Open Access Article
Open Access ArticleAza-BODIPY probe for selective visualization of cyclooxygenase-2 in cancer cells†
Thitima
Pewklang‡
a,
Kantapat
Chansaenpak‡
b,
Rung-Yi
Lai
a,
Parinya
Noisa
 c and
Anyanee
Kamkaew
c and
Anyanee
Kamkaew
 *a
*a
aSchool of Chemistry, Institute of Science, Suranaree University of Technology, Nakhon Ratchasima, Thailand 30000. E-mail: anyanee@sut.ac.th
bNational Nanotechnology Center, National Science and Technology Development Agency, Thailand Science Park, Pathum Thani, Thailand 12120
cLaboratory of Cell-Based Assays and Innovations, School of Biotechnology, Institute of Agricultural Technology, Suranaree University of Technology, Nakhon Ratchasima, Thailand 30000
First published on 30th April 2019
Abstract
AZB-IMC2 was developed as a COX-2 specific probe that exhibited a brighter fluorescence signal in cancer cells that overexpress COX-2 compared to normal cells. Oxidative stress agent-treated inflamed cell lines inducing high COX-2 levels revealed an enhanced fluorescence signal. Inhibitory studies showed a markedly reduced fluorescence intensity in cancer cells. The results suggested that AZB-IMC2 could be developed as a promising molecular tool for imaging guiding during surgery.
Introduction
Cyclooxygenase (COX), a prostaglandin-endoperoxide synthase, is a family of the two isoenzymes, COX-1 and COX-2, and plays an essential role in the formation of prostanoids.1 Among the isoenzymes, clinical data revealed that COX-2 is overexpressed in all stages of cancer, from the earliest premalignant phase to metastasis2 and high levels of COX-2 were found to be upregulated in various types of cancers, such as pancreatic, colon, gastric, stomach, breast, head/neck carcinoma, or inflammatory lesions.3–11 Therefore, COX-2 has been considered as a promising target for tumor-specific target-based therapeutics.6,12–18 In the past decades, some fluorescence-based COX-2-targeting probes have been developed,19–25 however, COX-2-specific cancer biomarker development is still needed for clinical use.Indomethacin (IMC), a commercially available nonsteroid anti-inflammatory drug (NSAID), exerts tumor-selective diagnosis and therapeutic potency by targeting COX-derived prostaglandin (PG) biosynthesis.26,27 Although IMC is a non-selective COX isoforms inhibitor (COX-1 and COX-2), it becomes more selective after conjugation with fluorescent dyes.6,24,28–31 For example, indomethacin (IMC) was linked to Nile Blue dye using a hexanediamine linker as a NIR fluorescent imaging probe.30 The probe showed Golgi localization in cancer cells that overexpressed COX-2. Moreover, this NIR probe preferentially labeled the tumors in vivo, suggesting potential tool for imaging guided surgery. Another study employed a quinolinium-based moiety conjugated to IMC using click chemistry and the conjugate showed remarkable targeted ability towards cancer cells over normal cells regarding the COX-2 levels.29
In general, dyes that absorb and emit light in NIR region are ideal to minimize interference from cells autofluorescence, which is a major background noise in cell imaging. However, most currently used NIR-emitting dyes in bioimaging, particularly cyanine-based fluorophores, suffer from inherently low chemical and photostability.32–34 These phenomena significantly limit the use of these dyes in long-term optical imaging that requires duration of light exposure in cellular environment. Borondipyrromethenes (BODIPYs) and their derivatives, in comparison, contain boron bridge that introduces rigidity to the system leading to prevention of trans–cis isomerization and twisting, enhancing fluorescent quantum yield and photostability of the dyes.35–38
According to the bivalent ligand approach, it was found that duplication pharmacophores leads to an increase in potency of binding recognition compared to the corresponding monovalent ligand.39–44 Therefore, in this study, we reported bivalent conjugation of indomethacin to an aza-borondipyrromethene (aza-BODIPY) dye, a class of BODIPYs with NIR emission,45 to enhance COX-2 selectivity (AZB-IMC2).
Aza-BODIPY dye was linked to two molecules of indomethacin using azidotriethyleneglycol linker (Scheme 1). AZB-IMC2 has advantages of optical properties with NIR absorption (λmax ∼ 700 nm) and emission (λmax ∼ 730 nm) which can detect cancer cells by fluorescence imaging. NIR fluorescent COX-2 inhibitors are attractive candidates as deep-seated tissue targeted imaging agents. Such compounds are nonradioactive and stable, thus, they can be developed conveniently for clinical imaging agents.
