Open Access Article
Open Access ArticleImportance of the biofilm matrix for the erosion stability of Bacillus subtilis NCIB 3610 biofilms†
M. Klotza,
M. Kretschmerb,
A. Goetza,
S. Ezendama,
O. Lieleg b and
M. Opitz
b and
M. Opitz *a
*a
aCenter for NanoScience, Faculty of Physics, Ludwig-Maximilians-Universität München, Munich, Germany. E-mail: opitz@physik.uni-muenchen.de
bMunich School of BioEngineering and Department of Mechanical Engineering, Technische Universität München, Garching, Germany
First published on 11th April 2019
Abstract
Production and secretion of biomolecules can provide new emergent functionalities to the synthesizing organism. In particular, the secretion of extracellular polymeric substances (EPS) by biofilm forming bacteria creates a biofilm matrix that protects the individual bacteria within the biofilm from external stressors such as antibiotics, chemicals and shear flow. Although the main matrix components of biofilms formed by Bacillus subtilis are known, it remains unclear how these matrix components contribute to the erosion stability of B. subtilis biofilms. Here, we combine different biophysical techniques to assess this relation. In particular, we quantify the importance of specific biofilm matrix components on the erosion behavior of biofilms formed by the well-studied Bacillus subtilis NCIB 3610. We find that the absence of biofilm matrix components decreases the erosion stability of NCIB 3610 biofilms in water, largely by abolishing the hydrophobic surface properties of the biofilm and by reducing the biofilm stiffness. However, the erosion resistance of NCIB 3610 biofilms is strongly increased in the presence of metal ions or the antibiotic ciprofloxacin. In the first case, unspecific ionic cross-linking of biofilm components or individual bacteria seems to be responsible for the observed effect, and in the second case there seems to be an unspecific interaction between the antibiotic and the biofilm matrix. Taken together, our results emphasize the importance of the biofilm matrix to reduce biofilm erosion and give insights into how the specific biomolecules interact with certain chemicals to fulfill this task.
Introduction
The production and secretion of extracellular polymeric substances (EPS) by biofilm forming bacteria provides the biofilm forming community with new emergent properties:1 the bacteria are embedded within these exopolymers (the biofilm matrix), the resulting clusters protect them from the environment, antibiotics2,3 and other chemicals,4 and the structural rigidity provided by the biofilm matrix enables the community to withstand high shear forces or other mechanical stresses5 and governs invasion resistance.6 Although biofilm formation can be beneficial for industrial applications,7,8 biofilms growing on heart valves or medical implants cause serious infections and device failure.9,10 The biofilm matrix can be composed of different exopolymeric substances such as proteins, polysaccharides, DNA or lipids.11–13 One finds an accumulation of multiple substances like eDNA, proteins, lipids, water and sugar within the assembled biofilms. However, not all bacterial cells produce these biopolymers that determine the structure of bacterial biofilms;14 instead, one observes a division of labor of secreting and non-secreting cells and a high physiological heterogeneity in biofilms.15 Although the biofilm matrix composition is understood for many bacterial species,11,12,14,16–18 it is largely unknown how the individual biopolymers contribute to the biofilm properties, e.g. which of them provide the biofilm with its high mechanical stability. Only recently, technical advances in high-resolution optical microscopy19 and scanning electron microscopy allowed the investigation on how specific matrix components affect biofilm structure. For biofilms formed by Escherichia coli it was shown, that cellulose serves as an architectural element and that a network of curli fibers forms the outer biofilm layer.17,20,21 Similarly, the function of specific proteins for cell–cell adhesion and cell encasement within Vibrio cholerae biofilms was described.16,22 However, further studies are needed to understand the role of single matrix components for certain (physical) biofilm properties. In this study, we focus on the soil bacterium Bacillus subtilis that forms biofilms on solid nutrient surfaces in air, or at liquid–air interfaces.23–25 The biofilm matrix of the B. subtilis strain NCIB 3610 used in this work is mainly composed of an exopolysaccharide produced by the gene products of the epsA-O operon26 and an amyloid fiber forming protein TasA.27,28 A second biofilm matrix protein, BslA,29,30 is a self-assembling hydrophobin that is primarily found on the surface of B. subtilis NCIB 3610 biofilms.In the past years many studies have investigated physical properties of B. subtilis biofilms including biofilm erosion,4 biofilm elasticity31,32 and surface properties.31,33–36 However, only recently direct correlations between the biofilm matrix composition and specific biofilm properties could be determined.31,34,35,37 Importantly, it was shown that the surface layer protein BslA is important for the surface stiffness and surface roughness31 of NCIB 3610 biofilms, and this protein also strongly affects the biofilm wettability.29,33,38 Both for B. subtilis B-1 biofilms and biofilms generated by Pseudomonas aeruginosa39 it was shown that multi-valent ions protect the biofilms from erosion. However, if and how these ions interact with biopolymers within the biofilm matrix to provide this stability remains elusive.
Using a set of deletion mutants, we here investigate the importance of selected biofilm matrix components on the erosion stability of B. subtilis NCIB 3610 biofilms. In addition, we show that – similar to biofilms created by strain B-1 – also biofilms generated by strain NCIB 3610 are protected from erosion in the presence of metal ions. In contrast to our expectations, this effect does not stem from interactions between a specific matrix component with these ions, but seems to be due to unspecific cross-linking effects – either between some matrix components or between the bacterial cells themselves. Finally, we report that the antibiotic ciprofloxacin increases the erosion resistance of NCIB 3610 biofilms – an effect that is surprising but appears to be specific rather than generic for antibiotics.
