Open Access Article
Open Access ArticleSmall DNA circles as bacterial topoisomerase I inhibitors†
Dawei Li *ac,
Qiang Wanga,
Bing Zhoua,
Qiang Zhugeac and
Bei Lv
*ac,
Qiang Wanga,
Bing Zhoua,
Qiang Zhugeac and
Bei Lv *b
*b
aCollege of Biology and the Environment, Nanjing Forestry University, 159 Longpan Road, Nanjing, 210037, China. E-mail: dwli@njfu.edu.cn
bJiangsu Key Laboratory for Biofunctional Molecules, College of Life Science and Chemistry, Jiangsu Second Normal University, Nanjing, 210013, China. E-mail: lvbei@jssnu.edu.cn
cThe Southern Modern Forestry Collaborative Innovation Center, Nanjing Forestry University, 159 Longpan Road, Nanjing, 210037, China
First published on 11th June 2019
Abstract
Bacterial topoisomerase I is a potential target during the course of antibacterial drug therapy. In our studies, specifically designed small DNA circles with high bending stress were synthesized. It is demonstrated that small DNA circles showed high inhibitory effect on the activity of bacterial topoisomerase I and the single-stranded regions associated with bending deformation in DNA circles are believed to be the crucial factor for trapping the enzymes and decreasing the effective concentration of the topoisomerases in the reaction solution. In addition, the DNA circles showed high thermal stability and excellent nuclease resistance. In consideration of the low cytotoxicity of DNA-based biopharmaceuticals, our results may provide a new idea for the future design and optimization of DNA-based therapeutic agents for antibacterial therapy.
Introduction
Topoisomerases are essential enzymes which have the special ability to change the topological structures of DNA by cleaving the DNA backbone and then rejoining it in the cells.1–3 Those ubiquitous proteins are reported to be involved in many cellular processes such as replication, transcription and recombination and critical for cell growth and proliferation.4–6 For the importance of their cellular role, some topoisomerases are reported to be selected as the targets of anti-cancer drugs and antibiotics.7–9 Bacterial type IIA topoisomerases, topoisomerase IV and DNA gyrase for example, are well utilized clinical targets for antibacterial chemotherapy.10,11 However, resistance of bacterial pathogens to current antibiotics has grown to be an urgent crisis. Compounds that can inhibit the activities of bacterial type IA topoisomerases have been extensively studied in recent years.12–14Bacterial topoisomerase I belongs to type IA topoisomerase, which can cause single-stranded break on substrate DNA and then re-seal it.15,16 During the course of its action, topoisomerase I can covalently attach to the 5′ end of the broken DNA strand. It has been confirmed that single-stranded regions in its substrate DNA are the crucial binding locations for the activity of bacterial topoisomerases I.17,18 Yang and co-workers reported that short linear duplex oligonucleotides containing single-stranded segments can serve as the inhibitory agent for bacterial topoisomerases I.19,20 However, those linear oligonucleotides exhibited low thermal stability and poor nuclease resistance. In addition, the helical structure of linear duplex oligonucleotides is different from the negatively supercoiled circular DNA (as the innate substrate of bacterial topoisomerases I), which may decrease the affinity between the inhibitor and enzyme. On the other hand, small DNA circles occur in nature, for example as protein-constrained loops, and can be synthesized by a number of methods.21,22 It has been reported that forcible curvature in small circular DNA can be recognized by DNA topoisomerase I23 More importantly, torsional stress can cause local unwinding of the double helix in DNA mini-circles,24–26 which is the indispensable binding location for the activity of type IA topoisomerases. Inspired by those previous discoveries, we therefore speculated that the disruptions of the regular helical structure caused by DNA bending in small circular DNA could be targeted by bacterial topoisomerase I, which may decrease the effective concentration of the topoisomerase in the reaction solution. In this work, the possibility of using small DNA circles as the irreversible inhibitors of bacterial topoisomerase I was investigated. A series of covalently closed mini-DNA circles with different size were designed and synthesized. Our results show that all the duplex circular oligonucleotides exhibited inhibitory effect on the activity of bacterial topoisomerase I and the most efficient one displayed an IC50 value of 10 nM. All the circular DNA in our studies display excellent thermal stability and nuclease resistance.
