Open Access Article
Open Access ArticleBifunctional nano-Ag3PO4 with capabilities of enhancing ceftazidime for sterilization and removing residues†
Yahui Zhang‡
ab,
Xiaochen Zhang‡a,
Ruiming Hu*c,
Yang Yanga,
Ping Lia and
Qingsheng Wu *a
*a
aSchool of Chemical Science and Engineering, School of Life Science and Technology, Shanghai Key Laboratory of Chemical Assessment and Sustainability, Tongji University, Shanghai 200092, China. E-mail: qswu@tongji.edu.cn; Tel: +86-21-65982620
bTaiyuan Environmental Science Research Institute, Taiyuan 030002, China
cHuashan Hospital, Fudan University, Shanghai 200040, China. E-mail: ruiminghu7@163.com
First published on 6th June 2019
Abstract
Since the efficacy of antibiotics towards bacteria is decreasing over time, the rising of antibiotic emission has become an increasingly grave issue. In this study, we proposed an integrated antibacterial nanotechnology without pollution residues, which synergistically enhances the antibacterial activity of ceftazidime by using the inorganic nano-Ag3PO4, and subsequently removes drug residues by photocatalysis. Ag3PO4 were synthesized using a simple ion-exchange method without any reducing agent or protectant. The combined antibacterial activity of Ag3PO4 and 22 kinds of antibiotics against Escherichia coli was first studied. The results showed that Ag3PO4 and ceftazidime exhibited the best synergistic effect. Next, the synergy mechanism was proposed, the non-chemical bond forces between Ag3PO4 and ceftazidime was determined by zeta potential analyzer, X-ray photoelectron spectroscopy (XPS) and infrared spectroscopy (IR). The interaction between antimicrobials and bacteria was further demonstrated by surface plasma resonance spectroscopy (SPR), scanning electron microscopy (SEM) and propidium iodide (PI) staining. In addition, the production of reactive oxygen species (ROS), the induction of oxidative stress and dissolution of silver ions in Ag3PO4 were studied and found out that only under light, could the ROS be generated. In conclusion, the synergistic effect of Ag3PO4 and ceftazidime is responsible for the joint destruction of cell wall.
Introduction
Since penicillin was discovered by the British bacteriologist in 1928 for the first time,1 there have been thousands of antibiotics, and hundreds of them are commonly used in the clinical practice. However, due to the resistance of bacteria, many of those antibiotics are no longer effective. However, the development of antibiotics is usually limited by genomic data, technology and innovation.2 At present, antibiotics were mostly abused in agriculture, aquaculture and animal husbandry.3 These antibiotics accumulated in animals, fish and crabs might further transmitted to human by consuming animal food. Excessive antibiotics might also transfer into the water cycle and further threat to public health.4The development of nanotechnology has led people to pay attention to the anti-infective effect of inorganic metal particles.5–7 Here, we proposed a synergistic system of inorganic nanoparticles enhancing the efficacy of traditional organic antibiotics. Since the synergistic antibacterial effect of silver and amoxicillin has been reported in 2005,8 the synergistic effect of nanoparticles and other antibiotics were reported successively.9–12 Ag3PO4 is a promising antibacterial agent,13,14 and more importantly is regarded as a significant breakthrough in the field of visible-light-driven photocatalysts.15,16 Wu et al.17 and Liu et al.18 synthesized Ag3PO4 with different morphologies and studied its antibacterial activity. Other researchers have improved the antibacterial activity of Ag3PO4 by doping graphene.19,20 Huang et al.21 and Gan et al.22 combined CoFe2O4 with Ag3PO4 to improve its photocatalytic property. Chen et al.23 and Yan et al.24 increased the photocatalytic performance of Ag3PO4 by adding In2O3 or In2S3. However, the combined antibacterial activities of Ag3PO4 and antibiotics have not been studied.
In this study, we designed a non-polluting synergistic antibacterial nanotechnology that not only synergistically enhances the efficacy of ceftazidime by Ag3PO4, but also photocatalytically degrades the drug residues. To elucidate the synergistic mechanism, the interaction of Ag3PO4 with ceftazidime was tested, as well as the interaction of antimicrobials (both Ag3PO4 and ceftazidime) with bacteria. This non-polluting antibacterial nanotechnology is expected to benefit human health and the water environment.