Materials and methods
Strains and growth conditions
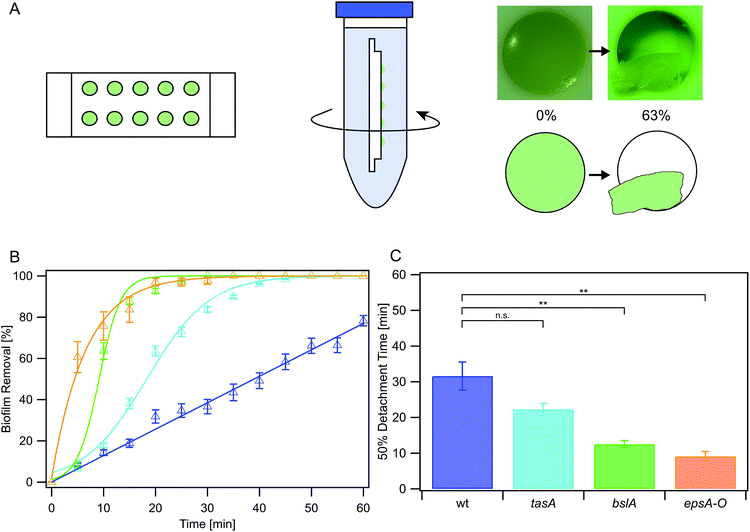
The B. subtilis strains used in this study are NCIB 3610, CA017, ZK3660 and N24, as described in Table 1. LB medium (Luria/Miller; Carl Roth GmbH, Karlsruhe, Germany) served as complex medium for all B. subtilis strains and contained the corresponding antibiotic (Table 1). Bacteria were cultivated overnight in 5 ml medium at 37 °C and 300 rpm agitation. The cultures were then diluted to an OD600 of 0.05 and grown until an OD600 of 0.1 was reached, representing the beginning of the exponential growth phase. The cultures were again diluted to an OD600 of 0.05 for the erosion assay. Sterile polytetrafluorethylene (PTFE) chips (Fig. 1a) containing 10 chambers (i.e., cylindrical holes of 6 mm diameter and 3 mm depth) were filled with hot LB-agar as growth medium for the biofilms. After cooling of the medium 5 μl of the diluted cultures were applied on each agar patch and the chips were then incubated at 37 °C lying flat in sterile Petri dishes without agitation for 24 h for biofilm growth. These chips were then transferred to the prefilled falcon tubes used in the erosion assays as described below.| Strain | Genotype | Remaining main matrix composition | Antibiotic and concentration | Reference |
|---|---|---|---|---|
| a These strains are non-isogenic derivatives of strain NCIB 3610. | ||||
| NCIB 3610 | Wild-type | Proteins TasA & BslA, exopolysaccharide | None | 53 |
| CA017a | tasA::kan | Protein BslA, exopolysaccharide | Kanamycin, 50 μg ml−1 | 28 |
| N24a | bslA::cat | Protein TasA, exopolysaccharide | Chloramphenicol, 5 μg ml−1 | 29 |
| ZK3660a | epsA-O::tet | Protein TasA | Tetracycline, 12.5 μg ml−1 | 54 |
| BD630 | Wild-type | Unable to produce a proper biofilm matrix | None | 55 |
Erosion assay
To conduct the erosion tests, 45 ml of a testing liquid was prepared and filled into falcon tubes. Depending on conditions specific for the different experiments, either double deionized water, ionic solution or antibiotic solution was used as testing liquid. The PTFE chips were placed into the falcon tubes (Fig. 1a) which in turn were mounted onto a lab shaker (Innova 4200, New Brunswick Scientific). The lab shaker was then set into rotation at 300 rpm for defined time intervals, which generated a shear stress of approximately 180 mPa. The shear stress was calculated by measuring the rotation speed inside the falcon tubes and using the appropriate calculation presented in Grumbein et al., 2014.4 After exposure to this shaking induced shear force, images of the biofilm-covered agar patches were acquired (Fig. 1a) using a stereomicroscope (Nikon SMZ1000).Images were then analyzed using the freeware ImageJ (version 1.48) and a graphic tablet (Wacom Intuos Art M) by manually marking the area of each patch that was covered with biofilm for all measured time steps. The detached area fraction was determined by dividing the area covered at each time step with the area covered at the beginning (t = 0 min).
Measurements with antibiotic agents were performed for three different antibiotics at the following concentrations: vancomycin 0.75 μg ml−1 (Van); ciprofloxacin 0.25 μg ml−1 (Cipro); spectinomycin 64 μg ml−1 (Spec). In order for the antibiotic agents to be able to penetrate the biofilm/the cells, the measurements protocols were adapted as follows: a reference image for time t = −15 min was taken. Then the biofilm slides were placed into a falcon tube filled with 45 ml of water (containing the antibiotic in the concentrations described above) for 15 minutes. After that, the slides were taken out of the falcon tubes and an image was taken of all biofilm patches (t = 0 min) and the measurements were then performed as described before.
Rheological characterization
Biofilms were cultivated in Petri dishes on LB-agar for 24 h using similar growth conditions as described above. The two following differences in the growth protocol were applied to obtain large amounts of biofilm as needed for rheology: cultivation overnight was executed in 10 ml LB medium, and 100 μl of this liquid culture was applied and homogeneously distributed on a Petri dish filled with LB-agar. The biofilms were incubated with different salt solutions (50 mM FeCl3, 50 mM CuSO4, 50 mM CaCl2, 250 mM NaCl, or double deionized water as a control) for one hour. For hydrophilic biofilms (epsA-O, bslA), 10 ml of the solutions were poured onto the biofilm; for hydrophobic biofilms (B. subtilis NCIB 3610, tasA) 20 ml were added, so that the liquids were able to cover the whole biofilm. After the incubation the solutions were discarded and the biofilms were harvested from the agar plates by manual scraping. Additionally, reference samples were prepared which were not incubated with liquids at all. Those reference biofilms were harvested directly after the growth period.Rheological measurements were performed using a commercial shear rheometer (MCR 302; Anton Paar GmbH) equipped with a 25 mm plate–plate geometry. The plate separation was set to 0.4 mm for almost all samples; only for the samples incubated with FeCl3 (which exhibited a very high stiffness) a larger plate separation of 0.7 mm had to be used. For a single measurement, biofilm from up to two agar plates had to be pooled to collect enough material to fill the measuring gap. A solvent trap was applied to prevent drying of the samples during the measurement. All measurements were conducted at 21 °C and in strain-controlled mode, and frequency sweeps were recorded in a frequency range of 0.1 Hz to 10 Hz. Small strains corresponding to a constant torque of 0.5 μNm were applied to guarantee linear response. For each condition, three different samples were tested.