Results and discussion
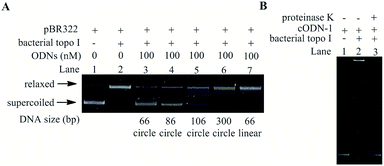
In order to investigate the possibility of using mini-DNA circles as the efficient inhibitors of bacterial topoisomerases I, a series of duplex circular oligodeoxyribonucleotides (cODNs) were designed and synthesized. All sequences of ODNs are shown in Table S1.† The covalently closed duplex cODNs were obtained by annealing two single-stranded ODNs and then sealing the breaks in DNA backbone24 (see detail in Materials and methods). The cODN-1, cODN-2 and cODN-3 possess 66, 86 and 106 base pairs (bp) in length respectively, in which high bending stress exists in their circular backbone.24 Another circular DNA of 300 bp (DC-300) was obtained by ligase-catalyzed ring closure of linear DNA with complementary cohesive ends according to our previous reports.27,28 It is clear that bending and torsional stress increase with decreasing of the ring size. It is our speculation that the disruptions and local unwinding of double helix caused by DNA bending and torsional deformations in the cODNs could be recognized by bacterial topoisomerases I. As shown in Fig. 1, in the absence of cODNs (left side), the plasmid DNA is relaxed by the topoisomerase. On the other hand, because of the existence of the similar structure with single-stranded region in both substrate plasmid and small DNA circles, bacterial topoisomerases I can be trapped by cODNs (right side) and the plasmid relaxation reaction is inhibited.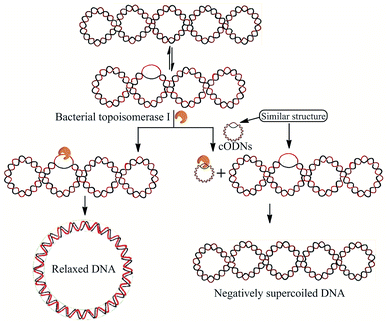 | ||
| Fig. 1 Illustration of the supercoiled plasmid relaxation catalyzed by bacterial topoisomerase I in the presence or absence of cODN. | ||
With the aim to verify whether the cODNs can inhibit the relaxation activity of bacterial topoisomerases I, negatively supercoiled plasmid pBR322 was used as the substrate of the relaxation reaction in the presence of cODNs. The inhibitory efficiency can be evaluated by measuring the variation in relaxation percentage of supercoiled pBR322.29 As shown in Lane 2 in Fig. 2A, all DNA products were in its relaxed conformation (upper band) after the negatively supercoiled plasmid (lower band in Lane 1 in Fig. 2) was treated with bacterial topoisomerases I within 30 minutes in the absence of cODNs. However, few relaxed products can be found when a 66 bp of cODN-1 (100 nM) was added into the relaxation reaction, which indicates that cODN-1 suppressed the activity of bacterial topoisomerases I and acted as an inhibitor in the relaxation reaction (Lane 3). With the purpose of investigating the correlation between inhibitory efficiency and degree of DNA bending, another three small circular DNA (86 bp, 106 bp and 300 bp) were used as the inhibitors in the relaxation assay.
As shown in Lane 4 to 6, the inhibitory efficiency decreased with the increasing of the size of DNA circles, which is consistent with the suggestion that the deformation of DNA double helix associated with DNA bending is the crucial factor for the inhibitory action on the activity of bacterial topoisomerases I. In order to further confirm the inhibition caused by the local unwinding of the double helix associated with the bending stress in cODN-1, a 66 bp linear duplex DNA was prepared and used as the inhibitor with the same concentration of cODN-1 in the relaxation assay. The linear duplex DNA has the same nucleotide sequence with cODN-1 but no forcible torsional stress accumulates in its backbone. As shown in Lane 7 in Fig. 2A, most of plasmids were relaxed and no apparent inhibitory effects can be found. It has been reported that single stranded regions existed in small DNA circles with highly bending stress but not in its linear counterpart that was proved by single-strand specific endonucleases assays.24 The observations in our studies indicate that the inhibition effect was indeed caused by the bending and torsional deformation of local double helix in highly bending cODNs.
