Open Access Article
Open Access ArticleSpecific purification of a single protein from a cell broth mixture using molecularly imprinted membranes for the biopharmaceutical industry†
Wenyuan Xie *ad,
Honglei Wangb,
Yen Wah Tong*bc,
Niranjani Sankarakumarb,
Ming Yinc,
Defeng Wu
*ad,
Honglei Wangb,
Yen Wah Tong*bc,
Niranjani Sankarakumarb,
Ming Yinc,
Defeng Wu d and
Xiaoli Duane
d and
Xiaoli Duane
aInstitute for Innovative Materials and Energy, Yangzhou University, Yangzhou 225002, Jiangsu, People's Republic of China. E-mail: xiewy@yzu.edu.cn
bDepartment of Chemical & Biomolecular Engineering, National University of Singapore, Block E5 #02-09, 4 Engineering Drive 4, Singapore 117585, Singapore
cYangzhou Zhongcheng Nanotech Co, Ltd., 7# Chuangye Road, Guangling District, Yangzhou 225000, Jiangsu, China
dSchool of Chemistry and Chemical Engineering, Yangzhou University, Yangzhou 225002, Jiangsu, People's Republic of China
eSchool of Chemistry and Materials Science, Ludong University, Yantai 264025, People's Republic of China
First published on 29th July 2019
Abstract
A surface imprinting method is presented herein for the development of a highly selective yet highly permeable molecularly imprinted membrane for protein separation and purification. The resultant protein imprinted membrane was shown to exhibit great potential for the efficient separation of the template protein from a binary mixture and a cell lysate solution, while maintaining high transport flux for complementary molecules. Bovine Serum Albumin (BSA) and Lysozyme (Lys) were individually immobilized on a cellulose acetate membrane as template molecules. In situ surface crosslinking polymerization was then used for protein imprinting on the membrane for a controlled duration. Both membranes showed high adsorption capacity towards template proteins in the competitive batch rebinding tests. In addition, the adsorption capacity could be greatly enhanced in a continuous permeation procedure, where the resultant membrane specifically adsorbed the template protein for more than 40 h. Moreover, this is the first report of purification of a specific protein from the cell broth mixture using a molecularly imprinted membrane. The protein imprinted membrane enables the transport of multiple non-template proteins with high permeation rate in a complex system, thus opening the way for high efficiency protein separation at a low cost for the industry.
Introduction
Molecularly imprinted polymers (MIPs) have a wide range of applications, such as in the field of separation,1 catalysis,2 drug design,3 analytical chemistry,4,5 biosensing6 and other materials-related areas.7,8 MIPs display molecular selectivity that is comparable to their biological counterparts such as enzymes and antibodies while possessing intrinsic strengths which are absent in biological molecules, such as robustness, reusability, and being inexpensive to prepare.9 As such, MIPs could potentially be used to mimic and replace their biological equivalents. In recent years, the molecular imprinting technique (MIT) has achieved great progress, especially in the separation of small molecules. Challenges still remain in the separation of biomacromolecules due to their flexible, complex and large-sized structures.10 A major challenge lies in the limited mass transfer of the biomolecules in the bulk polymer, which hinders the templates' removal and targets' rebinding. The surface imprinting technique is one of the most efficient strategies to overcome this obstacle.11 Surface imprinted thin films could be employed as shell layers of various kinds of substrates, for instance, nanoparticles,12–14 nanowires15–17 nanofibers,18 nanotubes19 and microspheres.20,21 However, further separation steps might be required, such as dispersion and recollection of core–shell particles in the protein solution.Membrane separation has been widely used in industrial purification due to its single-step operation procedure, low energy consumption and ease of scaling-up. Besides, it has great potential in protein separation and purification based on the size, charge and other properties of proteins.22–24 However, most of membranes were not tailored specially for a particular protein. Besides, membrane properties were adapted specifically for each separation system.25
In contrast, molecularly imprinted membranes (MIMs) offer researchers opportunities to design membranes that are targeted to specific molecules.26,27 Thereby, a generic MIM can be employed for separating different protein mixtures and systems as long as the desired target molecules are the same. MIMs combine advantages of both the molecular imprinting technology (i.e. high intrinsic selectivity) and the membrane technology (i.e. low energy consumption, ease of scale-up and the potential for continuous operation).26 Recently, MIMs for protein separation have been reported with several strategies, including electrospinning,28 surface grafting29–31 and nanopores incorporation.32 High specific recognition of the template proteins has been achieved. However, drawbacks such as weak binding force and low perm-selectivity, are still challenging the comprehensive membrane performance.33
Consequently, finding new approaches to construct the surface imprinted layer is of great importance to achieve new MIMs with excellent perm-selectivity for protein purification. Furthermore, the separation mechanism of MIMs in the continuous membrane separation process, which differs greatly from that of the static adsorption process, needs to be investigated systematically.