Results and discussion
Synthesis of AZB-IMC2
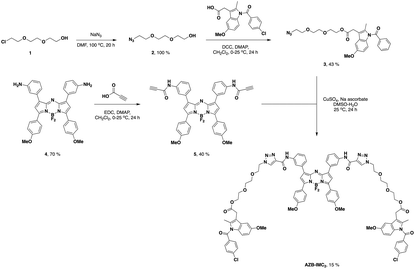
AZB-IMC2 was synthesized according to Scheme 1. Compound 2 was obtained from the reaction of sodium azide with 2-(2-(2-chloroethoxy)ethoxy)ethanol that give quantitative yield.46 After that, 2 was undergone esterification with indomethacin to give compound 3. In another part, amino aza-BODIPY 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 47 was amide coupled with propiolic acid to obtain alkyne substituted aza-BODIPY 5. Finally, the azide–alkyne Huisgen cycloaddition between the azide 3 and the alkyne 5 was performed to give AZB-IMC2 which was used as a COX-2 specific probe for cancer cells imaging.
47 was amide coupled with propiolic acid to obtain alkyne substituted aza-BODIPY 5. Finally, the azide–alkyne Huisgen cycloaddition between the azide 3 and the alkyne 5 was performed to give AZB-IMC2 which was used as a COX-2 specific probe for cancer cells imaging.
Optical properties
AZB-IMC2 showed strong UV-vis-NIR absorption peaking at 704 nm and emitted bright fluorescence peaking at 734 nm in DMSO (Fig. 1). The optical properties were determined under various conditions (DMSO, CHCl3, H2O + 3% Tween-80 and 0.01 M PBS pH 7.4 + 3% Tween-80) however, the absorption and emission maxima did not shift significantly. In water and PBS media, Tween-80, the biocompatible surfactant, was added to increase solubility of the dye (see ESI†). AZB-IMC2 exhibited high quantum yield in organic solvents, whereas low quantum yield was observed in aqueous media (Table 1). This might be the results from hydrophobic nature of the targeting probe leading to aggregation in aqueous medium. As AZB-IMC2 could absorb and emit lights in NIR region, this could be beneficial for deep tissue imaging.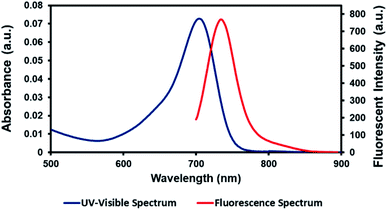 | ||
| Fig. 1 UV-vis-NIR absorption and fluorescent spectra of AZB-IMC2 (2.9 μM) excited at 670 nm in DMSO. | ||
| Solvent | λ max (nm) | ε (M−1 cm−1) | λ emiss a (nm) | Δλ (nm) | Φ f b |
|---|---|---|---|---|---|
| a Samples were excited at 670 nm. b Relative to Zn-phthalocyanine in pyridine (Φf = 0.30). c With 3% Tween-80. | |||||
| DMSO | 704 | 2.5 × 10−4 | 734 | 30 | 0.47 |
| CHCl3 | 694 | 2.5 × 10−4 | 722 | 28 | 0.80 |
| H2Oc | 710 | 1.7 × 10−4 | 725 | 15 | 0.01 |
| PBSc pH 7.4 | 710 | 1.5 × 10−4 | 725 | 15 | 0.05 |
Cell viability
Next, cell viability of AZB-IMC2 were tested in different cell lines including cancer (HeLa, HepG2 and MCF-7) and normal (Hek293, RAW 264.7 and HFF) cells. The cells were treated with various concentrations of AZB-IMC2 ranging from 0–50 μM for 24 h before quantifying cell viability. Fig. 2 showed all the cells remained more than 80% viability at the concentration up to 20 μM while at higher concentration (50 μM) the cell viability reduced to only 50%. This phenomenon might be due to the aggregation of the probe at high concentration. However, the highest concentration we used in the following experiments was 5 μM which all the cells maintained full viability.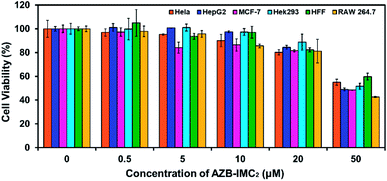 | ||
| Fig. 2 Effect of AZB-IMC2 on cell viability in different cell lines. All cells were treated with AZB-IMC2 at different concentrations for 24 h. | ||
Confocal imaging
COX-2-targeting ability of AZB-IMC2 was then investigated in living cells. We selected 6 different cell lines, cancer cells (HeLa, HepG2 and MCF-7) known to overexpress COX-2 and normal cells with lower expression of COX-2 (HEK293, HFF and RAW 264.7).19,24,25,30,48 After all the cells exposed to AZB-IMC2, increased fluorescent signals were observed when the time increased. Moreover, the brighter fluorescence was detected in cancer cells compared to normal cells and the differences were obviously observed after 6 h incubation (Fig. 3).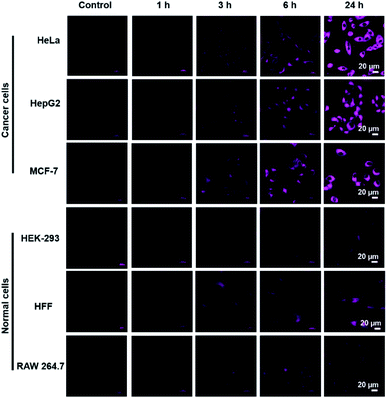 | ||
| Fig. 3 Confocal and fluorescence microscopy images of AZB-IMC2 (5 μM) depend on incubation time (1, 3, 6, and 24 h). | ||
In addition, in cancer cells, the fluorescence was found to be in dose-dependent manner (Fig. 4). We also noticed that 5 μM of AZB-IMC2 was enough for cell internalization and could be used as an indicator to distinguish cancer from normal cell lines.