In order to study the affinity between 66 bp of cODN-1 and bacterial topoisomerases I, electrophoretic mobility shift assay (EMSA) was used to study the binding of the proteins on the small DNA circles. It is clear that DNA–protein complex often shows slower mobility shift than the unbound DNA in native PAGE.30 As shown in Fig. 2B, comparing with free cODN-1 (Lane 1), a much more slower moving band (Lane 2) can be observed when the DNA circles were treated with bacterial topoisomerases I. However, the free DNA band was recovered after the mixture in Lane 2 was digested by proteinase K (Lane 3), which verified that the slower band in Lane 2 is caused by the formation of DNA–protein complex. It has been reported that the bacterial topoisomerase I weakly binds a relaxed DNA mini-circle of 235 bp and exhibits a stronger binding on its negatively supercoiled topoisomers.31 In the current studies, the strong binding affinity between the cODN-1 of 66 bp and bacterial topoisomerase I was observed. Although the cODN-1 was obtained by circularization of linear oligonucleotides, high bending and torsional stress accumulates along the backbone of cODN-1, which can cause the deformation and kinks of double helix in the very small DNA circles.24 The local disruption of DNA duplex in cODN-1 may be recognized by bacterial topoisomerase I. In addition, it has been studied that the multiple bend locations in the DNA mini-circles with high torsional stress can be determined by the sequence,32 which may provide a new method to optimize the sequence of the small DNA circle and increase the affinity between the mini-DNA circles and topoisomerase.
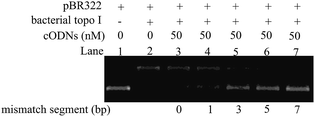
With the purpose to investigate the possibility to increase the inhibitory efficiency of small DNA circles by creating more single-stranded segments, a series of cODNs with different length of mismatched base pairs were designed and synthesized. Originally, the mismatched segments were designed in 66 bp of cODN-1. Unfortunately, we failed to produce those DNA mini-circles, possibly due to the obstruction of ligation caused by highly DNA bending.24 However, several new cODNs were obtained by making some mismatched regions in 86 bp of cODN-2. The cODN-2-M1, cODN-2-M3, cODN-2-M5, cODN-2-M7 contain 1, 3, 5 or 7 mismatched base pairs and were used as the inhibitors in the relaxation reaction catalyzed by bacterial topoisomerases I (see Table S1† for detail sequence information). The assay results were observed in the presence of 50 nM cODNs. As shown in Fig. 3, all the newly synthesized mismatch-containing cODNs exhibited high inhibitory effect on the activity of bacterial topoisomerases I. More importantly, the attenuation magnitudes of bacterial topoisomerases I activities are gradually suppressed when the length of single-stranded regions are increased from cODN-2-M1 to cODN-2-M7 by adding the amount of mismatch. The observation is consistent with the suggestion that the inhibition efficiency could be improved with increasing the single-stranded regions created by mismatch in cODNs.
IC50 is a quantitative measure of the potency of a substance in inhibiting a specific reaction and it indicates how much of a particular inhibitor is needed to inhibit a given biological process by half.33 In order to evaluate the inhibitory effect of the small DNA circles, the IC50 values of each cODNs were quantified. As shown in Fig. S1,† the relaxation percentages of supercoiled plasmid pBR322 were monitored by agarose gel in different concentration of cODNs. It is shown that relaxation efficiency decreased with the increase in concentration of each cODN. Inhibition percentages are plotted against the logarithms of cODNs concentration and are fitted to the sigmoid. IC50 is quantified as the concentration of cODNs at which 50% of enzyme activities are inhibited. As shown in Table 1, among cODNs without particular designed mismatch regions, the most efficient inhibitor is 66 bp of cODN-1 and the IC50 value is 36.4 nM which is about 10 folds lower than that of its linear counterpart (ODN-1-L, 396.2 nM). In addition, the IC50 values of cODNs increase with the increasing of ring size. The observation suggested that the smaller DNA circle with highly bending stress can act as better inhibitor in relaxation reaction. On the other hand, the most efficient inhibition is achieved by cODN-2-M7 and the IC50 value is 10 nM which is 8 folds lower than that of cODN-2. The ring size of cODN-2 and cODN-2-M7 is same and the only difference between the two DNA circle is that 7 mismatched base pairs exist in cODN-2-M7. The result indicated that mismatched regions in cODNs can facilitate the increasing of inhibition efficiency. The data shown in Table 1 are consistent with the results in Fig. 2 and 3. In addition, the single-stranded circular (scODN-1) and linear (ODN-1-f) oligonucleotides were used as the inhibitors of bacterial topoisomerase I and IC50 values were also measured (Table 1 and Fig. S2†). The results show that the single-stranded oligonucleotides exhibited high inhibitory effect on the activity of bacterial topoisomerases I no matter which conformation they take. Although single-stranded oligonucleotides showed excellent inhibition efficiency in our in vitro relaxation reaction assays, they may be easily degraded by nucleases or trapped by single-stranded specific binding proteins in vivo.