Experimental
Materials
Standard strain of Escherichia coli (ATCC 25922) was supplied from Shanghai Tenth People's Hospital. Silver nitrate (AgNO3) and disodium hydrogen phosphate (Na2HPO4) were purchased from Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China). 3-Mercaptopropionic acid (MPA) and N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) were purchased from Aladdin (Shanghai, China). N-Hydroxysuccinimide (NHS), propidium iodide (PI) and 2′,7′-dichlorofluorescin diacetate (DCFH-DA, 97%) were purchased from Sigma-Aldrich (Shanghai, China). Susceptibility paper and Luria–Bertani (LB) culture medium were purchased from Microbial Reagent Co. Ltd. (Hangzhou, China). Ceftazidime (CAZ) powder was purchased from Mengry Bio-Technology Co. Ltd. (Shanghai, China).Preparation of nano-Ag3PO4 and Ag3PO4–CAZ complex
Nano-Ag3PO4 was prepared by ion exchange method.25 10 mL 0.4 M AgNO3 was added into 10 mL 0.15 M Na2HPO4 dropwise. After 30 min reaction, the yellow precipitate was washed with deionized water and ethanol several times, and then dried in vacuum at 60 °C overnight. For synthesis of Ag3PO4–CAZ complex, 100 mg Ag3PO4 was mixed with 100 mg ceftazidime (CAZ) in 4 mL water. After ultrasonic oscillations for 30 min, the mixtures were washed with deionized water for several times, and then freeze drying overnight.Evaluate antibacterial activities by disk diffusion test
Several bacterial colonies were inoculated to 50 mL LB broth and incubate in shaker overnight. Then the bacteria solution was diluted to about 1 × 108 CFU mL−1 (OD600 = 0.1). 100 μL tenfold diluted suspension (107 CFU mL−1) was equably coated on every LB agar plate. The drug sensitive slips (diameter 7 mm) were attached on it. 10 μL Ag3PO4 (10, 100, 1000 mg L−1) solution was added on slip, respectively. After incubated at 37 °C for 20 h, the inhibition zone diameters were measured. All experiments were repeated at least three times to eliminate error.Characterization of the synergistic antibacterial system
X-ray diffraction (XRD) patterns were recorded by a Bruker D8 Advanced X-ray diffractometer with a Cu Kα (1.5 Å) source. Field scanning electron microscopy (SEM) images were obtained with a Hitachi S-4800 SEM. The absorbance (OD) values of bacteria solution were measured by Agilent UV-Vis 8453 Ultraviolet-visible spectrometer. Particles surface charges were analyzed by Malvern Zetasizer Nano ZS90 dynamic light scattering instrument (DLS). The surface elements of the samples were conducted by Kratos AXIS Ultra DLD X-ray Photoelectron Spectroscopy (XPS). Sample functional groups were recorded using Nicolet NEXUS 6700 Fourier transform infrared (FTIR) spectrometer. Fluorescence spectra of the samples were collected by Hitachi F-7000 Fluorescence spectrophotometer.Surface plasmon resonance (SPR) analysis of the interactions between micro/nanoparticles and bacteria
SPR experiments were performed with Reichert SR7500DC dual channel SPR instrument. Specific experimental procedures can be referred to the previous study.26 Briefly, the naked sensor chips were activated by piranha solution, then the carboxyl groups were linked through MPA solution, EDC/NHS solution was used to excite carboxyl terminal, finally Ag3PO4 and Ag3PO4–CAZ were attached. After finished these pretreatment process, a series of bacteria solution (1.25 × 107, 2.5 × 107, 5 × 107, and 1 × 108 CFU mL−1) successively flowed over the sensory chip surface. The equilibrium dissociation constant (KD) was calculated by ClampXP software.SEM analysis to observe cell morphology
The E. coli cells were transferred into 50 mL LB medium with optical density (OD) around 0.1. After incubation in the shaker at 37 °C, 150 rpm, until the OD600 value reached 0.6. The bacteria solution was dispensed into test tubes with 3 mL of each tube. 300 μL Ag3PO4 solution, 300 μL ceftazidime solution were added alone or in combination (with final concentration as 10, 100, 1000 mg L−1), incubated at 37 °C, 200 rpm for 4 h on a shaker bed. The above test tubes were centrifuged at 8000 rpm for 3 min, then collected cells were immobilized by 5 mL 2.5% glutaraldehyde (diluted with PB buffer) for 2 h, subsequently washed with PB three times. The dehydration was accomplished with graded ethanol (30%, 50%, 75%, 85%, 95%, and 100%) for 20 min each. Then the cells were dispersed in 1 mL ethanol, and 20 μL of the cells were dripped onto a silicon wafer. After freeze-drying overnight, cell morphology was observed by SEM.Assays for membrane integrity
The bacteria cells were transferred into 50 mL LB broth until the OD600 value reached 0.1. Then it was incubated in the shaker until the OD600 reached 0.5. The bacteria solution was dispensed into test tubes with 3 mL of each. After added with Ag3PO4, ceftazidime alone or jointly the tubes were incubated at 37 °C, 200 rpm for 3 h, and each sample was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 2 min. Harvested cells were stained with 5 mL 5 μM propidium iodide (PI) in the dark for 20 min. After centrifuged at 10
000 rpm for 2 min. Harvested cells were stained with 5 mL 5 μM propidium iodide (PI) in the dark for 20 min. After centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 2 min, 3 mL phosphate buffered saline (PBS) was used to disperse bacteria. Finally, the samples were analyzed on a spectrofluorometer with excitation wavelength of 535 nm, emission wavelength of 600 nm.
000 rpm for 2 min, 3 mL phosphate buffered saline (PBS) was used to disperse bacteria. Finally, the samples were analyzed on a spectrofluorometer with excitation wavelength of 535 nm, emission wavelength of 600 nm.