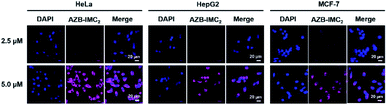 | ||
| Fig. 4 Confocal images of dose-dependent effect of AZB-IMC2 (2.5 and 5.0 μM) in HeLa, HepG2, and MCF-7 cells, which were incubated for 3 h. | ||
Fluorescence images of lipopolysaccharide-induced inflammatory cell lines
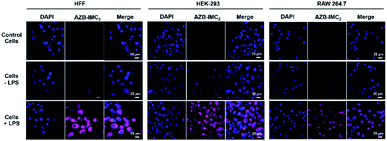
To expand an application of AZB-IMC2 to detect intracellular COX-2 inflamed cells, HEK-293, RAW 264.7 and fibroblast (HFF) cell lines were first induced with LPS (lipopolysaccharides) to upregulate COX-2 levels. After incubation with LPS for 12 h, followed by treatment with AZB-IMC2 for 2 h, an enhanced fluorescence was observed in all cell lines (Fig. 5), indicating that AZB-IMC2 could be used to monitor the oxidative stress in the living system.COX-2 inhibition experiment
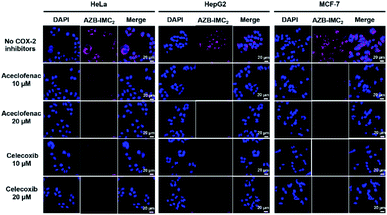
Moreover, to evaluate the selectivity towards cancer cells over normal cells via COX-2 mediated endocytosis upon binding to indomethacin units, the COX-2 blocking experiments were performed. Cancer cells were incubated with 10 μM and 20 μM of potent COX-2 specific inhibitors (aceclofenac and celecoxib)49,50 for 3 h to down-regulate COX-2 levels before adding AZB-IMC2. Fig. 6 shows that all cancer cells (HeLa, HepG2 and MCF-7) incubated with AZB-IMC2 in the absence of COX-2 inhibitors exhibited bright fluorescence, whereas, a significant reduction in the fluorescent intensity was observed when the cells exposed to COX-2 inhibitors prior to AZB-IMC2 treatment. Moreover, the fluorescence intensity decreased with increasing amount of celecoxib and aceclofenac, implying that celecoxib and aceclofenac prevented the labeling of cancer cells by AZB-IMC2. Therefore, the indomethacin-conjugated probe AZB-IMC2 displayed a clear relationship with COX-2 expression levels.Conclusions
In summary, we successfully developed the COX-2 specific probe (AZB-IMC2) for NIR cancer imaging. AZB-IMC2 contained two moieties of indomethacin showing strong absorption and fluorescence in the NIR region (above 700 nm). AZB-IMC2 was not harmful to both cancer (HeLa, HepG2 and MCF-7) and normal (Hek293, RAW 264.7 and HFF) cells at the concentration up to 20 μM. Moreover, as shown in confocal images, AZB-IMC2 exhibited high selectivity towards cancer cells where COX-2 is overexpressed after at least 3 h incubation. In addition, LPS-treated inflamed cell lines (Hek293, RAW 264.7 and HFF) showed enhanced fluorescence as COX-2 level was upregulated. On the other hand, inhibition of COX-2 levels in cancer cells caused significant decreasing of fluorescent signals. These results confirmed that AZB-IMC2 responded to COX-2 expression level of the cells, which can be useful in development of a COX-2-specific cancer biomarker for clinical applications.Experimental
Materials and instruments