Within each rheological experiment, the storage and loss modulus were determined; however, for simplicity (as the loss moduli exhibited the same trends we describe for the storage moduli), only the storage modulus is discussed in the manuscript. Since the recorded moduli showed only a very weak frequency dependence, the obtained storage moduli were averaged over the complete measured frequency spectrum to obtain the bar plots shown in the manuscript. Obvious outliers resulting from measuring artefacts were excluded from calculating these mean values. This averaged storage modulus is referred to as biofilm stiffness (G0) in the manuscript. Error bars denote the standard deviation calculated from the mean values of three frequency sweeps.
Wetting assay
Here, the above described day cultures were diluted to an optical density of 0.05 in a 1.5 ml reaction tube containing 500 μl of LB medium before being spotted onto LB-agar plates. On each plate, five 5 μl spots were applied at equal distance and the plates were incubated at 37 °C for 24 h. Then 10 μl drops of water were applied to the biofilm surface. An image of each droplet on the biofilm surface was acquired with a Canon PowerShot G12 digital camera and the contact angle was determined using the angle tool of ImageJ.Significance analysis
For significance analysis we performed a Wilcoxon rank test using the program Igor7Pro (version 7.04, WaveMetrics) as the samples were not normally distributed. The significance level was chosen as α = 0.05 (**) and plotted in the corresponding figures and ESI Table 2.†Results
Matrix components contribute to NCIB 3610 biofilm erosion stability
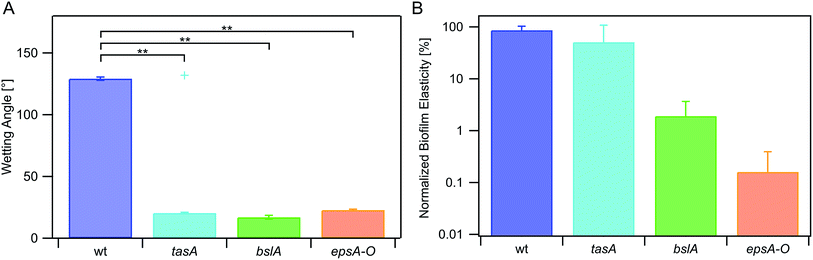
In a first set of experiments, we studied the erosion stability of biofilms formed by the Bacillus subtilis NCIB 3610 wild-type strain exposed to water (Fig. 1a). When challenged by fluid shear induced by shaking, biofilms formed by this strain detached from the agar patches on the PTFE chip within 65 min: the averaged erosion behavior of these wild-type strain biofilms was approximately linear (Fig. 1b, Table S1†), with 50% of the biofilm mass being detached from the agar surface after 32 ± 4 min (Fig. 1c). This was in contrast to submerged control samples investigated in the absence of shear forces, which did not detach (data not shown). In a next step, we investigated the erosion stability of biofilms formed by several mutant strains each lacking one or two components of the biofilm matrix (Fig. 1b). The first deletion mutant, tasA, lacks the amyloid fiber forming protein TasA.27,28 Biofilms formed by this mutant detached from the agar patches in a sigmoidal fashion (Fig. 1b and Table S1†) and faster than the wild-type strain biofilms: here 50% of the biofilm was already eroded after 22 ± 2 min (Fig. 1c). Biofilms generated by a deletion mutant lacking the surface layer protein BslA29,30 also detached in a sigmoidal fashion (Fig. 1b, Table S1†) with 50% of the biofilm mass being eroded within 13 ± 2 min. The lowest erosion stability was observed for biofilms formed by a deletion mutant unable to produce the exopolysaccharide.26 Here, biofilm removal was exponentially growing with time (Fig. 1b, Table S1†), with a 50% detachment value of 9 ± 2 min (Fig. 1c). In this context, it is important to note that expression of the exopolysaccharide is necessary for the production of the BslA protein.29 Consequently, this epsA-O mutant also lacks the BslA protein which might explain why this biofilm variant exhibits the highest sensitivity to erosion. From this data, we conclude that – among the conditions tested here – the absence of either BslA and/or the exopolysaccharide has a strong influence on the erosion stability of the biofilm.To better understand the mechanisms by which the biofilm matrix components contribute to the biofilm erosion stability, we investigated two physical biofilm properties that could be relevant. First, we analyzed if the lack of certain biofilm matrix components would affect the wettability of NCIB 3610 biofilms. This question was motivated by previous results on wild-type biofilms which demonstrated that a reduction of the biofilm surface hydrophobicity can enable faster erosion.40 To assess the biofilm wettability, we determined contact angles: values greater than 90° indicate a hydrophobic surface, while contact angles smaller than 90° represent a hydrophilic surface. We found that the surface of biofilms formed by the wild-type strain was hydrophobic with a contact angle of (129 ± 2)° (Fig. 2a). In contrast, bslA and epsA-O mutant strains formed biofilms with hydrophilic surfaces as demonstrated by measured contact angles smaller than 35° (Fig. 2a). For tasA biofilms, the wetting properties were a bit more complicated. Here, the biofilm initially exhibited large contact angles in the range of (132 ± 2)°; however, the droplet then spread and reached final contact angle values comparable to those obtained for the other mutant strain biofilms.