Determination of reactive oxygen species (ROS)
To determine the ROS content generated by Ag3PO4, 5,5-dimethyl-1-pyrroline N-oxide (DMPO) was used as the radical trap. Ag3PO4 suspension with DMPO was analyzed by Bruker EMX electron paramagnetic resonance (EPR) spectrometer. To simulate daylight environment, Ag3PO4 suspension was exposed to xenon lamp for 20 min before measurement. Next, intracellular ROS levels were also determined. The bacteria cells were transferred into 50 mL LB broth until the OD600 value reached 0.1. After incubated in the shaker at 37 °C, 150 rpm for 2.5 h, the suspension was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 2 min. Then 60 mL 10 μM 2′,7′-dichlorofluorescein diacetate (DCFH-DA) was used to stain cells for 20 min in the dark. After washed with PBS twice, harvest cells were resuspended in PBS with OD600 about 0.5. Next, the test tubes were added with 3 mL above-mentioned bacteria solution. The samples were then treated with Ag3PO4, ceftazidime alone or jointly for 2 h, at 37 °C, 200 rpm. At last, the samples were analyzed on a spectrofluorometer with an excitation wavelength of 488 nm, emission wavelength of 520 nm.
000 rpm for 2 min. Then 60 mL 10 μM 2′,7′-dichlorofluorescein diacetate (DCFH-DA) was used to stain cells for 20 min in the dark. After washed with PBS twice, harvest cells were resuspended in PBS with OD600 about 0.5. Next, the test tubes were added with 3 mL above-mentioned bacteria solution. The samples were then treated with Ag3PO4, ceftazidime alone or jointly for 2 h, at 37 °C, 200 rpm. At last, the samples were analyzed on a spectrofluorometer with an excitation wavelength of 488 nm, emission wavelength of 520 nm.
Detection of metal ion release
100 mg L−1 Ag3PO4 was mixed with 100 mg L−1 ceftazidime in LB broth. All tubes were incubated at 37 °C, 150 rpm. The samples were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min after 0.5, 2, 6, 12 and 24 h. 1 mL supernatants of each tube were digested with 1 mL HNO3, 1 mL H2O2 for 30 min at 100 °C. After the liquid cooled down, all samples were diluted with 2% HNO3 to 12 mL. The silver ion content was measured by Agilent 720ES inductive-coupled plasma (ICP) optical emission spectrometer.
000 rpm for 10 min after 0.5, 2, 6, 12 and 24 h. 1 mL supernatants of each tube were digested with 1 mL HNO3, 1 mL H2O2 for 30 min at 100 °C. After the liquid cooled down, all samples were diluted with 2% HNO3 to 12 mL. The silver ion content was measured by Agilent 720ES inductive-coupled plasma (ICP) optical emission spectrometer.
Photocatalytic degradation experiments
The prepared ceftazidime solution was added into 25 mL different concentrations of Ag3PO4 aqueous solution. After ultrasonic treatment for 10 min, it was placed in the XPA-Photochemical Reactor (Xujiang Electromechanical Plant, Nanjing, China), stirred in the dark for 30 min to achieve an adsorption equilibrium, and then switched on 500 W xenon lamp to simulate the solar environment (wavelength: 200–2500 nm) for photocatalytic reaction. 3.4 mL of each sample were collected at regular intervals. After centrifugation, these samples were filtered with a 0.22 μm microporous filter and the supernatant was collected. The concentration of ceftazidime was determined by measuring its absorbance at 255 nm with Agilent UV-Vis 8453 Ultraviolet-visible spectrometer.Results and discussion
The cooperative antibacterial effects