| Oligonucleotide | IC50 (nM) |
|---|---|
| cODN-1 (66 bp, circular) | 36.4 |
| cODN-2 (86 bp, circular) | 85.7 |
| cODN-3 (106 bp, circular) | 144.2 |
| DC-300 (300 bp, circular) | 281.1 |
| ODN-1-L (66 bp, linear) | 396.2 |
| cODN-2-M1 (circular) | 60.2 |
| cODN-2-M3 (circular) | 33.1 |
| cODN-2-M5 (circular) | 19.2 |
| cODN-2-M7 (circular) | 10.0 |
| scODN-1 (66 nt, single-stranded circular) | 9.3 |
| ODN-1-f (66 nt, single-stranded linear) | 8.8 |
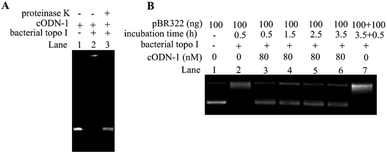
Yang and co-workers reported that linear oligonucleotides containing mismatch or bulge can act as the irreversible inhibitors of bacterial topoisomerase I,19,20 but no direct evidence such as the formation of covalent complex between DNA and proteins was provided in their studies. In order to investigate whether cODNs can cause the irreversible damages to bacterial topoisomerase I, the complex formed between cODN-1 and bacterial topoisomerase I was examined by the denatured PAGE. As shown in Lane 2 in Fig. 4A, the DNA–protein complex was observed. Since the non-covalent interactions between biomolecules can be disrupted in the gel running condition, the result suggests that the complex was formed through covalent interactions between DNA and protein. However, the band of cODN-1 was recovered after proteinase K digestion, which implies that the phosphate backbone of cODN-1 kept integrity during the course of DNA–protein complex formation. With the aim to study whether cODNs can inhibit the activity of bacterial topoisomerases I in a long time period, time-dependent assay of relaxation of pBR322 is performed in the presence of cODN-1. On one hand, in the absence of cODN-1, almost all pBR322 plasmids were relaxed after the reaction time reach 3.5 h. Then an extra 100 ng pBR322 was added and the reaction mixture was further incubated for another 0.5 h. It is shown in Lane 7 in Fig. 4B that the newly added pBR322 was relaxed, which indicated the activity of enzyme still remains in the reaction solution even it was incubated at 37 °C for 3.5 h. On the other hand, as shown in Lane 3–6, no apparent increase of relaxed products can be found when the reaction mixture was incubated up to 3.5 hours. The result suggests that the interaction between cODN-1 and bacterial topoisomerases I is relatively stable and inhibition effect of cODN-1 remains although the prolonged time is set in the reaction. In addition, the control experiment was performed to rule out the possibility that inhibition assays were affected by the bovine serum albumin in reaction buffer. As shown in Fig. S3,† no apparent difference was observed when the plasmid relaxation inhibition assays were performed in the reaction buffer with or without bovine serum albumin.
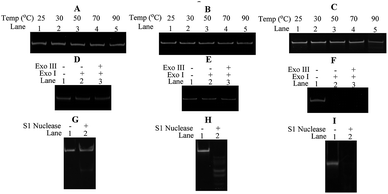
In order to evaluate the thermal stability of ODNs used in our studies, cODN-1, scODN-1 and ODN-1-f were incubated at different temperature in the same buffer condition. As shown in Fig. 5A–C, the DNA circles (cODN-1 and scODN-1) kept integrity and no decomposed product is observed. However, degradation products were found when ODN-1-f (single-stranded oligonucleotide) was heated to 90 °C for 30 minutes. The melting of the linear duplex ODN-1-L is 71.4 °C (based on calculation of OligoAnalyzer provided by IDT) and its duplex structure will be denatured at 90 °C. In addition, the exonuclease and endonuclease sensitivity of the three DNA molecules were also examined. As shown in Fig. 5D–F, cODN-1 and scODN-1 remained while ODN-1-f was completely degraded when those DNA molecules were treated with exonuclease I and exonuclease III. S1 nuclease, a type of endonuclease, was also used to examine the stability of the ODNs in our studies. As shown in Fig. 5G–I, single-stranded oligonucleotides (scODN-1 and ODN-1-f) were completely degraded by the S1 nuclease within 15 minutes. However, only a faint band was observed when cODN-1 was treated with S1 nuclease in the same reaction condition. This happens because the deformation of double helix exists in the backbone of cODN-1 and it can be recognized and cleaved by the S1 nuclease to produce the linear duplex DNA.