All glassware was oven-dried prior to use. All the reagents have been purchased from the commercially sources (Sigma Aldrich, TCI, Carlo Erba, Acros, Merck) and used without further purification. Column chromatography purifications were performed using silica gel for chromatography (Carlo Erba) as a stationary phase. Analytical thin layer chromatography (TLC) was performed on TLC Silica gel 60 F254 (Merck) and visualized with UV cabinet. 1H NMR and 13C NMR spectra were recorded on Bruker-500 MHz spectrometer at room temperature. Chemical shifts of 1H NMR spectra were recorded and reported in ppm from the solvent resonance (CDCl3 at 7.24 ppm). Data are reported as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, m = multiplet), coupling constants, and number of protons. 13C NMR spectra were also recorded in ppm from the solvent resonance (CDCl3 at 77.23 ppm). MS were measured under ESI and MALDI conditions.Synthetic procedures
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (100
MeOH (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 to 90
0 to 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) to yield 0.3062 g (43%) of 3 as a yellow sticky oil. 1H NMR (500 MHz, CDCl3): δ = 7.62 (d, J = 8.5 Hz, 2H), 7.43 (d, J = 8.4 Hz, 2H), 6.94 (d, J = 2.4 Hz, 1H), 6.85 (d, J = 9.0 Hz, 1H), 6.64–6.62 (m, 1H), 4.24 (t, J = 9.5 Hz, 2H), 3.80 (s, 3H), 3.66 (s, 2H), 3.60 (t, J = 10.1 Hz, 2H), 3.56 (s, 4H), 3.32 (t, J = 10.0 Hz, 2H), 2.34 (s, 3H), 2.19 (s, 2H). 13C NMR (500 MHz, CDCl3): δ = 170.9, 168.3, 156.1, 139.3, 136.1, 134.0, 131.2, 131.2, 129.2, 115.0, 112.6, 111.7, 101.5, 70.7, 70.6, 70.1, 69.2, 64.2, 55.8, 50.7, 30.3, 13.5 ppm. MS (ESI+) m/z: the calculated value (calcd) for ([M + Na]+): 537.15, found 537.15.
10) to yield 0.3062 g (43%) of 3 as a yellow sticky oil. 1H NMR (500 MHz, CDCl3): δ = 7.62 (d, J = 8.5 Hz, 2H), 7.43 (d, J = 8.4 Hz, 2H), 6.94 (d, J = 2.4 Hz, 1H), 6.85 (d, J = 9.0 Hz, 1H), 6.64–6.62 (m, 1H), 4.24 (t, J = 9.5 Hz, 2H), 3.80 (s, 3H), 3.66 (s, 2H), 3.60 (t, J = 10.1 Hz, 2H), 3.56 (s, 4H), 3.32 (t, J = 10.0 Hz, 2H), 2.34 (s, 3H), 2.19 (s, 2H). 13C NMR (500 MHz, CDCl3): δ = 170.9, 168.3, 156.1, 139.3, 136.1, 134.0, 131.2, 131.2, 129.2, 115.0, 112.6, 111.7, 101.5, 70.7, 70.6, 70.1, 69.2, 64.2, 55.8, 50.7, 30.3, 13.5 ppm. MS (ESI+) m/z: the calculated value (calcd) for ([M + Na]+): 537.15, found 537.15.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc (1
EtOAc (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) to yield 69 mg (40%) of 5 as a dark green solid. 1H NMR (500 MHz, CDCl3): δ = 8.50 (s, 1H), 8.03–8.01 (m, 2H), 7.90 (d J = 8.7 Hz, 1H), 7.74–7.68 (m, 1H), 7.50 (s, 1H), 7.39–7.37 (m, 1H), 6.96 (d, J = 8.7 Hz, 2H), 6.64 (s, 1H), 3.86 (s, 3H), 2.97 (s, 1H). 13C NMR (500 MHz, CDCl3): δ = 162.3, 161.2, 150.0, 145.4, 142.2, 137.6, 133.5, 132.0, 131.4, 129.5, 126.1, 121.2, 119.4, 114.5, 114.1, 75.4, 74.8, 55.7 ppm. MS (MALDI-TOF) m/z: the calculated value (calcd) for C40H28BF2N5NaO4 ([M + Na]+): 714.21, C40H28BF2KN5O4 ([M + K]+): 730.18, found 714.27, 730.23.