Overall, this agreed with previous results from the literature29 and demonstrated that the absence of one or two biofilm matrix components strongly alters the wetting behavior of the biofilm, shifting it from hydrophobic to hydrophilic. Moreover, the differences in biofilm erosion stability described above agree well with the differences in biofilm wettability: the hydrophobic wild-type biofilm showed the strongest erosion resistance, the hydrophilic bslA and epsA-O biofilms (which both lack the hydrophobic surface protein BslA) showed the weakest erosion resistance, and the tasA biofilms exhibited both intermediate wetting and intermediate erosion properties.
A second possible biofilm property which could affect the biofilm erosion process is the biofilm bulk stiffness. Although previous experiments31 showed that the stiffness of untreated wild-type biofilm is comparable to that of biofilms formed by the individual mutant strains, this could change after prolonged biofilm exposure to water (as it is the case in our erosion tests) – especially since the different biofilm variants exhibited differences in wettability. Indeed, after incubation with water, biofilms formed by the three mutant strains showed a decreased biofilm stiffness (Fig. 2b): this drop in biofilm stiffness was strong for the epsA-O mutant biofilm, weaker for the bslA mutant biofilms and virtually absent for (hydrophobic) tasA and wild-type biofilms. From this data, we conclude that softening of the biofilm matrix by prolonged exposure to water may also contribute to the erosion behavior of the biofilm – and that this softening effect is directly related to the biofilm wettability.
Metal ions increase the erosion stability of NCIB 3610 biofilms
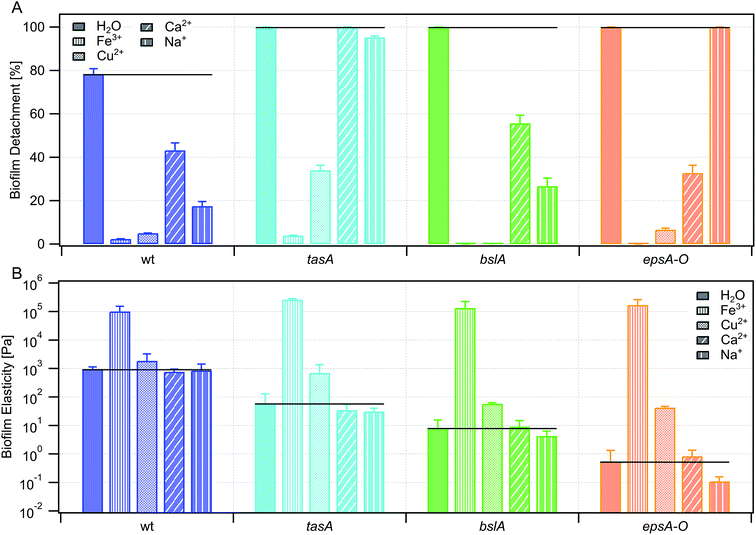
In Grumbein et al., 2014![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 it was shown that selected metal ions increase the erosion stability of Bacillus subtilis B-1 biofilms, whose biofilm matrix is mainly composed of γ-polyglutamate. Consequently, we assessed if metal ions are also able to increase the erosion stability of NCIB 3610 biofilms which – as described above – possess a completely different biofilm matrix. We therefore tested the erosion behavior of NCIB 3610 biofilms in the presence of Fe3+, Cu2+ and Ca2+; those ions were chosen since they have been shown to induce a strong increase in erosion stability of B-1 biofilms.4 In addition, we also tested the influence of the mono-valent ion Na+, which had shown no effect on the erosion behavior of B-1 biofilms.
4 it was shown that selected metal ions increase the erosion stability of Bacillus subtilis B-1 biofilms, whose biofilm matrix is mainly composed of γ-polyglutamate. Consequently, we assessed if metal ions are also able to increase the erosion stability of NCIB 3610 biofilms which – as described above – possess a completely different biofilm matrix. We therefore tested the erosion behavior of NCIB 3610 biofilms in the presence of Fe3+, Cu2+ and Ca2+; those ions were chosen since they have been shown to induce a strong increase in erosion stability of B-1 biofilms.4 In addition, we also tested the influence of the mono-valent ion Na+, which had shown no effect on the erosion behavior of B-1 biofilms.
Indeed, we found that all three multi-valent ions Fe3+, Cu2+, and Ca2+ strongly decreased erosion of NCIB 3610 biofilms (Fig. 3a); in case of the former two, biofilm erosion was almost totally inhibited. However, biofilm erosion was also decreased in the presence of the mono-valent ion Na+ (Fig. 3a). In a set of control experiments, we ensured that the pH of the ion solutions alone was not responsible for the observed increase in erosion stability (Fig. 4). Hence, we believe that the presence of the metal ions in our experiments is the main reason for the decreased biofilm erosion of the tested biofilms. Interestingly, the effect of metal ions to decrease biofilm erosion is not exclusively due to an increase in biofilm stiffness (Fig. 3b), although we did find an effect on both properties for Fe3+ and – albeit weaker – for Cu2+.
