Open Access Article
Open Access ArticleSialyl LewisX mimic-decorated liposomes for anti-angiogenic everolimus delivery to E-selectin expressing endothelial cells†
Chanikarn Chantarasrivong a,
Yuriko Higuchia,
Masahiro Tsudaa,
Yuuki Yamanea,
Mitsuru Hashidab,
Miku Konishic,
Naoko Komurac,
Hiromune Ando
a,
Yuriko Higuchia,
Masahiro Tsudaa,
Yuuki Yamanea,
Mitsuru Hashidab,
Miku Konishic,
Naoko Komurac,
Hiromune Ando c and
Fumiyoshi Yamashita*a
c and
Fumiyoshi Yamashita*a
aGraduate School of Pharmaceutical Sciences, Kyoto University, 46-29 Yoshidashimoadachi-cho, Sakyo-ku, Kyoto 606-8501, Japan. E-mail: yama@pharm.kyoto-u.ac.jp
bInstitute for Advanced Study, Kyoto University, Yoshidaushinomiya-cho, Sakyo-ku, Kyoto 606-8501, Japan
cCenter for Highly Advanced Integration of Nano and Life Sciences (G-CHAIN), Gifu University, 1-1 Yanagido, Gifu-shi, Gifu 501-1193, Japan
First published on 2nd July 2019
Abstract
In this study, we developed novel E-selectin-targeting liposomes, i.e., 3′-(1-carboxy)ethyl sialyl LewisX (3′-CE sLeX) mimic liposomes, for targeted delivery of everolimus (EVE) in anti-angiogenic therapy. We investigated the uptake and efficacy of these E-selectin targeting liposomes in inflammatory cytokine-treated human umbilical vein endothelial cells (HUVECs). The uptake of EVE in 3′-CE sLeX mimic liposomes increased steadily and almost caught up with the uptake of plain EVE at 3 h, which was higher than that in PEGylated liposomes (PEG-liposomes). Inhibition of uptake by anti-E-selectin antibody suggested involvement of E-selectin-mediated endocytotic processes. Migration in cells treated with EVE/3′-CE sLeX mimic liposomes was suppressed by more than half when compared to the control. This treatment was also seen to significantly inhibit the formation of capillary tubes and networks. In addition, Thr389 phosphorylation of pS6 kinase, as a marker of mTOR activity, was remarkably suppressed to less than endogenous levels by EVE/3′-CE sLeX mimic liposomes. In conclusion, the present study demonstrated that EVE/3′-CE sLeX mimic liposomes were intracellularly taken up by E-selectin and prompted anti-angiogenic effects of EVE involved in the mTOR signaling pathway. However, moderate retention of EVE in the liposomes might limit the targeting ability of 3′-CE sLeX mimic liposomes.
Introduction
Anti-angiogenic therapy is one of the most effective approaches for the treatment of various types of cancers; this therapy causes damage to the surrounding endothelial cells, which limits oxygen/nutrient supply to tumor cells and results in tumor necrosis. This advantage has led to many studies on anti-angiogenic drugs and their subsequent approval by the FDA for cancer therapy.1–3 However, anti-angiogenic therapy may induce adaptive tumor microenvironmental defense mechanisms, leading to drug resistance or invasion.4,5 While combinatorial therapies using different drug targets are suggested to overcome this resistance,4,5 attention has also been paid to targeted drug delivery to improve the efficacy of drugs in anti-angiogenic therapy.6 For example, nanoparticles coated with collagen IV-binding peptides7 or cyclic RGD peptides8 were delivered to tumor endothelial cells, thereby improving the anti-tumorigenic and anti-metastatic activity of cytotoxic drugs in tumor-bearing mice. It has also been demonstrated that paclitaxel conjugated with factor VIIa, which binds to tissue factors expressed aberrantly in the tumor endothelium, suppresses tumor growth in paclitaxel-resistant cancer and inhibits its metastasis.9E-selectin is one of the most attractive targets expressed on the tumor endothelium,10–12 whose expression is strongly induced by cytokines and angiogenic growth factors secreted from tumors to facilitate angiogenesis and subsequent tumor growth/penetration.13–16 So far, E-selectin-directed drug delivery systems have mostly utilized sialyl LewisX (sLeX), a natural ligand of E-selectin,17–20 although there are a few exceptions such as anti-E-selectin antibodies,21 E-selectin-binding peptides,22 and aptamers.23 Previously, we synthesized novel sLeX analogs and conjugated them with 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-polyethylene glycol-2000 (DSPE-PEG2000) for targeted liposomal delivery. Despite being a structurally simplified trisaccharide analog, the 3′-(1-carboxy)ethyl sLeX (3′-CE sLeX) mimic exhibited higher potency than native sLeX in terms of enhancement of liposome uptake in E-selectin-expressing endothelial cells.24 The 3′-CE sLeX mimic liposomes appeared promising for the delivery of anti-angiogenic drugs to tumor endothelial cells.