3) to yield 69 mg (40%) of 5 as a dark green solid. 1H NMR (500 MHz, CDCl3): δ = 8.50 (s, 1H), 8.03–8.01 (m, 2H), 7.90 (d J = 8.7 Hz, 1H), 7.74–7.68 (m, 1H), 7.50 (s, 1H), 7.39–7.37 (m, 1H), 6.96 (d, J = 8.7 Hz, 2H), 6.64 (s, 1H), 3.86 (s, 3H), 2.97 (s, 1H). 13C NMR (500 MHz, CDCl3): δ = 162.3, 161.2, 150.0, 145.4, 142.2, 137.6, 133.5, 132.0, 131.4, 129.5, 126.1, 121.2, 119.4, 114.5, 114.1, 75.4, 74.8, 55.7 ppm. MS (MALDI-TOF) m/z: the calculated value (calcd) for C40H28BF2N5NaO4 ([M + Na]+): 714.21, C40H28BF2KN5O4 ([M + K]+): 730.18, found 714.27, 730.23.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc (1
EtOAc (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), followed by DCM
3), followed by DCM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (95
MeOH (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) to yield 20 mg (15%) of AZB-IMC2 as a dark green solid. 1H NMR (500 MHz, CDCl3): δ = 8.97 (s, 1H), 8.35 (s, 1H), 8.29 (s, 1H), 8.08 (d, J = 8.8 Hz, 2H), 7.84 (d, J = 7.8 Hz, 1H), 7.68 (d, J = 7.9 Hz, 2H), 7.59 (s, 1H), 7.57 (s, 1H), 7.45–7.40 (m, 1H), 7.07 (s, 1H), 6.98 (d, J = 8.8 Hz, 2H), 6.92 (s, 1H), 6.81 (d, J = 9.0 Hz, 2H), 6.60 (d, J = 8.9 Hz, 1H), 4.58 (s, 2H), 4.26 (t, J = 4.6 Hz, 2H), 4.05–4.04 (m, 2H), 3.87 (s, 3H), 3.81 (m, 2H), 3.76 (s, 3H), 3.66–3.63 (m, 2H), 3.54–3.52 (m, 2H), 3.45 (t, J = 3.7 Hz, 2H), 2.31 (s, 3H). 13C NMR (500 MHz, CDCl3): δ = 171.2, 168.6, 162.3, 158.5, 158.4, 157.1, 156.3, 143.6, 139.6, 138.2, 136.4, 134.2, 133.6, 132.1, 131.5, 131.0, 129.6, 129.4, 127.4, 125.8, 124.5, 121.1, 120.7, 119.4, 115.2, 114.6, 112.8, 112.0, 111.8, 101.9, 70.9, 70.8, 69.5, 64.4, 56.0, 34.3, 34.3, 30.5, 30.0, 13.7 ppm. MS (MALDI-TOF) m/z: the calculated value (calcd) for C90H83BCl2FN13NaO16 ([M − F + H]+): 1701.55, C90H82BCl2F2N13NaO16 ([M + Na]+): 1742.53, C90H82BCl2F2KN13O16 ([M + K]+): 1758.51, found 1701.83, 1742.87, 1758.84.
5) to yield 20 mg (15%) of AZB-IMC2 as a dark green solid. 1H NMR (500 MHz, CDCl3): δ = 8.97 (s, 1H), 8.35 (s, 1H), 8.29 (s, 1H), 8.08 (d, J = 8.8 Hz, 2H), 7.84 (d, J = 7.8 Hz, 1H), 7.68 (d, J = 7.9 Hz, 2H), 7.59 (s, 1H), 7.57 (s, 1H), 7.45–7.40 (m, 1H), 7.07 (s, 1H), 6.98 (d, J = 8.8 Hz, 2H), 6.92 (s, 1H), 6.81 (d, J = 9.0 Hz, 2H), 6.60 (d, J = 8.9 Hz, 1H), 4.58 (s, 2H), 4.26 (t, J = 4.6 Hz, 2H), 4.05–4.04 (m, 2H), 3.87 (s, 3H), 3.81 (m, 2H), 3.76 (s, 3H), 3.66–3.63 (m, 2H), 3.54–3.52 (m, 2H), 3.45 (t, J = 3.7 Hz, 2H), 2.31 (s, 3H). 13C NMR (500 MHz, CDCl3): δ = 171.2, 168.6, 162.3, 158.5, 158.4, 157.1, 156.3, 143.6, 139.6, 138.2, 136.4, 134.2, 133.6, 132.1, 131.5, 131.0, 129.6, 129.4, 127.4, 125.8, 124.5, 121.1, 120.7, 119.4, 115.2, 114.6, 112.8, 112.0, 111.8, 101.9, 70.9, 70.8, 69.5, 64.4, 56.0, 34.3, 34.3, 30.5, 30.0, 13.7 ppm. MS (MALDI-TOF) m/z: the calculated value (calcd) for C90H83BCl2FN13NaO16 ([M − F + H]+): 1701.55, C90H82BCl2F2N13NaO16 ([M + Na]+): 1742.53, C90H82BCl2F2KN13O16 ([M + K]+): 1758.51, found 1701.83, 1742.87, 1758.84.