Disk diffusion test was used to study the combined antibacterial activity of Ag3PO4 and 22 kinds of antibiotics against E. coli. The results show that Ag3PO4 with gentamicin, tetracycline, minocycline and ceftazidime have better synergistic effect (Table 1). Their maximum inhibition zone increase by 55%, 58%, 52% and 131% respectively. However, when Ag3PO4 combines with other antibiotics, the inhibition zones do not increase significantly, and even the antagonistic effect is observed for ampicillin (−33%) and polymyxin B (−52%). Ag3PO4 shows the best synergistic effect on ceftazidime, so their synergistic mechanism was systematically studied. In addition, when Ag nanoparticles were added, the inhibition zone of ceftazidime remains unchanged, indicating that they could not produce synergistic effect (Table S1†).| Antibiotics (μg per disc) | Inhibition zone diameters (mm) | Increasing rate of inhibition zone area (%) | |||||
|---|---|---|---|---|---|---|---|
| Without Ag3PO4 | Ag3PO4 0.01 μg per disc | Ag3PO4 0.1 μg per disc | Ag3PO4 1 μg per disc | Ag3PO4 0.01 μg per disc | Ag3PO4 0.1 μg per disc | Ag3PO4 1 μg per disc | |
| a All experiments were done in triplicate, and standard deviations were negligible. The inhibition zone diameter of Ag3PO4 alone is 7 mm. | |||||||
| Aminoglycosides | |||||||
| Kanamycin 30 | 21.5 | 20.3 | 18.8 | 21.3 | −11 | −24 | −2 |
| Neomycin 30 | 19.8 | 21.3 | 22.8 | 19.5 | 16 | 33 | −3 |
| Amikacin 30 | 21.5 | 24.5 | 24 | 21.8 | 27 | 25 | 2 |
| Gentamicin 10 | 17.25 | 21.5 | 20.5 | 19.5 | 55 | 41 | 28 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Tetracyclines | |||||||
| Tetracycline 30 | 15.5 | 19.5 | 17 | 17 | 58 | 20 | 20 |
| Minocycline 30 | 14 | 15.3 | 16.5 | 17.3 | 19 | 39 | 52 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Quinolones | |||||||
| Norfloxacin 10 | 26 | 28.8 | 29.3 | 28.3 | 22 | 27 | 18 |
| Ofloxacin 5 | 29.5 | 31 | 31 | 31 | 10 | 10 | 10 |
| Ciprofloxacin 5 | 29 | 29.8 | 31 | 32 | 5 | 14 | 22 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Penicillins | |||||||
| Piperacillin 100 | 23.1 | 24.9 | 25.8 | 27.3 | 16 | 24 | 39 |
| Ampicillin 10 | 11 | 10 | 11 | 9 | −17 | 0 | −33 |
| Carbenicillin 100 | 23.8 | 27.5 | 26.5 | 25 | 34 | 24 | 11 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Cephalosporin | |||||||
| Ceftazidime 30 | 19.25 | 27.25 | 29.25 | 22.5 | 100 | 131 | 37 |
| Cefradine 30 | 16.8 | 19.3 | 18.3 | 17.5 | 32 | 19 | −8 |
| Ceftriaxone 30 | 29.8 | 34.5 | 29.8 | 33 | 34 | 0 | 23 |
| Cefazolin 30 | 25.5 | 25.3 | 25.5 | 27 | −2 | 0 | 12 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Cefuroxime 30 | 20 | 21.5 | 22 | 21.5 | 16 | 21 | 16 |
| Cefoperazone 75 | 27.5 | 31.8 | 28.3 | 29 | 33 | 6 | 11 |
| Cephalexin 30 | 21 | 19.3 | 21.8 | 21 | −16 | 7 | 0 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Others | |||||||
| Chloramphenicol 30 | 23.5 | 27 | 22.25 | 27 | 32 | −10 | 32 |
| Polymyxin B 300 | 16.25 | 16 | 16 | 11.25 | −3 | −3 | −52 |
| Furazolidone 300 | 20.8 | 24 | 22.3 | 22.5 | 34 | 15 | 18 |
Characterization of the antibacterial systems
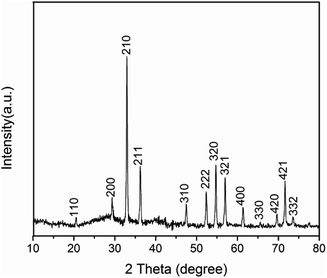
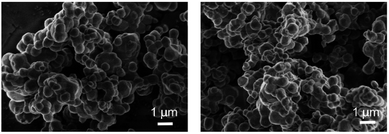
XRD analysis of Ag3PO4 (Fig. 1) reveals a cubic crystal structure (JCPDS no. 06-0505), no other miscellaneous peaks appears, demonstrating the formation of pure Ag3PO4 crystals. As shown in Fig. 2, Ag3PO4 particles have an irregular spherical shape, particle size distribution is between 400 to 800 nm, with an average size of about 650 nm. The mean zeta potential of Ag3PO4 and Ag3PO4–CAZ complexes in distilled water is −40.2 mV and −44.2 mV, the increased number of negative charges is due to COO− on ceftazidime. The surface charge is an important factor in influencing the particles to “see” and react to the cells.27The interaction between antibiotics and inorganic particles
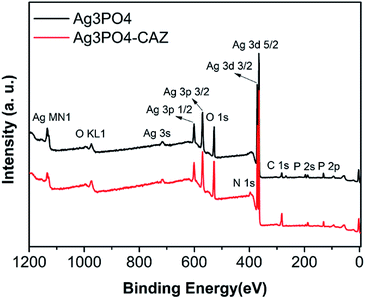
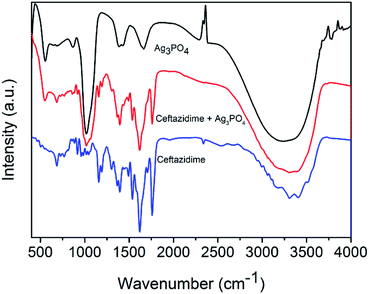