Conclusions
In conclusion, a series of cODNs were synthesized and used as the inhibitors of bacterial topoisomerases I. The cODNs showed high inhibitory efficiency and the most efficient one displayed an IC50 value of 10 nM. It is believed that the single-stranded region associated with bending deformation in cODNs is the crucial factor for the binding of bacterial topoisomerases I and the irreversible damages to the enzyme can be generated by forming the covalent cODN–protein complex. In addition, the cODNs showed high thermal stability and excellent nuclease resistance. Although DNA-based biopharmaceuticals have the advantages of low cytotoxicity and can be prepared easily,34,35 the risk of host genome integration should be taken into consideration during the further design of the new type of DNA-based antibacterial agents. In addition, it should be pointed out that the small DNA circles alone can hardly penetrate into bacteria and exhibit bacteria growth inhibition effect in the current stage. However, we believe that it is possible to facilitate the delivery of oligonucleotides into bacteria with the development of the drug-delivery carriers based on nanotechnology and pharmaceutics. It is our hope that the results reported in the current studies could benefit our understanding of the mechanism of type I topoisomerase catalyzed DNA relaxation and provide a new idea for the future design and optimization of DNA based therapeutic agents for antibacterial therapy.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Natural Science Foundation of Jiangsu Province (No. BK20181028), the Startup Foundation (GXL2014038), the Jiangsu Innovative Research Program for Talent from the World’s Famous Universities, and the Priority Academic Program Development (PAPD) program of Jiangsu Province at Nanjing Forestry University. This work was also funded by the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (17KJB150011), the Startup Foundation (JSNU2016YB02) at Jiangsu Second Normal University.Notes and references
- N. Gilbert and J. Allan, Curr. Opin. Genet. Dev., 2014, 25, 15–21 CrossRef CAS PubMed.
- J. J. Champoux, Annu. Rev. Biochem., 2001, 70, 369–413 CrossRef CAS PubMed.
- J. C. Wang, Annu. Rev. Biochem., 1996, 65, 635–692 CrossRef CAS PubMed.
- J. B. Schvartzman, M. L. Martinez-Robles, P. Hernandez and D. B. Krimer, Biochem. Soc. Trans., 2013, 41, 646–651 CrossRef CAS PubMed.
- A. D. Bates and A. Maxwell, DNA Topology, Oxford University Press, Oxford, New York, 2nd edn, 2005 Search PubMed.
- R. Madabhushi, Int. J. Mol. Sci., 2018, 19, 1917 CrossRef PubMed.
- X. Liang, Q. Wu, S. Luan, Z. Yin, C. He, L. Yin, Y. Zou, Z. Yuan, L. Li, X. Song, M. He, C. Lv and W. Zhang, Eur. J. Med. Chem., 2019, 171, 129–168 CrossRef CAS PubMed.
- Y. Pommier, Chem. Rev., 2009, 109, 2894–2902 CrossRef CAS PubMed.
- B. J. Bradbury and M. J. Pucci, Curr. Opin. Pharmacol., 2008, 8, 574–581 CrossRef CAS PubMed.
- S. L. Badshah and A. Ullah, Eur. J. Med. Chem., 2018, 152, 393–400 CrossRef CAS PubMed.
- D. E. Ehmann and S. D. Lahiri, Curr. Opin. Pharmacol., 2014, 18, 76–83 CrossRef CAS PubMed.
- Y. C. Tse-Dinh, Nucleic Acids Res., 2009, 37, 731–737 CrossRef CAS PubMed.
- Y. C. Tse-Dinh, Future Med. Chem., 2015, 7, 459–471 CrossRef CAS PubMed.