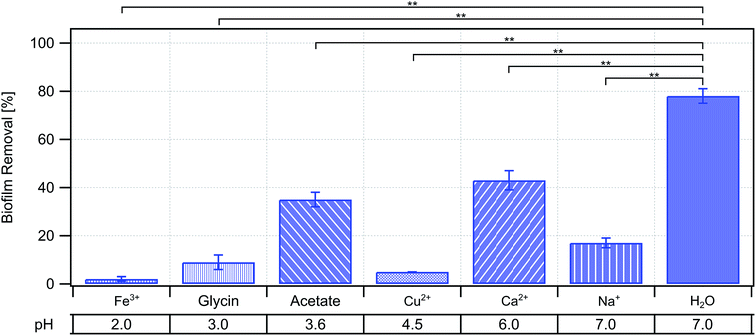 | ||
| Fig. 4 Influence of pH on biofilm erosion. The pH of each tested ionic solution is given underneath the corresponding biofilm removal data of the wild-type strain. | ||
Interaction of metal ions with biofilm matrix components
In a next set of experiments, we assessed the question if the tested metal ions interacted with specific components of the biofilm matrix, and if such an interaction was responsible for the observed increase in biofilm erosion stability. To do so, we repeated the erosion experiments with the ion solutions, but this time we applied the ion solutions to biofilms created by the mutant strains introduced above. We found that, in the presence of Fe3+, biofilm erosion was almost completely inhibited for all mutant strains tested (Fig. 3a). At the same time, biofilm stiffness was increased to similar levels for all mutant strain biofilms as for the wild-type biofilm (Fig. 3b). Interestingly, a bacterial colony formed by a B. subtilis strain unable to produce any biofilm matrix (ESI Fig. 1†) was also completely protected from erosion in the presence of Fe3+. From those experiments, we conclude that the Fe3+ ions do not interact with a single specific biofilm matrix component. Instead, the observed increase in biofilm stiffness and erosion resistance seems to be due to unspecific ionic cross-linking effects. Of course, the source of this unspecific cross-linking effect is difficult to pinpoint as it could occur either between different matrix components, between matrix components and biofilm bacteria, or between individual bacterial cells themselves and/or the agar substrate.In the presence of Cu2+, we obtained a similar picture as for Fe3+: biofilm erosion was decreased for all mutant strains; however, this effect was not always as strong as for the wild-type biofilm (Fig. 3a). As for Fe3+, also for Cu2+, we detected an increase in biofilm stiffness for all biofilm variants – albeit less pronounced than for Fe3+. Taken together, this seemed to suggest that erosion sensitivity and biofilm stiffness are directly related for B. subtilis NCIB 3610 biofilms, and both can be affected by unspecific cross-linking effects induced by multi-valent ions.
Experiments conducted with solutions containing either Ca2+ or Na+, challenged this picture. Here, we did not find combinations of ions and biofilms where the biofilm stiffness was increased compared to the control performed with pure water. Nevertheless, there were conditions where biofilm erosion was significantly decreased – even for Na+ (Fig. 3).
The antibiotic ciprofloxacin increases erosion stability of NCIB 3610 biofilms
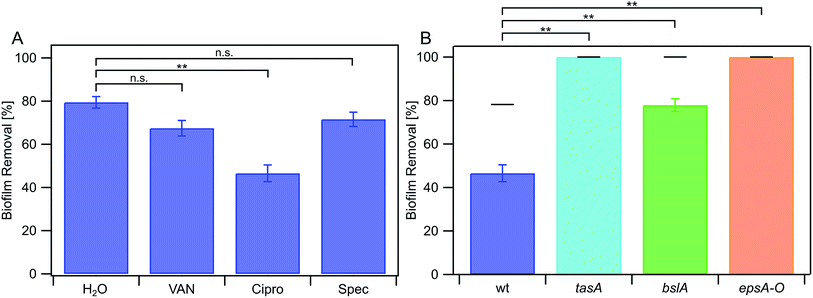
In a last set of experiments, we investigated if specific antibiotics also affect the erosion stability of NCIB 3610 biofilms. Antibiotics represent an important medical treatment to counteract bacterial growth and are often applied in an aqueous environment. We tested three different antibiotics that differ in their mode of action: (i) vancomycin, which inhibits the production of the bacterial cell membrane41 (ii) ciprofloxacin, that inhibits DNA replication42 and (iii) spectinomycin, that inhibits protein synthesis.43 All antibiotics where applied at concentrations that are above the individual minimal inhibitory concentration (MIC) proven to have an effect on single bacterial cells (ESI Fig. 2†). We find that biofilm erosion of the wild-type strain NCIB 3610 is not significantly affected in the presence of vancomycin and spectinomycin (Fig. 5a). The presence of ciprofloxacin, however, increased the erosion stability of NCIB 3610 biofilms (Fig. 5a). Surprisingly, the presence of this antibiotic lead to a small decrease in biofilm stiffness (ESI Fig. 3†), which is the opposite trend as we described above for the metal ions. When we studied the erosion stability of the mutant strain biofilms lacking one or two biofilm matrix components, and found that in the absence of one or two biofilm matrix components the protective effect of ciprofloxacin was strongly reduced (Fig. 5b).Discussion
In this study, we presented a quantitative time-resolved analysis on the erosion stability of Bacillus subtilis NCIB 3610 biofilms. In particular, we demonstrated the importance of the biofilm matrix in this regard and showed that the erosion kinetics differ for all strains studied ranging from linear erosion observed for the wild-type strain, over sigmoidal erosion kinetics for strains lacking the TasA protein in the biofilm matrix, to exponential erosion kinetics for the strain not able to produce the exopolysaccharide. Linear erosion kinetics were previously also found for a different Bacillus subtilis variant, i.e. B. subtilis B-1,4 whose biofilm matrix is mainly composed of γ-polyglutamate.7Different molecular factors might be important for the erosion stability of bacterial biofilms44–46 such as biofilm composition or the distribution of molecules throughout the extracellular matrix. In this study, we addressed two physical biofilm properties influencing biofilm erosion. We showed that B. subtilis NCIB 3610 biofilms with a hydrophobic biofilm surface, as well as biofilms with a high biofilm stiffness exhibit an increased biofilm erosion stability, and that this erosion stability is strongly reduced if one or two biofilm matrix components are missing in the biofilm matrix. Thereby, the TasA protein seems to be of minor importance, while the surface layer protein BslA and the exopolysaccharide have a strong impact. However, the presence of all biofilm matrix components is necessary to achieve the full erosion stability of NCIB 3610 biofilms. It appears reasonable that a less hydrophobic biofilm would be more susceptible to softening induced by water ingress, and that a softer biofilm can be removed from a surface by shear forces more easily than a stiff biofilm variant. However, how the different biofilm matrix molecules work together to fine-tune both of those properties at the same time, cannot be disentangled at this point.