The content of ceftazidime on the surface of Ag3PO4 was analyzed by XPS. The results (Fig. 3) demonstrate the existence of Ag, P, C, O and N elements. Comparing of the signal intensity of N with Ag, the molar ratio of ceftazidime molecules to Ag atoms in Ag3PO4–CAZ is 0.03. Ag3PO4 takes negative charges, so that the amino groups in ceftazidime determine its adsorption capacity on the surface of Ag3PO4. For further investigating their bonding type, the infrared spectra were analyzed. In Fig. 4, the absorption bands at 556 cm−1 and 1015 cm−1 are the characteristic peaks of Ag3PO4 due to the vibrations of PO43− groups. The characteristic peaks of ceftazidime appear at 1759 cm−1 and 1709 cm−1 due to the stretching vibrations of four-membered lactam carbonyl and secondary amide carbonyl groups. The N–H bending vibrations are observed at 1620 cm−1. The band at 1531 cm−1 is due to torsional vibrations of aromatic ring. Compared with the single infrared spectra, there are no obvious changes in characteristic peaks of Ag3PO4–CAZ spectrum, indicating that the combination of Ag3PO4 and ceftazidime is a physical effect and no chemical bond is formed or dissociated, thus no chemical reaction occurs. Therefore, it can be considered that the synergistic effect of Ag3PO4 with ceftazidime is not due to the formation of new compounds.The interaction of antibacterial agents and bacteria
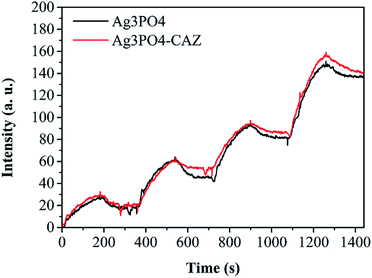
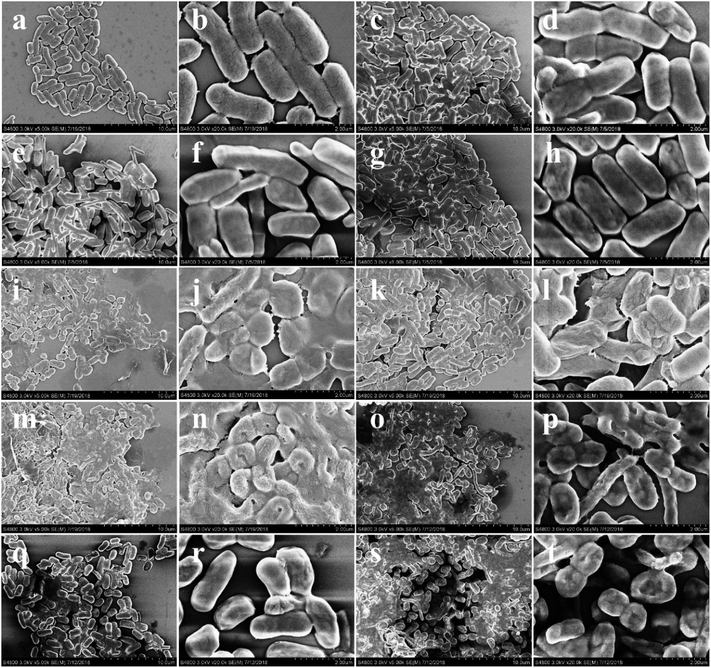
SPR biosensor was used to study how the adsorption of ceftazidime on Ag3PO4 affects the interaction between Ag3PO4 and bacteria. SPR biosensor is commonly used to detect the interaction of biological macromolecules in real time. We extended its function24 to determine the interaction between nanoparticles and organisms dynamically and quantitatively. As shown in Fig. 5, the ascending curves represent the binding process, the faster the curve rises, the faster the combination, and the greater the association constant (Ka). The descending curves indicate dissociation process, the faster the curve drops, the faster the dissociation, and the larger the dissociation constant (Kd). The equilibrium dissociation constant (KD) was calculated by the ratio of Kd to Ka. Smaller KD value indicates stronger affinity and less dissociation, which means a greater interaction force. The KD values of E. coli with Ag3PO4 and Ag3PO4–CAZ are 4.17 mg L−1 and 3.92 mg L−1, suggesting that the interaction of bacteria with Ag3PO4–CAZ is slightly stronger than Ag3PO4, though the Ag3PO4 surface negative charges increase due to its combination with ceftazidime, resulting in the increase of electrostatic repulsion between particles and bacteria. However, because of biochemical reaction between ceftazidime and trans-peptidase in cell wall, the increase of chemical force is more pronounced than electrostatic repulsion.The SEM analysis28,29 was used to study the impact of changes in the interaction between nanoparticles and bacteria on membrane morphology of the bacteria. As shown in Fig. 6d, j and p, while the concentration of Ag3PO4 increased from 10 mg L−1 to 1000 mg L−1, the cell wall damage aggravated, and pits appear in the center of the bacteria. When Ag3PO4 is combined with ceftazidime, cell wall breakage is worse than that of bacteria treated with drugs alone. As seen in Fig. 6j, l and n, when 100 mg L−1 Ag3PO4 combined with 100 mg L−1 ceftazidime, there are obvious holes in the cell wall. Comparing with Fig. 6h, n and t, it is found that as the concentration of the combination drug increases, the pore size on the cell wall gradually increases, and no obvious hole was observed in the bacteria under the action of 10 mg L−1 combined drug, while the cell wall completely collapsed after treated with 1000 mg L−1 combined drug.