UV/vis and fluorescence spectroscopic methods
All the UV/vis absorption spectra and fluorescence were recorded on UV-vis Spectrophotometer (Agilent Technologies Cary 300) and Spectrofluorometer (PerkinElmer LS55), respectively. In both experiments, stock solutions (350 μM) of AZB-IMC2 probe was prepared in DMSO. 10 μM solutions of probes in chloroform, DMSO, DI water, and 0.01 M PBS buffer (pH 7.4) with 3% DMSO were prepared as working solutions. For fluorescence experiments, the emission spectra were recorded at excitation wavelength of 670 nm.Cell culture
Human cervical cancer (HeLa), human hepatoma cell cancer (HepG2), Michigan Cancer Foundation-7 (MCF-7), human embryonic kidney 293 (HEK-293), Human Foreskin Fibroblast (HFF), and RAW264.7 cell lines were cultured on 75 cm3 culture flasks in Dulbecco's Modified Eagle's Media (Hyclone) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin–streptomycin (Corning). All the cells were cultured at 37 °C in a humidified 95% air, 5% CO2 atmosphere.Cell viability assay
The cells were seeded on 96-well plate approximately 7 × 103 cells per well and incubated in completed media for 24 h. Thereafter, the cells were treated with 0, 0.5, 5, 10, 20, 50 μM of AZB-IMC2 and continued culturing for 24 h. After incubation, the cells were washed with 0.01 M PBS (3 times) and treated with 20 μL of methylthiazolyldiphenyl-tetrazolium bromide (MTT reagent, 0.5 mg mL−1, Sigma-Aldrich) for 2 h. After media removal, DMSO was added to dissolve formazan product. The cell's viabilities were detected through UV-vis absorption of formazan at wavelength 560 nm.Time dependent internalization
HeLa, HepG2, MCF-7, HEK-293, HFF and RAW 294.7 cells, approximately 1 × 104 cells, were seeded on 8-well chambered coverglass (LabTek, Nunc) and incubated in completed media for 24 h. After that, the cells were treated with 5 μM of AZB-IMC2 for 0, 1, 3, 6, 24 h. After incubation, the cells were washed with 0.01 M PBS (3 times) and treated with 0.5 μg mL−1 of Hoechst 33342 containing media. The cells were imaged by Laser Scanning Confocal Microscope (Nikon A1Rsi).Dose dependent internalization
HeLa, HepG2, MCF-7 cells, approximately 1 × 104 cells, were seeded on 8-well chambered coverglass and incubated in completed media for 24 h. After that, the cells were treated with 2.5 and 5.0 μM of AZB-IMC2 probe for 3 h. After incubation, the cells were washed with 0.01 M PBS (3 times) and treated with 0.5 μg mL−1 of Hoechst 33342 containing media. The cells were visualized by Laser Scanning Confocal Microscope (Nikon A1Rsi).LPS-induced inflammation experiment
To study inflammation effects, HEK-293, HFF and RAW 294.7 were seeded on 8-well chambered coverglass and incubated for 24 h. After that, the cells were treated with 1 μg mL−1 lipopolysaccharides (LPS, O111:B4, Sigma) in DMEM for 12 h. After incubation in LPS containing media, the cells were washed three times with 0.01 M PBS and treated with 5 μM of AZB-IMC2 for 2 h. Before imaging, the cells were washed three times with 0.01 M PBS and treated with 0.5 μg mL−1 of Hoechst 33342 containing media.COX-2 blocking experiment
To study inhibition effect of COX-2 with blocking, HeLa, HepG2, and MCF-7 cells were incubated with 10 μM and 20 μM of aceclofenac and celecoxib (Tokyo Company Industry) for 3 h before being treated with 5 μM of AZB-IMC2 and incubated for another 3 h. Thereafter, the cells were washed with 0.01 M PBS three times and treated with 0.5 μg mL−1 of Hoechst 33342 containing media before visualization by Laser Scanning Confocal Microscope (Nikon A1Rsi).Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by SUT Research and Development Fund.Notes and references
- F. A. Fitzpatrick, Curr. Pharm. Des., 2004, 10, 577–588 CrossRef CAS.