Everolimus (EVE) is an mTOR (mammalian target of rapamycin) inhibitor, which inhibits complex formation of serine/threonine kinase (mTOR) to raptor and mLST8 by binding to the cyclophilin FK binding protein-12. mTOR is generally activated in cancers and plays important roles in multiple cellular processes, especially tumor-relevant angiogenesis, endothelial cell proliferation, survival, and migration.25–29 While several studies showed that EVE inhibits angiogenesis and tumor growth in tumor models,30,31 the low bioavailability and low water solubility of the drug have limited its use. There have been attempts to overcome these drawbacks through the use of liposomes or nanocarriers.32–34 In this study, we used human umbilical vein endothelial cells (HUVECs) activated with inflammatory cytokines as E-selectin-expressing models that imitate the mechanisms promoting tumorigenesis in the tumor microenvironment.35–39 The everolimus-encapsulated 3′-CE sLeX mimic liposomes were prepared and the physicochemical properties, cellular uptake, and pharmacological effects were then investigated.
Experimental
Materials
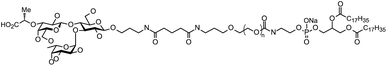
1,2-Distearoyl-sn-glycero-3-phosphocholine (DSPC) was purchased from Avanti Polar Lipids (Alabaster, AL, USA). Cholesterol, ethanol, and dimethyl sulfoxide (DMSO) were purchased from Nacalai Tesque (Kyoto, Japan). N-(methylpolyoxyethylene oxycarbonyl)-1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE-PEG2000) was purchased from NOF Corporation (Tokyo, Japan). 3′-CE sLeX-DSPE-PEG (Fig. 1) were synthesized by the method reported previously.24 EVE was purchased from Chem Scene (South Brunswick Township, NJ, USA). Phosphate-buffered saline (PBS) was purchased from Nissui Pharmaceutical (Tokyo, Japan). TNF-α was purchased from Life Technologies (Carlsbad, CA, USA) and IL-1β was from Sigma-Aldrich (St. Louis, MO, USA).Preparation of EVE-loaded liposomes
EVE-loaded liposomes were prepared by ethanol injection method.32 DSPC (5.5 μmol), cholesterol (4 μmol), and DSPE-PEG or 3′-CE sLeX-DSPE-PEG (0.5 μmol) were dissolved in 1 mL ethanol heated to 70 °C in a glass tube. Twenty-four microliters of DMSO containing 0.5 μmol EVE was added to the lipid solution and incubated for 3 min at 70 °C. The mixture was rapidly added to 2 mL PBS in a glass vial and stirred at 70 °C for 30 s. The ethanol in the mixture was removed under reduced pressure using a centrifugal evaporator (SpeedVac Concentrator SPD131DDA, Thermo Fisher Scientific, Waltham, MA, USA). The suspension was sonicated in a bath-type sonicator (ASU-3M, AS ONE, Osaka, Japan) at 70 °C for 10 min and then with a tip-type sonicator (Ultrasonic generator US 300, Nissei, Tokyo, Japan) at an intensity of 200 W for 4 min. The liposome solution was filtered through a 0.45 μm membrane filter (Cosmonice Filter W, Nacalai Tesque, Kyoto, Japan) and was purified using a PD-10 gel filtration column (GE Healthcare, Buckinghamshire, UK) equilibrated with PBS.Characterization of EVE-loaded liposomes