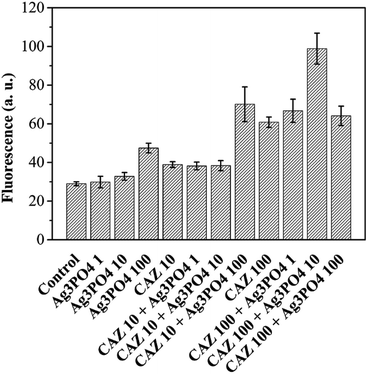
The cell permeability was analyzed by propidium iodide (PI) stain.30,31 As shown in Fig. 7, when 100 mg L−1 CAZ combined with 10 mg L−1 Ag3PO4, cell permeability increases 62.6% than 100 mg L−1 CAZ. Most obviously, when 10 mg L−1 CAZ combined with 100 mg L−1 Ag3PO4, cell permeability increases 80.2% than 10 mg L−1 CAZ. CAZ can inhibit trans-peptidase and interfere with bacterial cell wall synthesis, resulting in increased interaction between Ag3PO4 and bacteria, as well as increased the membrane damage and the cell permeability.
The study of ROS generated by Ag3PO4, intracellular ROS levels and silver ions release
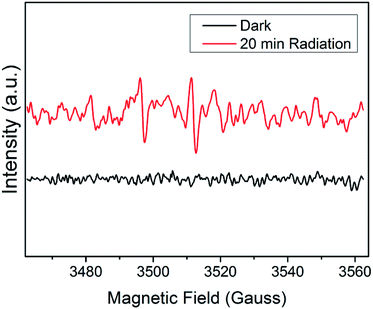
To further investigate the synergistic mechanisms, the ROS generated by Ag3PO4, intracellular ROS levels32,33 and silver ions release34 were tested. EPR is a commonly used method28 for detecting ROS produced by semiconductor materials in a solution environment. Unpaired electrons of different free radicals can be captured by trapping agent DMPO, causing a change in field intensity. As shown in Fig. 8, Ag3PO4 suspension is not able to produce free radical signals in the dark (i.e. antibacterial experimental conditions). However, after irradiation with a 500 W xenon lamp for 20 minutes, a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 hydroxyl radical signal appears. Therefore, it can be concluded that light irradiation can make Ag3PO4 produce ROS, which is beneficial to its subsequent photocatalytic degradation of antibiotic residues. In the antibacterial experiment (in the dark), although Ag3PO4 itself does not produce ROS, it can cause an increase in the intracellular ROS levels. When E. coli cells were treated by 10 mg L−1 and 100 mg L−1 Ag3PO4, intracellular ROS levels increased by 23% and 60% (Fig. 9). Besides, the addition of antibiotics makes no difference to ROS levels.
1 hydroxyl radical signal appears. Therefore, it can be concluded that light irradiation can make Ag3PO4 produce ROS, which is beneficial to its subsequent photocatalytic degradation of antibiotic residues. In the antibacterial experiment (in the dark), although Ag3PO4 itself does not produce ROS, it can cause an increase in the intracellular ROS levels. When E. coli cells were treated by 10 mg L−1 and 100 mg L−1 Ag3PO4, intracellular ROS levels increased by 23% and 60% (Fig. 9). Besides, the addition of antibiotics makes no difference to ROS levels.
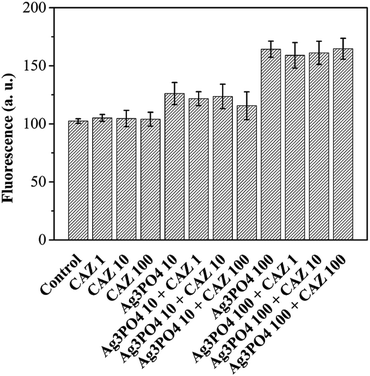 | ||
| Fig. 9 Intracellular reactive oxygen species (ROS) probed by DCFH-DA (concentration units are mg L−1). | ||
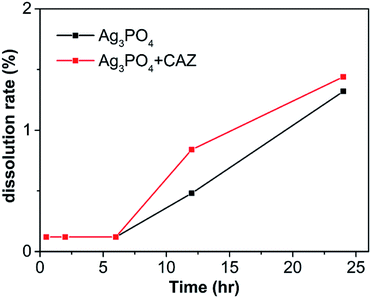
The solubility of Ag3PO4 in LB broth is shown in Fig. 10. The dissolution rate of Ag3PO4 alone increases from 0.12% to 1.32% when soaked from 0.5 h to 24 h. In the presence of ceftazidime, it increases from 0.12% to 1.44%. It is found that the addition of antibiotics has little effect on Ag3PO4 solubility. Therefore, the synergistic mechanisms between them cannot be explained by more silver ions release when antibiotics were added.