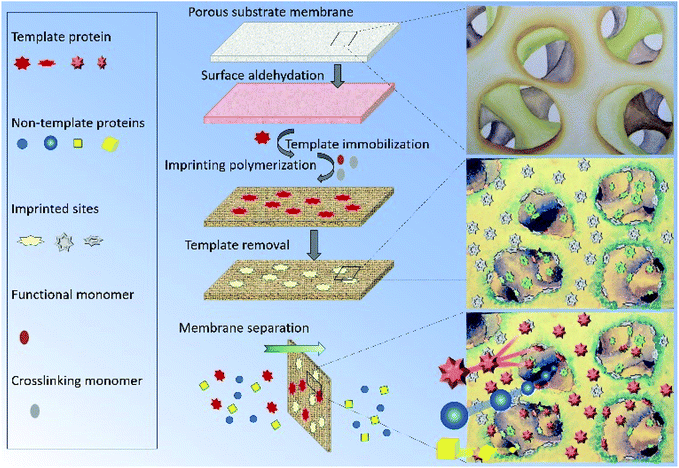
To address the above problems, we herein report a novel protein surface imprinting technique for the preparation of a protein imprinted membrane with high binding capacity to the template protein and high permeability to the non-template protein. Following a similar molecularly imprinting technique based on our previous works,34–36 the template protein was first covalently immobilized on a porous cellulose acetate membrane. Subsequently, redox initiators were spread onto the porous substrate by spin-coating, and then in situ crosslinking polymerization was carried out. Finally, the immobilized template protein was removed by hydrolysis to leave imprinted sites on the membrane (as shown in Fig. 1). In this study, two molecularly imprinted membranes, MIMBSA and MIMLys, were prepared by using bovine serum albumin (BSA) and lysozyme (Lys) as template proteins respectively. The effectiveness of this method was examined through the separation of template proteins from the BSA–Lys binary mixture. In addition, membrane performance was tested in a continuous separation setup, with a complex mixture to simulate an industrial system, consisting of BSA, lysozyme and bacterial cell lysate. Furthermore, membrane separation mechanism of MIMs was investigated.
Experimental section
Materials
Cellulose acetate (CA) (CA-389-30) was purchased from Eastman Chemical Company, USA. Phosphate buffered saline (PBS, 0.1 M), sodium periodate, bovine serum albumin (BSA), lysozyme (Lys), fluorescein isothiocyanate (FITC), methyl methacrylate (MMA), ethylene glycol dimethacrylate (EGDMA), ammonium persulfate (APS), sodium bisulfite (SBS), sodium dodecyl sulfate (SDS) and trifluoroacetic acid (TFA) were purchased from Sigma Aldrich, USA. N-methyl-2-pyrrolidone (NMP, 99.5%) was purchased from Merck KGaA, Darmstadt, Germany. Tryptone, yeast extract and agar were purchased from BactoTM. Escherichia coli ATCC®19853 was purchased from the American Type Culture Collection, USA.Preparation of MIMs and NIMs
Structural characterization of membranes
Membrane structures before and after aldehyde functionalization were monitored using a FTIR Spectrum 2000 (PerkinElmer) with an attenuated total reflection (ATR-FTIR) technique. The surface elemental composition of the membranes after each surface modification step were determined through X-ray photoelectron spectroscopy (XPS) (AXIS HIS, 165 Ultra, Shimadzu). Surface roughness changes of membranes after each modification step were analyzed using a Nanoscope IIIa AFM (Veeco) in tapping mode. Morphological observation of the membranes was conducted using a field emission scanning electron microscopy (FESEM) (JSM-6700F, JEOL). Membranes were first freeze-dried and then fractured in liquid nitrogen. The fractured membranes were then mounted onto the sample holder using conductive carbon adhesive tape. The samples were sputtered with platinum before FESEM observation. The nitrogen distribution of the protein-adsorbed membranes was measured using an energy dispersive X-ray (EDX, Oxford Instruments, Aztec X-MaxN). All EDX analyses were performed using the line scan mode for 150 s along the cross-section of the MIMs. The pore size of the original CA membrane, MIMs and NIMs were characterized via neutral solute rejection by using a dead-end permeation cell, and detailed technical descriptions can be found in the ESI.