- K. Subbaramaiah and A. J. Dannenberg, Trends Pharmacol. Sci., 2003, 24, 96–102 CrossRef CAS.
- M. Breinig, P. Schirmacher and M. A. Kern, Curr. Pharm. Des., 2007, 13, 3305–3315 CrossRef CAS.
- K. Echizen, O. Hirose, Y. Maeda and M. Oshima, Cancer Sci., 2016, 107, 391–397 CrossRef CAS.
- M. J. Edelman, L. Hodgson, X. Wang, R. A. Kratzke and E. E. Vokes, J. Clin. Oncol., 2012, 30, 2019–2020 CrossRef CAS.
- H. S. Kim, A. Sharma, W. X. Ren, J. Han and J. S. Kim, Biomaterials, 2018, 185, 63–72 CrossRef CAS.
- M. L. Nasi and M. Castiglione, Ann. Oncol., 2002, 13, 1169–1171 CrossRef CAS.
- S. Nath, L. D. Roy, P. Grover, S. Rao and P. Mukherjee, Pancreas, 2015, 44, 909–917 CrossRef CAS PubMed.
- A. Thiel, J. Mrena and A. Ristimaki, Cancer Metastasis Rev., 2011, 30, 387–395 CrossRef CAS PubMed.
- D. Wang and R. N. Dubois, Oncogene, 2010, 29, 781–788 CrossRef CAS PubMed.
- Z. Khan, N. Khan, R. P. Tiwari, N. K. Sah, G. Prasad and P. S. Bisen, Curr. Drug Targets, 2011, 12, 1082–1093 CrossRef CAS.
- R. de Souza Pereira, Recent Pat. Anticancer Drug Discov., 2009, 4, 157–163 CrossRef.
- E. F. de Vries, Curr. Pharm. Des., 2006, 12, 3847–3856 CrossRef CAS PubMed.
- J. F. Evans and S. L. Kargman, Curr. Pharm. Des., 2004, 10, 627–634 CrossRef CAS PubMed.
- H. R. Herschman, J. J. Talley and R. DuBois, Mol. Imaging Biol., 2003, 5, 286–303 CrossRef.
- L. Milas, Am. J. Clin. Oncol., 2003, 26, S66–S69 Search PubMed.
- K. Chansaenpak, M. Wang, S. Liu, Z. Wu, H. Yuan, P. S. Conti, Z. Li and F. P. Gabbaï, RSC Adv., 2016, 6, 23126–23133 RSC.
- K. Chansaenpak, B. Vabre and F. P. Gabbaï, Chem. Soc. Rev., 2016, 45, 954–971 RSC.
- A. Bhardwaj, J. Kaur, F. Wuest and E. E. Knaus, ChemMedChem, 2014, 9, 109–116 CrossRef CAS PubMed.
- Y. Ti, L. Yu, Y. Tang, T. Jin, M. Yang, R. Wang, Y. Xu and W. Zhu, Sens. Actuators, B, 2018, 265, 582–590 CrossRef CAS.
- M. J. Uddin, B. C. Crews, A. L. Blobaum, P. J. Kingsley, D. L. Gorden, J. O. McIntyre, L. M. Matrisian, K. Subbaramaiah, A. J. Dannenberg, D. W. Piston and L. J. Marnett, Cancer Res., 2010, 70, 3618–3627 CrossRef CAS.
- M. J. Uddin, B. C. Crews, K. Ghebreselasie and L. J. Marnett, Bioconjugate Chem., 2013, 24, 712–723 CrossRef CAS PubMed.
- H. Zhang, J. Fan, J. Wang, B. Dou, F. Zhou, J. Cao, J. Qu, Z. Cao, W. Zhao and X. Peng, J. Am. Chem. Soc., 2013, 135, 17469–17475 CrossRef CAS PubMed.
- H. Zhang, J. Fan, J. Wang, S. Zhang, B. Dou and X. Peng, J. Am. Chem. Soc., 2013, 135, 11663–11669 CrossRef CAS.
- Q. Zhang, Z. Han, J. Tao, W. Zhang, P. Li, L. Tang and Y. Gu, J. Biophotonics, 2018, 11, e201700339 CrossRef PubMed.
- R. J. Flower, Nat. Rev. Drug Discovery, 2003, 2, 179–191 CrossRef CAS.