Photocatalytic degradation of ceftazidime residues
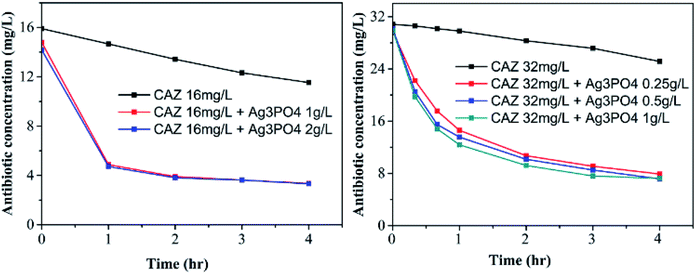
After ceftazidime fulfilled its antibacterial mission, the photocatalytic degradation of CAZ by Ag3PO4 under simulated solar radiation was conducted to eliminate environment pollution. As shown in Fig. 11, after 2 h of simulated sun exposure in presence of 1 g L−1 Ag3PO4, 16 mg L−1 CAZ degrades by 75.6% and 32 mg L−1 CAZ degrades by 71.2%. CAZ degradation slows down between two and four hours. Ag3PO4 has different degradation efficiencies for different types of antibiotics, it could remove about 30% of ciprofloxacin after 2.5 h,35 72% of tetracycline after 1 h,36 87% of sulfamethazine and 73% of cloxacillin after 1.5 h37 under visible light irradiation. Combining the synergistic antibacterial activity with the successive highly photocatalytic degradation of CAZ, the non-polluting nanotechnology for CAZ using nano-Ag3PO4 was established.Conclusions
Ag3PO4 exhibits the best synergistic effect on ceftazidime against E. coli among 22 antibiotics and can further photocatalytically remove the residues. This non-polluting antibacterial nanotechnology is expected to benefit human health and the water environment. XPS and IR tests indicated that ceftazidime binded to the surface of Ag3PO4 by non-chemical bond forces. SPR experiments showed that the adsorption of ceftazidime on the surface of Ag3PO4 produced a stronger interaction between Ag3PO4 and bacteria. SEM and PI dye tests further showed that Ag3PO4 combined with ceftazidime caused more serious damage to cell wall and greater cell permeability (Fig. 12). In addition, Ag3PO4 produced very low levels of ROS and Ag+, so their contributions to antimicrobial activities were almost negligible. The application of nanotechnology in the field of biomedicine is expected to provide strong protection against infection treatment.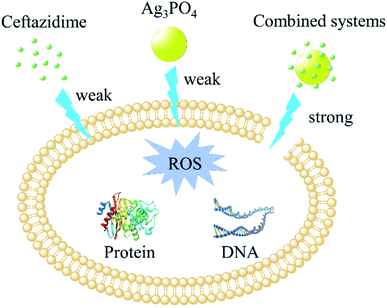 | ||
| Fig. 12 Schematic drawing of the synergistic mechanisms of Ag3PO4 with ceftazidime against Escherichia coli. | ||
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the National Natural Science Foundation of China (no. 51771138, 21471114, 21671132), the State Major Research Plan (973) of China (no. 2011CB932404).References
- A. Fleming, Bull. W. H. O., 2001, 79, 780–790 CAS.
- E. D. Brown and G. D. Wright, Nature, 2016, 529, 336–343 CrossRef CAS.
- R. Laxminarayan, P. Matsoso, S. Pant, C. Brower, J. Rottingen, K. Klugman and S. Davies, Lancet, 2016, 387, 168–175 CrossRef.
- M. Qiao, G. Ying, A. C. Singer and Y. Zhu, Environ. Int., 2018, 110, 160–172 CrossRef CAS.
- S. Singh, R. K. Gundampati, K. Mitra, K. Ramesh, M. V. Jagannadham, N. Misra and B. Ray, RSC Adv., 2015, 5, 81994–82004 RSC.
- L. Wang, C. Hu and L. Shao, Int. J. Nanomed., 2017, 12, 1227–1249 CrossRef CAS PubMed.
- P. K. Mishra, H. Mishra, A. Ekielski, S. Talegaonkar and B. Vaidya, Drug Discovery Today, 2017, 22, 1825–1834 CrossRef CAS PubMed.
- P. Li, J. Li, C. Z. Wu, Q. S. Wu and J. Li, Nanotechnology, 2005, 16, 1912–1917 CrossRef CAS.
- K. Zheng, M. I. Setyawati, T. Lim, D. T. Leong and J. Xie, ACS Nano, 2016, 10, 7934–7942 CrossRef CAS.
- L. Zou, J. Lu, J. Wang, X. Ren, L. Zhang, Y. Gao, M. E. Rottenberg and A. Holmgren, EMBO Mol. Med., 2017, 9, 1165–1178 CrossRef CAS.
- H. Deng, D. McShan, Y. Zhang, S. S. Sinha, Z. Arslan, P. C. Ray and H. Yu, Environ. Sci. Technol., 2016, 50, 8840–8848 CrossRef CAS PubMed.
- P. Prema, S. Thangapandiyan and G. Immanuel, Carbohydr. Polym., 2017, 158, 141–148 CrossRef CAS PubMed.
- J. J. Buckley, A. F. Lee, L. Olivi and K. Wilson, J. Mater. Chem., 2010, 20, 8056–8063 RSC.