†Membrane performance characterization
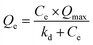 | (1) |
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Qe = lg Qe = lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) A + m × lg A + m × lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ce Ce
| (2) |
The separation factor of MIMBSA and NIMBSA was expressed as eqn (3)38 and the separation factor of MIMLys and NIMLys was expressed as eqn (4)39
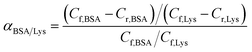 | (3) |
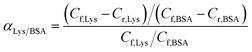 | (4) |
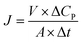 | (5) |
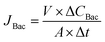 | (6) |
Results and discussion
Synthesis of membranes
Cellulose acetate (CA) membrane was used as the substrate for surface chemical modification. The fabrication of the MIM began by immobilizing the template protein onto the CA membrane substrate. The appearance of the characteristic aldehyde peak at ∼1720 cm−1 in the FTIR spectrum proved the successful modification of the membrane using NaIO4-generated aldehyde groups from the hydroxyl groups of the CA (Fig. S2†). The maximum number of aldehyde groups on the CA membrane surface was generated to maximize the density of the immobilized proteins on the CA substrates (Fig. S3, see ESI† for details). Subsequently, an in situ surface crosslinking polymerization on the protein-immobilized CA membrane was applied and followed by hydrolysis to remove the template protein. The duration of the polymerization reaction was used to control the thickness of the imprinting polymer layer. Specifically, if the polymerization time was too short, the thickness of the polymer film may not be sufficient to imprint the shape of the template protein. On the contrary, an excessive polymerization imprinting time would form denser polymer monoliths that could completely cover and permanently entrap the template proteins. The AFM images of original CA membrane and a series of non-imprinted membranes (NIMs) in Fig. 2 indicate average roughness values of 1.7 nm before polymerization and 2.4, 3.2, 5.9 and 13.3 nm after polymerization for 5, 8, 14 and 18 h respectively. The thickness of the imprinted polymer layer after 14 h of polymerization was estimated to be around 4.2 nm by comparing with the average roughness of the original CA membrane, which suggests a high possibility of fully covering the BSA template as the hydrodynamic radius of BSA is 4.5 nm.41 Therefore, the 8 h polymerization time was chosen in subsequent studies, to balance between efficiency of the imprinting