- M. G. Papich, in Saunders Handbook of Veterinary Drugs, ed. M. G. Papich and W. B. Saunders, St. Louis, 2016, pp. 396–397, DOI:10.1016/b978-0-323-24485-5.00307-7.
- J. H. Jang, H. Lee, A. Sharma, S. M. Lee, T. H. Lee, C. Kang and J. S. Kim, Chem. Commun., 2016, 52, 9965–9968 RSC.
- H. S. Kim, T. Park, W. X. Ren, J. Y. Lim, M. Won, J. S. Heo, S. G. Lee and J. S. Kim, Dyes Pigm., 2018, 150, 261–266 CrossRef CAS.
- B. Wang, J. Fan, X. Wang, H. Zhu, J. Wang, H. Mu and X. Peng, Chem. Commun., 2015, 51, 792–795 RSC.
- H. Zhang, J. Fan, K. Wang, J. Li, C. Wang, Y. Nie, T. Jiang, H. Mu, X. Peng and K. Jiang, Anal. Chem., 2014, 86, 9131–9138 CrossRef CAS PubMed.
- C. Shi, J. B. Wu and D. Pan, Review on near-infrared heptamethine cyanine dyes as theranostic agents for tumor imaging, targeting, and photodynamic therapy, SPIE, 2016 Search PubMed.
- S. Luo, E. Zhang, Y. Su, T. Cheng and C. Shi, Biomaterials, 2011, 32, 7127–7138 CrossRef CAS.
- S. A. Soper and Q. L. Mattingly, J. Am. Chem. Soc., 1994, 116, 3744–3752 CrossRef CAS.
- A. Kamkaew, S. H. Lim, H. B. Lee, L. V. Kiew, L. Y. Chung and K. Burgess, Chem. Soc. Rev., 2013, 42, 77–88 RSC.
- A. Loudet and K. Burgess, Chem. Rev., 2007, 107, 4891–4932 CrossRef CAS PubMed.
- N. Boens, V. Leen and W. Dehaen, Chem. Soc. Rev., 2012, 41, 1130–1172 RSC.
- R. Ziessel, G. Ulrich and A. Harriman, New J. Chem., 2007, 31, 496–501 RSC.
- A. P. Tamiz, J. Zhang, M. Zhang, C. Z. Wang, K. M. Johnson and A. P. Kozikowski, J. Am. Chem. Soc., 2000, 122, 5393–5394 CrossRef CAS.
- M. Rosini, E. Simoni, M. Bartolini, E. Soriano, J. Marco-Contelles, V. Andrisano, B. Monti, M. Windisch, B. Hutter-Paier, D. W. McClymont, I. R. Mellor and M. L. Bolognesi, ChemMedChem, 2013, 8, 1276–1281 CrossRef CAS PubMed.
- I. Raji, F. Yadudu, E. Janeira, S. Fathi, L. Szymczak, J. R. Kornacki, K. Komatsu, J. D. Li, M. Mrksich and A. K. Oyelere, Bioorg. Med. Chem., 2017, 25, 1202–1218 CrossRef CAS PubMed.
- P. S. Portoghese, J. Med. Chem., 2001, 44, 2259–2269 CrossRef CAS PubMed.
- S. Howorka, J. Nam, H. Bayley and D. Kahne, Angew. Chem., Int. Ed. Engl., 2004, 43, 842–846 CrossRef CAS PubMed.
- T. Birnkammer, A. Spickenreither, I. Brunskole, M. Lopuch, N. Kagermeier, G. Bernhardt, S. Dove, R. Seifert, S. Elz and A. Buschauer, J. Med. Chem., 2012, 55, 1147–1160 CrossRef CAS PubMed.
- Y. Ge and D. F. O'Shea, Chem. Soc. Rev., 2016, 45, 3846–3864 RSC.
- Y. R. Baker, W. R. Galloway, J. T. Hodgkinson and D. R. Spring, Molecules, 2013, 18, 11783–11796 CrossRef CAS.
- A. Kamkaew and K. Burgess, Chem. Commun., 2015, 51, 10664–10667 RSC.
- G. Totzke, K. Schulze-Osthoff and R. U. Janicke, Oncogene, 2003, 22, 8021–8030 CrossRef PubMed.
- S. S. Solanki and R. Dahima, J. Adv. Pharm. Technol. Res., 2011, 2, 128–131 CrossRef CAS PubMed.
- J. L. Mateos, Drugs Today, 2010, 46(suppl. A), 1–25 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Detailed synthesis and characterizations of AZB-IMC2. See DOI: 10.1039/c9ra01948k |
| ‡ These authors contributed equally to this article. |
| This journal is © The Royal Society of Chemistry 2019 |