In our study, we also investigated the importance of certain chemicals on the erosion stability of Bacillus subtilis NCIB 3610 biofilms. First, we addressed the presence of metal ions on biofilm erosion stability. Both, mono- and multi-valent metal ions are present in many fluid environments such as the human blood,47 or in pipes and tubes of the food industry.48 In contrast to biofilms formed by the Bacillus subtilis strain B-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 that were protected from erosion by multi-valent ions only, we here found that the erosion properties of biofilms formed by strain NCIB 3610 can also be modulated by the mono-valent ion Na+. At this point, we attribute this difference in biofilm response to differences in the composition of the biofilm matrix of these strains.7,14 Similar to previous results obtained for B. subtilis B-1 biofilms,4 here we also detected a stiffening of the biofilm upon exposure to Fe3+ and Cu2+ ions. However, specific interactions between a certain biofilm matrix component and these metal ions seem not to be required to obtain this effect. This finding appears to fit into a broader picture,49,50 as an effect of selected metal ions on the erosion stability and/or stiffness was also described for Pseudomonas aeruginosa39,51 (where the main biofilm component is alginate) and Staphylococcus epidermidis biofilms.52 This suggests that protecting themselves from erosion by absorbing metal ions from the liquid environment might be a more generic principle that biofilms developed to optimize their survival in the presence of mechanical shear forces, and that different bacterial species make use of a distinct set of matrix molecules to achieve this effect.
4 that were protected from erosion by multi-valent ions only, we here found that the erosion properties of biofilms formed by strain NCIB 3610 can also be modulated by the mono-valent ion Na+. At this point, we attribute this difference in biofilm response to differences in the composition of the biofilm matrix of these strains.7,14 Similar to previous results obtained for B. subtilis B-1 biofilms,4 here we also detected a stiffening of the biofilm upon exposure to Fe3+ and Cu2+ ions. However, specific interactions between a certain biofilm matrix component and these metal ions seem not to be required to obtain this effect. This finding appears to fit into a broader picture,49,50 as an effect of selected metal ions on the erosion stability and/or stiffness was also described for Pseudomonas aeruginosa39,51 (where the main biofilm component is alginate) and Staphylococcus epidermidis biofilms.52 This suggests that protecting themselves from erosion by absorbing metal ions from the liquid environment might be a more generic principle that biofilms developed to optimize their survival in the presence of mechanical shear forces, and that different bacterial species make use of a distinct set of matrix molecules to achieve this effect.
Second, we investigated the effect of selected antibiotics on the erosion stability of biofilms. While the antibiotics vancomycin and spectinomycin had no effect, the presence of the antibiotic ciprofloxacin increased erosion stability of mature NCIB 3610 biofilms, that is attributed to an unspecific interaction with the biofilm matrix. However, biofilm stiffness was not considerably altered by the antibiotic ciprofloxacin, so we speculate the increase in biofilm erosion stability being due to an increase in biofilm adhesion. An effect of certain antibiotics on biofilm stiffness was also described for S. epidermidis and P. aeruginosa.49,50,52 In the latter case, the antibiotic ciprofloxacin decreased biofilm stiffness. To our knowledge, no information is yet available on how these different types of chemicals (ions or antibiotics) interact with the single biofilm matrix components to achieve the effects described above.
In summary, we have shown that the biofilm matrix composition is important for the erosion stability of NCIB 3610 biofilms and that this stability can be altered by certain chemicals such as mono- and multi-valent ions or the antibiotic ciprofloxacin.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank R. Kolter for Bacillus subtilis strains NCIB 3610, CA017 and ZK3660 and K. Kobayashi for strain N24. For technical assistance, we thank L. Kern, B. v. Bronk, M. Jung and N. Schäffler. This study was supported by the Deutsche Forschungsgemeinschaft through project B11 in the framework of SFB863. Additional financial support by the Center for Nanoscience (CeNS) is gratefully acknowledged.References
- H. C. Flemming, J. Wingender, U. Szewzyk, P. Steinberg, S. A. Rice and S. Kjelleberg, Biofilms: an emergent form of bacterial life, Nat. Rev. Microbiol., 2016, 14(9), 563 CrossRef CAS PubMed.
- P. S. Stewart, Mechanisms of antibiotic resistance in bacterial biofilms, Int. J. Med. Microbiol., 2002, 292, 107–113 CrossRef CAS PubMed.
- N. Høiby, T. Bjarnsholt, M. Givskov, S. Molin and O. Ciofu, Antibiotic resistance of bacterial biofilms, Int. J. Antimicrob. Agents, 2010, 35, 322–332 CrossRef PubMed.
- S. Grumbein, M. Opitz and O. Lieleg, Selected metal ions protect Bacillus subtilis biofilms from erosion, Metallomics, 2014, 6, 1441–1450 RSC.
- M. Irsfeld, B. M. Prüβ and S. J. Stafslien, Screening the mechanical stability of Escherichia coli biofilms through exposure to external, hydrodynamic shear forces, J. Basic Microbiol., 2014, 54, 1403–1409 CrossRef CAS PubMed.
- C. D. Nadell, K. Drescher, N. S. Wingreen and B. L. Bassler, Extracellular matrix structure governs invasion resistance in bacterial biofilms, ISME J., 2015, 9, 1700–1709 CrossRef PubMed.
- M. Morikawa, S. Kagihiro, M. Haruki, K. Takano, S. Branda, R. Kolter and S. Kanaya, Biofilm formation by a Bacillus subtilis strain that produces γ-polyglutamate, Microbiology, 2006, 152, 2801–2807 CrossRef CAS PubMed.
- T. L. Wood, R. Guha, L. Tang, M. Geitner, M. Kumar and T. K. Wood, Living biofouling-resistant membranes as a model for the beneficial use of engineered biofilms, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E2802–E2811 CrossRef CAS PubMed.
- L. Hall-Stoodley, J. W. Costerton and P. Stoodley, Bacterial biofilms: from the natural environment to infectious diseases, Nat. Rev. Microbiol., 2004, 2, 95–108 CrossRef CAS PubMed.