process and the surface imprinting coverage for fabrication of the MIMBSA. By using a similar estimation, the 5 h polymerization provided a 0.7 nm imprinting polymer layer which was used to imprint the Lys template molecules with a hydrodynamic radius of 1.9 nm.42 The reduction in nitrogen content detected after protein removal indicated that 83% and 75% of the immobilized template protein molecules was removed via acidic hydrolysis for both MIMBSA and MIMLys respectively (Table S1†).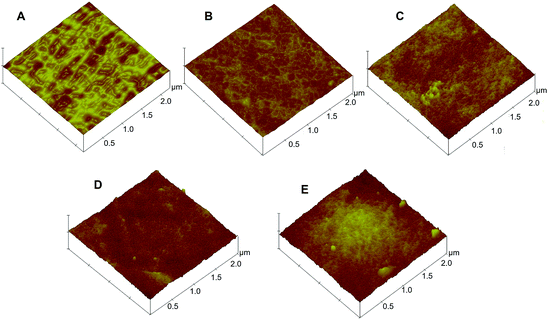 | ||
| Fig. 2 Surface morphology of membranes by atomic force microscopy (AFM): (A) original CA membrane, and NIMs with various polymerization duration of (B) 5 h; (C) 8 h; (D) 14 h; (E) 18 h. | ||
Structural characterization
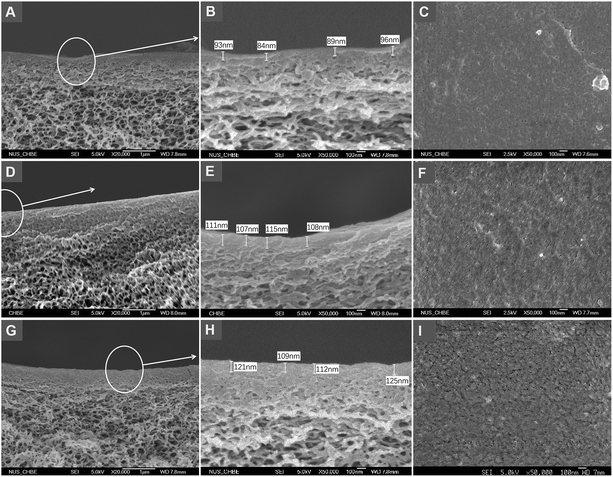
FESEM images in Fig. 3 show the aldehyde functionalized membranes and of those after in situ crosslinking polymerization. The comparison among the more detailed morphologies in Fig. 3B, E and H suggests that both the substrate underlayer and the top skin layer of the post-imprinted membrane are slightly thicker and denser than those before imprinting. Such morphological change is more pronounced in MIMBSA, indicating that the imprinting layer on MIMBSA is thicker than MIMLys. The comparison among Fig. 3C, F and I shows higher roughness on the surfaces of the membranes after imprinting. These morphological changes observed in the post-imprinted membrane suggests that in situ crosslinking polymerization might have taken place on both the skin layer and the substrate under layer due to the porous structural nature of the membrane.Membrane performance
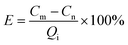 | (7) |
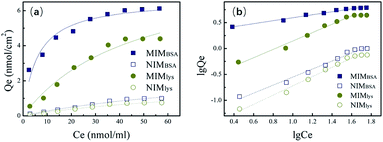
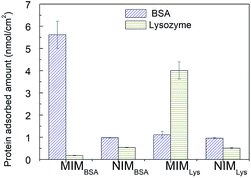
| Membrane | Langmuir isotherm, Qe = Ce × Qmax/(kd + Ce) | Freundlich isotherm, ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Qe = ln Qe = ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) A + m × ln A + m × ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ce Ce |
||||
|---|---|---|---|---|---|---|
| Qmax (nmol cm−2) | kd (nmol ml−1) | R2 | m | A (nmol cm−2) | R2 | |
| MIMBSA | 6.66 | 5.88 | 0.925 | 0.277 | 2.10 | 0.974 |
| NIMBSA | 2.79 | 92.63 | 0.976 | 0.851 | 0.04 | 0.984 |
| MIMlys | 8.60 | 46.33 | 0.972 | 0.815 | 0.21 | 0.988 |
| NIMlys | 2.03 | 86.77 | 0.975 | 0.952 | 0.02 | 0.991 |
The Freundlich isotherm fits better than the Langmuir isotherm especially for MIMs at lower concentration range. The Freundlich parameter m of MIMBSA and MIMlys are 0.277 and 0.815 respectively.
Permeation performance and mechanism
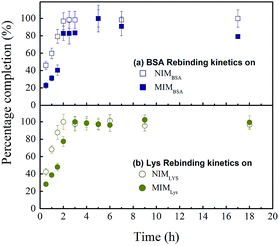
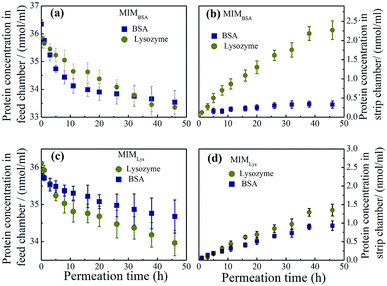
On the other hand, the separation performance of MIMLys was not as good as MIMBSA. Intrinsically, due to the lower imprinting efficiency of the MIMLys, the adsorption of lysozyme was not significantly higher than BSA as also found in the batch competitive rebinding test.