The concentration of phospholipids in the eluate was determined using a Phospholipids C-test Wako (Wako Pure Chemical Industry, Osaka, Japan). An aliquot of the stock solution was diluted in distilled water to a concentration of 0.2 mg total lipids per mL, and the particle size and zeta potential of the liposomes were measured in a Zetasizer Nano ZS (Malvern, Worcester, UK). On the other hand, another aliquot of the stock solution was diluted 40 times with methanol and vortexed for 3 min to disrupt liposomes. Following filtration of the samples using 0.45 μm filter, EVE entrapment efficiency of each liposome was determined using an HPLC system (Prominence UFLC, Shimadzu Corporation, Kyoto, Japan) equipped with a ZORBAX column (SB-C8 4.6 I.D. × 75 mm, 3.5 μm, Agilent Technologies, Santa Clara, CA, USA), which was maintained at 60 °C. The mobile phase comprised distilled water with 0.1% trifluoroacetic acid and acetonitrile with 20 mM triethylamine and 0.156% trifluoroacetic acid (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 v/v) and was maintained at a flow rate of 1 mL min−1. The wavelength of detection of EVE was set to 278 nm.
60 v/v) and was maintained at a flow rate of 1 mL min−1. The wavelength of detection of EVE was set to 278 nm.
Cell culture
Primary HUVECs were cultured in HuMedia-EG2 culture medium, according to the protocol supplied by the manufacturer (Kurabo Industry, Osaka, Japan). When the cells reached 70–80% confluence, they were harvested using trypsin–EDTA, suspended in the culture medium, and plated on a dish. The rest of the cells was maintained in a flask for up to 3 generations. On day 1 after plating, the cells were pretreated with 100 ng mL−1 TNF-α and 10 ng mL−1 IL-1β for 5 h, to induce E-selectin expression.Cellular toxicity
HUVECs were plated at a density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells/100 μL (equivalent to 30
000 cells/100 μL (equivalent to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2) in a 96-well plate. The seeding density was lower than that in other experiments (∼50
000 cells per cm2) in a 96-well plate. The seeding density was lower than that in other experiments (∼50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2), to make the cells more sensitive in toxicity detection.40 On day 1, following a 5 h cytokine treatment, the cells were treated with EVE or EVE-loaded liposomes at the indicated concentration. After incubating for 3 h, 10 μL of Cell Count Reagent SF (Nacalai Tesque, Kyoto, Japan) was added to the culture medium and the cells were incubated for 2 h. The cell viability was detected colorimetrically using BioTek Eon Microplate Spectrophotometer (Biotek, Winooski, VT, USA) at a wavelength of 450 nm.
000 cells per cm2), to make the cells more sensitive in toxicity detection.40 On day 1, following a 5 h cytokine treatment, the cells were treated with EVE or EVE-loaded liposomes at the indicated concentration. After incubating for 3 h, 10 μL of Cell Count Reagent SF (Nacalai Tesque, Kyoto, Japan) was added to the culture medium and the cells were incubated for 2 h. The cell viability was detected colorimetrically using BioTek Eon Microplate Spectrophotometer (Biotek, Winooski, VT, USA) at a wavelength of 450 nm.