- S. Sandhaus, T. Annamalai, G. Welmaker, R. A. Houghten, C. Paz, P. K. Garcia, A. Andres, G. Narula, C. R. Felix, S. Geden, M. Netherton, R. Gupta, K. H. Rohde, M. A. Giulianotti and Y. C. Tse-Dinh, Antimicrob. Agents Chemother., 2016, 60, 4028–4036 CrossRef CAS PubMed.
- C. J. Dorman and C. P. Corcoran, Nucleic Acids Res., 2009, 37, 672–678 CrossRef CAS PubMed.
- Z. Zhang, B. Cheng and Y. C. Tse-Dinh, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 6939–6944 CrossRef CAS PubMed.
- N. M. Baker, R. Rajan and A. Mondragon, Nucleic Acids Res., 2009, 37, 693–701 CrossRef CAS PubMed.
- K. Kirkegaard and J. C. Wang, J. Mol. Biol., 1985, 185, 625–637 CrossRef CAS PubMed.
- Z. Q. Yang, T. Y. Jiang, H. S. Zhong and Y. F. Liu, RSC Adv., 2016, 6, 107572–107576 RSC.
- Z. Q. Yang, T. Y. Jiang, H. S. Zhong and Y. Kang, J. Enzyme Inhib. Med. Chem., 2018, 33, 319 CrossRef CAS PubMed.
- J. Lukes, D. L. Guilbride, J. Votypka, A. Zikova, R. Benne and P. T. Englund, Eukaryotic Cell, 2002, 1, 495–502 CrossRef CAS PubMed.
- L. Saiz and J. M. G. Vilar, Curr. Opin. Struct. Biol., 2006, 16, 344–350 CrossRef CAS PubMed.
- Z. Q. Yang, D. W. Li, S. H. Hiew, M. T. Ng, W. X. Yuan, H. B. Su, F. W. Shao and T. H. Li, Chem. Commun., 2011, 47, 11309–11311 RSC.
- Q. Du, A. Kotlyar and A. Vologodskii, Nucleic Acids Res., 2008, 36, 1120–1128 CrossRef CAS PubMed.
- A. D. Bates, A. Noy, M. M. Piperakis, S. A. Harris and A. Maxwell, Biochem. Soc. Trans., 2013, 41, 565–570 CrossRef CAS PubMed.
- R. Kumar, A. El-Sagheer, J. Tumpane, P. Lincoln, L. M. Wilhelmsson and T. Brown, J. Am. Chem. Soc., 2007, 129, 6859–6864 CrossRef CAS PubMed.
- D. W. Li, B. Lv, Q. Wang, Y. Liu and Z. G. Qiang, Bioorg. Med. Chem. Lett., 2017, 27, 4086–4090 CrossRef CAS PubMed.
- D. Li, Z. Yang, G. Zhao, Y. Long, B. Lv, C. Li, S. Hiew, M. T. T. Ng, J. Guo, H. Tan, H. Zhang and T. Li, Chem. Commun., 2011, 47, 7479–7481 RSC.
- J. P. Laine, P. L. Opresko, F. E. Indig, J. A. Harrigan, C. von Kobbe and V. A. Bohr, Cancer Res., 2003, 63, 7136–7146 CAS.
- M. M. Garner and A. Revzin, Nucleic Acids Res., 1981, 9, 3047–3060 CrossRef CAS PubMed.
- C. Saintome and E. Delagoutte, PLoS One, 2018, 13, e0202138 CrossRef PubMed.
- Q. Wang, R. N. Irobalieva, W. Chiu, M. F. Schmid, J. M. Fogg, L. Zechiedrich and B. M. Pettitt, Nucleic Acids Res., 2017, 45, 7633–7642 CrossRef CAS PubMed.
- E. Damiani, J. A. Solorio, A. P. Doyle and H. M. Wallace, Toxicol. Lett., 2019, 302, 28–34 CrossRef CAS PubMed.
- S. Cansiz, L. Zhang, C. Wu, Y. Wu, I. T. Teng, W. Hou, Y. Wang, S. Wan, R. Cai, C. Jin, Q. Liu and W. Tan, Chem.–Asian J., 2015, 10, 2084–2094 CrossRef CAS PubMed.
- J. Li, L. Mo, C. H. Lu, T. Fu, H. H. Yang and W. Tan, Chem. Soc. Rev., 2016, 45, 1410–1431 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Material and methods, sequences of oligonucleotides and IC50 calculation. See DOI: 10.1039/c9ra02398d |
| This journal is © The Royal Society of Chemistry 2019 |