The other possible factor was that positive charges on lysozyme under a neutral pH were unfavorable for adsorption. Comparing the two permeation systems, the electrical charge density of lysozyme was much higher than that of BSA at the neutral permeation condition considering the huge differences of the isoelectric point (pI) and the molecular weight (MW) between BSA (pI: 5.1, MW: 67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000)45 and Lys (pI: 11, MW: 14
000)45 and Lys (pI: 11, MW: 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400).39 Consequently, a higher electron repulsion could be expected to exist among the lysozyme molecules in solution and those adsorbed on the MIMLys compared with that of the BSA and MIMBSA.46
400).39 Consequently, a higher electron repulsion could be expected to exist among the lysozyme molecules in solution and those adsorbed on the MIMLys compared with that of the BSA and MIMBSA.46
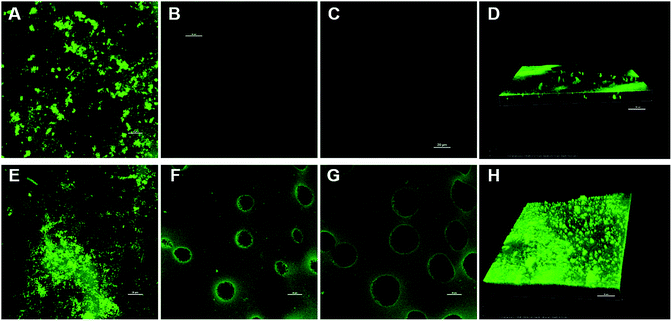
To further verify the above hypothesis of existence of the imprinted sites beneath the skin layer of MIMs, two pieces of MIMBSA bound with BSA-FITC conjugates were imaged with a confocal laser scanning microscope. One MIMBSA was after 48 h in the competitive rebinding test; and the other MIMBSA was after 48 h in the permeation test with BSA–Lys binary solution. From Fig. 8, it can be observed that more BSA-FITC conjugates were adsorbed on the top layer of MIMBSA after permeation than after rebinding. In addition, BSA-FITC conjugates adsorbed onto the inside layer of the porous substrates after permeation test. These sites were less accessible to BSA-FITC conjugates during the rebinding test.
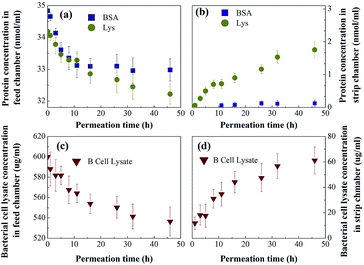
Permeation purification from bacterial lysate
To better mimic the complexity and diversity of a real industrial protein purification, MIMBSA was applied to a complex mixture consisting of BSA, lysozyme and bacterial lysate which was composed of proteins, lipids and nucleic acids. From Fig. 9, both the concentration of lysozyme and bacterial lysate increased in the strip chamber with a permeation rate of 0.85 nmol cm−2 h−1 and 27 μg cm−2 h−1 respectively, and only negligible amount of BSA could be found in the strip chamber even after 48 h. These results illustrated that template BSA was successfully separated from a multicomponent mixture with a high flux of other components. Thus, the protein imprinted membrane possesses a wide potential in the application of protein purification due to its high specification and selectivity in a complex system.Reusability of the protein imprinted membranes was also tested. The adsorbed protein after the rebinding tests and the protein residue after protein removal experiments were characterized by XPS. As shown in Fig. S7,† the adsorbed amounts of proteins remained the same even after the third rebinding cycle. Specifically, after the third rebinding or removal, the nitrogen element contents on the surface of MIM can still go up to or decrease to almost the same amount as the first rebinding, indicating good reusability of the MIM.