- D. Chudobova, L. Nejdl, J. Gumulec, O. Krystofova, M. A. M. Rodrigo, J. Kynicky, B. Ruttkay-Nedecky, P. Kopel, P. Babula, V. Adam and R. Kizek, Int. J. Mol. Sci., 2013, 14, 13592–13614 CrossRef PubMed.
- D. J. Martin, G. Liu, S. J. A. Moniz, Y. Bi, A. M. Beale, J. Ye and J. Tang, Chem. Soc. Rev., 2015, 44, 7808–7828 RSC.
- X. Chen, Y. Dai and X. Wang, J. Alloys Compd., 2015, 649, 910–932 CrossRef CAS.
- A. Wu, C. Tian, W. Chang, Y. Hong, Q. Zhang, Y. Qu and H. Fu, Mater. Res. Bull., 2013, 48, 3043–3048 CrossRef CAS.
- J. Liu, C. Luo, J. Wang, X. Yang and X. Zhong, CrystEngComm, 2012, 14, 8714–8721 RSC.
- X. Xie, C. Mao, X. Liu, L. Tan, Z. Cui, X. Yang, S. Zhu, Z. Li, X. Yuan, Y. Zheng, K. W. K. Yeung, P. K. Chu and S. Wu, ACS Cent. Sci., 2018, 4, 724–738 CrossRef CAS PubMed.
- L. Liu, J. Liu and D. D. Sun, Catal. Sci. Technol., 2012, 2, 2525–2532 RSC.
- S. Huang, Y. Xu, Q. Liu, T. Zhou, Y. Zhao, L. Jing, H. Xu and H. Li, Appl. Catal., B, 2017, 218, 174–185 CrossRef CAS.
- L. Gan, L. Xu and K. Qian, Mater. Des., 2016, 109, 354–360 CrossRef CAS.
- X. Chen, R. Li, X. Pan, X. Huang and Z. Yi, Chem. Eng. J., 2017, 320, 644–652 CrossRef CAS.
- T. Yan, J. Tian, W. Guan, Z. Qiao, W. Li, J. You and B. Huang, Appl. Catal., B, 2017, 202, 84–94 CrossRef CAS.
- Z. Yi, J. Ye, N. Kikugawa, T. Kako, S. Ouyang, H. Stuart-Williams, H. Yang, J. Cao, W. Luo, Z. Li, Y. Liu and R. L. Withers, Nat. Mater., 2010, 9, 559–564 CrossRef CAS.
- Y. Zhang, L. Wang, X. Xu, F. Li and Q. Wu, Nanomedicine, 2018, 13, 339–351 CrossRef CAS.
- K. W. Powers, S. C. Brown, V. B. Krishna, S. C. Wasdo, B. M. Moudgil and S. M. Roberts, Toxicol. Sci., 2006, 90, 296–303 CrossRef CAS PubMed.
- Y. H. Leung, A. M. C. Ng, X. Xu, Z. Shen, L. A. Gethings, M. T. Wong, C. M. N. Chan, M. Y. Guo, Y. H. Ng, A. B. Djurisic, P. K. H. Lee, W. K. Chan, L. H. Yu, D. L. Phillips, A. P. Y. Ma and F. C. C. Leung, Small, 2014, 10, 1171–1183 CrossRef CAS.
- I. N. Ghosh, S. D. Patil, T. K. Sharma, S. K. Srivastava, R. Pathania and N. K. Navani, Int. J. Nanomed., 2013, 8, 4721–4731 Search PubMed.
- Y. Gao, J. Wu, X. Ren, X. Tan, T. Hayat, A. Alsaedi, C. Cheng and C. Chen, Environ. Sci.: Nano, 2017, 4, 1016–1024 RSC.
- J. Ruben Morones-Ramirez, J. A. Winkler, C. S. Spina and J. J. Collins, Sci. Transl. Med., 2013, 5, 190ra81 Search PubMed.
- L. Ma, J. Wu, S. Wang, H. Yang, D. Liang and Z. Lu, J. Inorg. Biochem., 2017, 168, 38–45 CrossRef CAS PubMed.
- J. H. Priester, A. Singhal, B. Wu, G. D. Stucky and P. A. Holden, Analyst, 2014, 139, 954–963 RSC.
- T. Wu, C. Liu, B. Kong, J. Sun, Y. Gong, K. Liu, J. Xie, A. Pei and Y. Cui, ACS Cent. Sci., 2019, 5, 719–726 CrossRef CAS PubMed.
- Y. Liu, Q. Wu and Y. Zhao, Dalton Trans., 2017, 46, 6425–6432 RSC.
- L. Xie, Z. Yang, W. Xiong, Y. Zhou, J. Cao, Y. Peng, X. Li, C. Zhou, R. Xu and Y. Zhang, Appl. Surf. Sci., 2019, 465, 103–115 CrossRef CAS.
- B. Shao, X. Liu, Z. Liu, G. Zeng, Q. Liang, C. Liang, Y. Cheng, W. Zhang, Y. Liu and S. Gong, Chem. Eng. J., 2019, 368, 730–745 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra01969c |
| ‡ Both authors contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |