crystal X-ray structure analysis as shown in Fig. S2.†
5 to collect target product) to yield (Fe-Ph4Pc) μ-oxodimer as a black-purple solid (40 mg, 71.4%). HRMS (FAB, m/z): calcd (%) for C90H64Fe2N8O [M]+ 1348.3902; found for [M/2-O]+ mono Fe-Ph4Pc 668.1662. The μ-oxodimer was characterized by single crystal X-ray structure analysis as shown in Fig. S2.†Data collection and refinement
A crystal of the (Fe-Ph4Pc) μ-oxodimer was mounted on a loop. Data from X-ray diffraction was collected at 93 K by a Rigaku XtaLAB mini CCD diffractometer equipped with graphite monochromated Mo-Kα radiation (λ = 0.71073 Å). Collected data was integrated, corrected, and scaled using the program CrysAlisPro.28 The structure was refined using SHELXT29 Intrinsic Phasing and SHELXL.30 All non-hydrogen atoms were refined anisotropically. Hydrogen atoms were located at calculated positions and included in the structure factor calculation but were not refined. The program Olex2 was used as a graphical interface.31 Crystallographic data was deposited with the Cambridge Crystallographic Data Centre (CCDC) under deposition No. CCDC-1908492.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by Grant-in-Aid for Young Scientists (A) from the Ministry of Education, culture, Sports, Science, and Technology to O. S. (26708018) and Core Research for Evolutional Science and Technology, Japan Science and Technology Agency to O. S. (JPMJCR15P3). This work was also supported by KAKENHI, Japan Society for the Promotion of Science (JP15H05806, JP18H02084 and JP18H04265).Notes and references
- K. Oohora, Y. Kihira, E. Mizohata, T. Inoue and T. Hayashi, J. Am. Chem. Soc., 2013, 135, 17282–17285 CrossRef CAS PubMed.
- H. M. Key, P. Dydio, D. S. Clark and J. F. Hartwig, Nature, 2016, 534, 534–537 CrossRef CAS PubMed.
- K. Omura, Y. Aiba, H. Onoda, J. K. Stanfield, S. Ariyasu, H. Sugimoto, Y. Shiro, O. Shoji and Y. Watanabe, Chem. Commun., 2018, 54, 7892–7895 RSC.
- E. W. Reynolds, M. W. McHenry, F. Cannac, J. G. Gober, C. D. Snow and E. M. Brustad, J. Am. Chem. Soc., 2016, 138, 14506 CrossRef CAS PubMed.
- M. B. Winter, E. J. McLaurin, S. Y. Reece, C. Olea Jr, D. G. Nocera and M. A. Marletta, J. Am. Chem. Soc., 2010, 132, 5582–5583 CrossRef CAS PubMed.
- T. Hayashi, H. Dejima, T. Matsuo, H. Sato, D. Murata and Y. Hisaeda, J. Am. Chem. Soc., 2002, 124, 11226–11227 CrossRef CAS PubMed.
- V. S. Lelyveld, E. Brustad, F. H. Arnold and A. Jasanoff, J. Am. Chem. Soc., 2011, 133, 649–651 CrossRef CAS PubMed.
- T. Ono, Y. Hisaoka, A. Onoda, K. Oohora and T. Hayashi, Chem.–Asian J., 2016, 11, 1036–1042 CrossRef CAS PubMed.
- K. Oohora and T. Hayashi, Curr. Opin. Chem. Biol., 2014, 19, 154–161 CrossRef CAS PubMed.
- A. R. Fanelli, E. Antonini and A. Caputo, Adv. Protein Chem., 1964, 19, 73–222 CrossRef.
- P. R. Ortiz de Montellano, Cytochrome P450: structure, mechanism, and biochemistry, Plenum Press, New York, 3rd edn, 2005 Search PubMed.
- R. Tenhunen, H. S. Marver and R. Schmid, J. Biol. Chem., 1969, 244, 6388–6394 CAS.
- C. Shirataki, O. Shoji, M. Terada, S.-i. Ozaki, H. Sugimoto, Y. Shiro and Y. Watanabe, Angew. Chem., Int. Ed., 2014, 53, 2862–2866 CrossRef CAS PubMed.
- H. Uehara, Y. Shisaka, T. Nishimura, H. Sugimoto, Y. Shiro, Y. Miyake, H. Shinokubo, Y. Watanabe and O. Shoji, Angew. Chem., Int. Ed., 2017, 56, 15279–15283 CrossRef CAS PubMed.
- S. Letoffe, V. Redeker and C. Wandersman, Mol. Microbiol., 1998, 28, 1223–1234 CrossRef CAS PubMed.
- S. Letoffe, C. Deniau, N. Wolff, E. Dassa, P. Delepelaire, A. Lecroisey and C. Wandersman, Mol. Microbiol., 2001, 41, 439–450 CrossRef CAS PubMed.
- G. Jepkorir, J. C. Rodriguez, H. A. Rui, W. Im, S. Lovell, K. P. Battaile, A. Y. Alontaga, E. T. Yukl, P. Moenne-Loccoz and M. Rivera, J. Am. Chem. Soc., 2010, 132, 9857–9872 CrossRef CAS PubMed.
- A. Y. Alontaga, J. C. Rodriguez, E. Schoenbrunn, A. Becker, T. Funke, E. T. Yukl, T. Hayashi, J. Stobaugh, P. Moenne-Loccoz and M. Rivera, Biochemistry, 2009, 48, 96–109 CrossRef CAS PubMed.
- T. Ono, N. Xu, D. Koga, T. Ideo, M. Sugimoto and Y. Hisaeda, RSC Adv., 2018, 8, 39269–39273 RSC.
- W. Kabsch, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 133–144 CrossRef CAS PubMed.
- A. Vagin and A. Teplyakov, J. Appl. Crystallogr., 1997, 30, 1022–1025 CrossRef CAS.
- P. Emsley and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 2126–2132 CrossRef PubMed.
- P. D. Adams, P. V. Afonine, G. Bunkoczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L.-W. Hung, G. J. Kapral, R. W. Grosse-Kunstleve, A. J. McCoy, N. W. Moriarty, R. Oeffner, R. J. Read, D. C. Richardson, J. S. Richardson, T. C. Terwilliger and P. H. Zwart, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 213–221 CrossRef CAS PubMed.
- G. N. Murshudov, P. Skubak, A. A. Lebedev, N. S. Pannu, R. A. Steiner, R. A. Nicholls, M. D. Winn, F. Long and A. A. Vagin, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 355–367 CrossRef CAS PubMed.
- A. W. Schuttelkopf and D. M. F. van Aalten, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 1355–1363 CrossRef PubMed.
- A. A. Vagin, R. A. Steiner, A. A. Lebedev, L. Potterton, S. McNicholas, F. Long and G. N. Murshudov, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 2184–2195 CrossRef PubMed.
- W. L. DeLano, Pymol Mol. Graph. Syst., 2002, http://www.pymol.org Search PubMed.
- Rigaku Corporation, CrysAlisPro, Tokyo, Japan, 2015 Search PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71, 3–8 CrossRef PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 Search PubMed.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, J. Appl. Crystallogr., 2009, 42, 339–341 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1908492. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ra02872b |
| This journal is © The Royal Society of Chemistry 2019 |