Conclusions
We have shown a well-controlled molecular imprinting method for MIM fabrication. It is based on a simple yet elegant modification of a conventional cellulose acetate membrane, thus enabling common membranes to be used for highly effective protein separation. The above results showed that the protein separation was characterized in detail using adsorption and also continuous assays to elucidate the mechanism of this membrane imprinting method. This work documents that a combination of the specific recognition ability of a molecular imprinted polymer and the size-exclusion effect of a membrane during permeation synergistically enhance protein separation performance in terms of purity. Finally, we have shown that this novel membrane has the ability to separate protein from a complex protein mixture with a high flux thus making it suitable for industrial applications in protein purification. Although only BSA and lysozyme are used as target template in this study, this surface imprinting technique has demonstrated the great potential and viability for the preparation of protein-imprinted membranes using a wide range of other target template molecules.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
The authors appreciate the National Natural Science Foundation of China (NSFC No. 51803176) and NUS Foundation (R279000249646) for financially supporting this research. Special thanks are given to Dr Jingnan Luo and Dr Shirlaine Koh for their suggestions and help in this work.References
- L. Chen, X. Wang, W. Lu, X. Wu and J. Li, Chem. Soc. Rev., 2016, 45, 2137–2211 RSC.
- Y. Zhang and S. N. Riduan, Chem. Soc. Rev., 2012, 41, 2083–2094 RSC.
- C. Alvarez-Lorenzo and A. Concheiro, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2004, 804, 231–245 CrossRef CAS PubMed.
- R. Kecili and C. M. Hussain, Int. J. Anal. Chem., 2018, 8503853 Search PubMed.
- J. Wang, Q. Xu, W. W. Xia, Y. Shu, D. Jin, Y. Zang and X. Hu, Sens. Actuators, B, 2018, 271, 215–224 CrossRef CAS.
- W. S. P. Carvalho, M. Wei, N. Ikpo, Y. Gao and M. J. Serpe, Anal. Chem., 2018, 90, 459–479 CrossRef CAS.
- J. Wackerlig and R. Schirhagl, Anal. Chem., 2016, 88, 250–261 CrossRef CAS PubMed.
- H. Wang, Q. Xu, J. Wang, W. Du, F. Liu and X. Hu, Biosens. Bioelectron., 2018, 100, 105–114 CrossRef CAS PubMed.
- A. Katz and M. E. Davis, Nature, 2000, 403, 286–289 CrossRef CAS PubMed.
- M. J. Whitcombe, I. Chianella, L. Larcombe, S. A. Piletsky, J. Noble, R. Porter and A. Horgan, Chem. Soc. Rev., 2011, 40, 1547–1571 RSC.
- G. De Middeleer, P. Dubruel and S. De Saeger, TrAC, Trends Anal. Chem., 2016, 76, 71–85 CrossRef CAS.
- C. Yang, X. Yan, H. Guo and G. Fu, Biosens. Bioelectron., 2016, 75, 129–135 CrossRef CAS PubMed.
- Y. Hoshino, T. Miyoshi, M. Nakamoto and Y. Miura, J. Mater. Chem. B, 2017, 5, 9204–9210 RSC.
- J. Wackerlig and P. A. Lieberzeit, Sens. Actuators, B, 2015, 207, 144–157 CrossRef CAS.
- H.-H. Yang, S.-Q. Zhang, F. Tan, Z.-X. Zhuang and X.-R. Wang, J. Am. Chem. Soc., 2005, 127, 1378–1379 CrossRef CAS PubMed.
- T. Chen, M. Shao, H. Xu, S. Zhuo, S. Liu and S.-T. Lee, J. Mater. Chem., 2012, 22, 3990–3996 RSC.
- X. Xu, P. Guo, Z. Luo, Y. Ge, Y. Zhou, R. Chang, W. Du, C. Chang and Q. Fu, RSC Adv., 2017, 7, 18765–18774 RSC.
- Y. Li, Q. Bin, Z. Lin, Y. Chen, H. Yang, Z. Cai and G. Chen, Chem. Commun., 2015, 51, 202–205 RSC.
- Z. Luo, W. Du, P. Guo, P. Zheng, R. Chang, J. Wang, A. Zeng, C. Chang and Q. Fu, RSC Adv., 2015, 5, 72610–72620 RSC.
- L. Qin, X.-W. He, X. Yuan, W.-Y. Li and Y.-K. Zhang, Anal. Bioanal. Chem., 2011, 399, 3375–3385 CrossRef CAS PubMed.
- J. Wang, L. Tian, Y. Yan, Y. Liu, Y. Zhang and C. Yang, J. Braz. Chem. Soc., 2018, 29, 2–10 CAS.