- P. Gupta, S. Sarkar, B. Das, S. Bhattacharjee and P. Tribedi, Biofilm, pathogenesis and prevention—a journey to break the wall: a review, Arch. Microbiol., 2016, 198, 1–15 CrossRef CAS PubMed.
- Q. Wei and L. Z. Ma, Biofilm matrix and its regulation in Pseudomonas aeruginosa, Int. J. Mol. Sci., 2013, 14, 20983–21005 CrossRef PubMed.
- M. Marvasi, P. T. Visscher and L. Casillas Martinez, Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis, FEMS Microbiol. Lett., 2010, 313, 1–9 CrossRef CAS PubMed.
- H. C. Flemming and J. Wingender, The biofilm matrix, Nat. Rev. Microbiol., 2010, 8, 623–633 CrossRef CAS PubMed.
- L. Hobley, C. Harkins, C. E. MacPhee and N. R. Stanley-Wall, Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes, FEMS Microbiol. Rev., 2015, 39, 649–669 CrossRef CAS PubMed.
- P. S. Stewart and M. J. Franklin, Physiological heterogeneity in biofilms, Nat. Rev. Microbiol., 2008, 6, 199–210 CrossRef CAS PubMed.
- V. Berk, J. C. N. Fong, G. T. Dempsey, O. N. Develioglu, X. Zhuang, J. Liphardt, F. H. Yildiz and S. Chu, Molecular architecture and assembly principles of Vibrio cholerae biofilms, Science, 2012, 337, 236–239 CrossRef CAS PubMed.
- C. Hung, Y. Zhou, J. S. Pinkner, K. W. Dodson, J. R. Crowley, J. Heuser, M. R. Chapman, M. Hadjifrangiskou, J. P. Henderson and S. J. Hultgren, Escherichia coli Biofilms Have an Organized and Complex Extracellular Matrix Structure, mBio, 2013, 4, 1–10 CrossRef.
- J. Valle, C. Latasa, C. Gil, A. Toledo-Arana, C. Solano, J. R. Penadés and I. Lasa, Bap, a Biofilm Matrix Protein of Staphylococcus aureus Prevents Cellular Internalization through Binding to GP96 Host Receptor, PLoS Pathog., 2012, 8(8), e1002843 CrossRef CAS PubMed.
- K. Drescher, J. Dunkel, C. D. Nadell, S. van Teeffelen, I. Grnja, N. S. Wingreen, H. A. Stone and B. L. Bassler, Architectural transitions in Vibrio cholerae biofilms at single-cell resolution, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E2066–E2072 CrossRef CAS PubMed.
- D. O. Serra, A. M. Richter and R. Hengge, Cellulose as an architectural element in spatially structured Escherichia coli biofilms, J. Bacteriol., 2013, 195, 5540–5554 CrossRef CAS PubMed.
- D. O. Serra, A. M. Richter, G. Klauck, F. Mika and R. Hengge, Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm, mBio, 2013, 4, 1–12 CrossRef PubMed.
- J. K. Teschler, D. Zamorano-Sánchez, A. S. Utada, C. J. A. Warner, G. C. L. Wong, R. G. Linington and F. H. Yildiz, Living in the matrix: assembly and control of Vibrio cholerae biofilms, Nat. Rev. Microbiol., 2015, 13, 255–268 CrossRef CAS PubMed.
- H. Vlamakis, Y. Chai, P. Beauregard, R. Losick and R. Kolter, Sticking together: Building a biofilm the Bacillus subtilis way, Nat. Rev. Microbiol., 2013, 11, 157–168 CrossRef CAS PubMed.
- L. S. Cairns, L. Hobley and N. R. Stanley-Wall, Biofilm formation by Bacillus subtilis: new insights into regulatory strategies and assembly mechanisms, Mol. Microbiol., 2014, 93, 587–598 CrossRef CAS PubMed.
- R. Gallegos-Monterrosa, E. Mhatre and Á. T. Kovács, Specific Bacillus subtilis 168 variants form biofilms on nutrient-rich medium, Microbiology, 2016, 162, 1922–1932 CrossRef CAS PubMed.
- S. S. Branda, F. Chu, D. B. Kearns, R. Losick and R. Kolter, A major protein component of the Bacillus subtilis biofilm matrix, Mol. Microbiol., 2006, 59, 1229–1238 CrossRef CAS PubMed.
- D. Romero, C. Aguilar, R. Losick and R. Kolter, Amyloid fibers provide structural integrity to Bacillus subtilis biofilms, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 2230–2234 CrossRef CAS PubMed.
- H. Vlamakis, C. Aguilar, R. Losick and R. Kolter, Control of cell fate by the formation of an architecturally complex bacterial community, Genes Dev., 2008, 22, 945–953 CrossRef CAS PubMed.
- K. Kobayashi and M. Iwano, BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms, Mol. Microbiol., 2012, 85, 51–66 CrossRef CAS PubMed.
- L. Hobley, A. Ostrowski, F. V. Rao, K. M. Bromley, M. Porter, A. R. Prescott, C. E. MacPhee, D. M. F. van Aalten and N. R. Stanley-Wall, BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 13600–13605 CrossRef CAS PubMed.
- S. Kesel, S. Grumbein, I. Gümperlein, M. Tallawi, A. K. Marel, O. Lieleg and M. Opitz, Direct comparison of physical properties of Bacillus subtilis NCIB 3610 and B-1 biofilms, Appl. Environ. Microbiol., 2016, 82, 2424–2432 CrossRef CAS PubMed.
- S. Grumbein, D. Minev, M. Tallawi, K. Boettcher, F. Prade, F. Pfeiffer, C. U. Grosse and O. Lieleg, Hydrophobic Properties of Biofilm-Enriched Hybrid Mortar, Adv. Mater., 2016, 8138–8143 CrossRef CAS PubMed.
- M. Werb, C. F. García, N. C. Bach, S. Grumbein, S. A. Sieber, M. Opitz and O. Lieleg, Surface topology affects wetting behavior of Bacillus subtilis biofilms, npj Biofilms and Microbiomes, 2017, 3(1), 11 CrossRef PubMed.