- X. Q. Cheng, Z. X. Wang, X. Jiang, T. Li, C. H. Lau, Z. Guo, J. Ma and L. Shao, Prog. Mater. Sci., 2018, 92, 258–283 CrossRef CAS.
- D. Xu, S. Hein and K. Wang, Mater. Sci. Technol., 2008, 24, 1076–1087 CrossRef CAS.
- D. Y. Xing, S. Y. Chan and T.-S. Chung, Green Chem., 2012, 14, 1405–1412 RSC.
- N. F. Ishak, N. A. Hashim, M. H. D. Othman, P. Monash and F. M. Zuki, Ceram. Int., 2017, 43, 915–925 CrossRef CAS.
- M. Yoshikawa, K. Tharpa and Ş.-O. Dima, Chem. Rev., 2016, 116, 11500–11528 CrossRef CAS PubMed.
- R. I. Boysen, L. J. Schwarz, D. V. Nicolau and M. T. W. Hearn, J. Sep. Sci., 2017, 40, 314–335 CrossRef CAS PubMed.
- T. Zhu, D. Xu, Y. Wu, J. Li, M. Zhou, T. Tian, Y. Jiang, F. Li and G. Li, J. Mater. Chem. B, 2013, 1, 6449–6458 RSC.
- D. Liu and M. Ulbricht, RSC Adv., 2017, 7, 11012–11019 RSC.
- Y. Wu, M. Yan, J. Cui, Y. Yan and C. Li, Adv. Funct. Mater., 2015, 25, 5823–5832 CrossRef CAS.
- R.-R. Chen, L. Qin, M. Jia, X.-W. He and W.-Y. Li, J. Membr. Sci., 2010, 363, 212–220 CrossRef CAS.
- Z. Luo, W. Du, P. Guo, P. Zheng, R. Chang, J. Wang, A. Zeng, C. Chang and Q. Fu, RSC Adv., 2015, 5, 72610–72620 RSC.
- W. Li and J. Y. Walz, Sci. Rep., 2014, 4, 4418 CrossRef PubMed.
- W. Xie, F. He, B. Wang, T.-S. Chung, K. Jeyaseelan, A. Armugam and Y. W. Tong, J. Mater. Chem. A, 2013, 1, 7592–7600 RSC.
- N. Sankarakumar and Y. W. Tong, J. Mater. Chem. B, 2013, 1, 2031–2037 RSC.
- N. Sankarakumar and Y. W. Tong, RSC Adv., 2013, 3, 1519–1527 RSC.
- R. J. Ansell, in Molecularly Imprinted Polymers in Biotechnology, ed. B. Mattiasson and L. Ye, Springer International Publishing, Cham, 2015, vol. 150, pp. 51–93 Search PubMed.
- C. J. Tan, H. G. Chua, K. H. Ker and Y. W. Tong, Anal. Chem., 2008, 80, 683–692 CrossRef CAS PubMed.
- K. P. Wilson, B. A. Malcolm and B. W. Matthews, J. Biol. Chem., 1992, 267, 10842–10849 CAS.
- Y. Xiao and T.-S. Chung, J. Membr. Sci., 2007, 290, 78–85 CrossRef CAS.
- U. Böhme and U. Scheler, Chem. Phys. Lett., 2007, 435, 342–345 CrossRef.
- A. Bonincontro, V. Calandrini and G. Onori, Colloids Surf., B, 2001, 21, 311–316 CrossRef CAS PubMed.
- Z. Zhang and B. Wang, J. Appl. Polym. Sci., 2009, 113, 1050–1062 CrossRef CAS.
- P. R. Van Tassel, L. Guemouri, J. J. Ramsden, G. Tarjus, P. Viot and J. Talbot, J. Colloid Interface Sci., 1998, 207, 317–323 CrossRef CAS PubMed.
- B. Jachimska, M. Wasilewska and Z. Adamczyk, Langmuir, 2008, 24, 6866–6872 CrossRef CAS PubMed.
- A. Vinu, M. Miyahara and K. Ariga, J. Phys. Chem. B, 2005, 109, 6436–6441 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra02805f |
| This journal is © The Royal Society of Chemistry 2019 |