- M. Asally, M. Kittisopikul, P. Rue, Y. Du, Z. Hu, T. Cagatay, A. B. Robinson, H. Lu, J. Garcia-Ojalvo and G. M. Suel, Localized cell death focuses mechanical forces during 3D patterning in a biofilm, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 18891–18896 CrossRef CAS PubMed.
- W. Zhang, Y. Wang, S. Bougouffa, R. Tian, H. Cao, Y. Li, L. Cai, Y. H. Wong, G. Zhang, G. Zhou, X. Zhang, V. B. Bajic, A. Al-Suwailem and P. Y. Qian, Synchronized dynamics of bacterial niche-specific functions during biofilm development in a cold seep brine pool, Environ. Microbiol., 2015, 17, 4089–4104 CrossRef CAS PubMed.
- A. K. Epstein, B. Pokroy, A. Seminara and J. Aizenberg, Bacterial biofilm shows persistent resistance to liquid wetting and gas penetration, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 995–1000 CrossRef CAS PubMed.
- S. Kesel, B. von Bronk, C. Falcón García, A. Götz, O. Lieleg and M. Opitz, Matrix composition determines the dimensions of Bacillus subtilis NCIB 3610 biofilm colonies grown on LB agar, RSC Adv., 2017, 7, 31886–31898 RSC.
- S. Arnaouteli, A. S. Ferreira, M. Schor, R. J. Morris, K. M. Bromley, J. Jo, K. L. Cortez, T. Sukhodub, A. R. Prescott, L. E. P. Dietrich, C. E. MacPhee and N. R. Stanley-Wall, Bifunctionality of a biofilm matrix protein controlled by redox state, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E6184–E6191 CrossRef CAS PubMed.
- O. Lieleg, M. Caldara, R. Baumgärtel and K. Ribbeck, Mechanical robustness of Pseudomonas aeruginosa biofilms, Soft Matter, 2011, 7, 3307–3314 RSC.
- C. Falcón García, F. Stangl, A. Götz, W. Zhao, S. A. Sieber, M. Opitz and O. Lieleg, Topographical alterations render bacterial biofilms susceptible to chemical and mechanical stress, Biomater. Sci., 2019, 7(1), 220–232 RSC.
- C. Watanakunakorn, Mode of action and in vitro activity of vancomycin, J. Antimicrob. Chemother., 1984, 14, 7–18 CrossRef CAS.
- M. L. Fisher, J. M. Lawrence, I. C. Josty, R. Hopwell, E. E. C. Margerrison and M. E. Cullen, Ciprofloxacin and the fluoroquinolones, Am. J. Med., 1989, 87, 2–8 CrossRef.
- J. Davies, P. Anderson and B. D. Davis, Inhibition of Protein Synthesis by Spectinomycin, Science, 1965, 149, 5–8 CrossRef.
- L. C. Simões, M. Simões and M. J. Vieira, Biofilm interactions between distinct bacterial genera isolated from drinking water, Appl. Environ. Microbiol., 2007, 73, 6192–6200 CrossRef PubMed.
- O. E. Petrova and K. Sauer, Escaping the biofilm in more than one way: desorption, detachment or dispersion, Curr. Opin. Microbiol., 2016, 30, 67–78 CrossRef CAS PubMed.
- H. Jang, R. Rusconi and R. Stocker, Biofilm disruption by an air bubble reveals heterogeneous age-dependent detachment patterns dictated by initial extracellular matrix distribution, npj Biofilms and Microbiomes, 2017, 3, 0–1 CrossRef PubMed.
- G. E. Jackson and M. J. Byrne, Metal Ion Speciation in Blood Plasma: Gallium-67-Citrate and MRJ Contrast Agents, J. Nucl. Med., 1996, 379–386 CAS.
- A. Z. Aris, R. C. Y. Kam, A. P. Lim and S. M. Praveena, Concentration of ions in selected bottled water samples sold in Malaysia, Appl. Water Sci., 2013, 3, 67–75 CrossRef CAS.
- W. L. Jones, M. P. Sutton, L. McKittrick and P. S. Stewart, Chemical and antimicrobial treatments change the viscoelastic properties of bacterial biofilms, Biofouling, 2011, 27, 207–215 CrossRef CAS PubMed.
- M. Tallawi, M. Opitz and O. Lieleg, Modulation of the mechanical properties of bacterial biofilms in response to environmental challenges, Biomater. Sci., 2017, 5, 887–900 RSC.
- V. Körstgens, H. Flemming, J. Wingender and W. Borchard, Influence of calcium ions on the mechanical properties of a model biofilm of mucoid Pseudomonas aeruginosa, Water Sci. Technol., 2001, 43(6), 49–57 CrossRef.
- E. R. Brindle, D. A. Miller and P. S. Stewart, Hydrodynamic deformation and removal of Staphylococcus epidermidis biofilms treated with urea, chlorhexidine, iron chloride, or dispersinB, Biotechnol. Bioeng., 2011, 108, 2968–2977 CrossRef CAS PubMed.
- S. S. Branda, J. E. González-Pastor, S. Ben-Yehuda, R. Losick and R. Kolter, Fruiting body formation by Bacillus subtilis, Proc. Natl. Acad. Sci. U.S.A., 2001, 98, 11621–11626 CrossRef CAS PubMed.
- Y. Chai, P. B. Beauregard, H. Vlamakis, R. Losick and R. Kolter, Galactose Metabolism Plays a Crucial Role in Biofilm Formation by Bacillus subtilis, mBio, 2012, 3(4), e00184-12 CrossRef PubMed.
- M. Albano, J. Hahn and D. Dubnau, Expression of competence genes in Bacillus subtilis, J. Bacteriol., 1987, 169, 3110–3117 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra01955c |
| This journal is © The Royal Society of Chemistry 2019 |