Cellular uptake and inhibition by anti E-selectin antibody
HUVECs were plated at a density of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells/1 mL (50
000 cells/1 mL (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2) in a 12-well plate. On day 1, the cells were 80–90% confluent and used for uptake experiment. After cytokine treatment, EVE or EVE-loaded liposomes were added to the plates at a concentration equivalent to 1 μM EVE. Following incubation for the indicated time periods, the cells were washed with ice cold PBS, scraped off in 100 μL of 10 mM ammonium acetate buffer, and transferred to 1.5 mL tubes. One-hundred microliters of acetonitrile was added to the cell suspension and vortexed for 1 min. After centrifugation at 10
000 cells per cm2) in a 12-well plate. On day 1, the cells were 80–90% confluent and used for uptake experiment. After cytokine treatment, EVE or EVE-loaded liposomes were added to the plates at a concentration equivalent to 1 μM EVE. Following incubation for the indicated time periods, the cells were washed with ice cold PBS, scraped off in 100 μL of 10 mM ammonium acetate buffer, and transferred to 1.5 mL tubes. One-hundred microliters of acetonitrile was added to the cell suspension and vortexed for 1 min. After centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 5 min, the supernatant was collected and filtered with 0.45 μm membrane filter. EVE concentration was determined using an LC-MS/MS system (LC-MS-8030, Shimadzu Corporation, Kyoto, Japan). Separation of EVE was achieved on a COSMOSIL 5C18-MS-II column (4.6 mm I.D. × 150 mm, Nacalai Tesque, Kyoto, Japan), which was maintained at 40 °C, using 10 mM ammonium acetate and acetonitrile (20
000 × g for 5 min, the supernatant was collected and filtered with 0.45 μm membrane filter. EVE concentration was determined using an LC-MS/MS system (LC-MS-8030, Shimadzu Corporation, Kyoto, Japan). Separation of EVE was achieved on a COSMOSIL 5C18-MS-II column (4.6 mm I.D. × 150 mm, Nacalai Tesque, Kyoto, Japan), which was maintained at 40 °C, using 10 mM ammonium acetate and acetonitrile (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80, v/v) at a flow of 1.0 mL min−1. A post column splitter 1
80, v/v) at a flow of 1.0 mL min−1. A post column splitter 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 was installed before the MS interface. EVE was detected using the mass transition m/z 976.45 × 909.60. In contrast, the pellet from the cell lysate was dissolved in 50 μL RIPA buffer (20 mM Tris HCl, 150 mM NaCl, 1 mM EDTA, 1% Triton-X 100, 0.1% SDS, 0.1% Na-deoxycholate), and the protein concentration was measured using Pierce BCA protein assay kit (Thermo Scientific, Rockford, IL, USA) for normalization of cellular uptake.
4 was installed before the MS interface. EVE was detected using the mass transition m/z 976.45 × 909.60. In contrast, the pellet from the cell lysate was dissolved in 50 μL RIPA buffer (20 mM Tris HCl, 150 mM NaCl, 1 mM EDTA, 1% Triton-X 100, 0.1% SDS, 0.1% Na-deoxycholate), and the protein concentration was measured using Pierce BCA protein assay kit (Thermo Scientific, Rockford, IL, USA) for normalization of cellular uptake.
Inhibition of cell migration
HUVECs were plated at a density of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells/500 μL in a 4-well, 35 mm dish (Cellvis, Sunnyvale, CA). In this experiment, the cells were seeded at the higher density to become fully confluent on day 1. On day 1, after cytokine treatment, the cell layer was scratched with a 200 μL pipette tip and the culture medium was replaced with medium containing EVE or EVE-loaded liposomes at a concentration equivalent to 1 μM EVE. Live cell imaging was performed every hour for 3 h at 10× magnification using TD (transmitted image) scanning by a confocal microscope (Nikon A1RMP/Ti-E/PFS, Nikon Instruments Inc., Melville, NY, USA) equipped with NIS-elements AR 4.13.00 software. The scratch wound area was determined using Fiji package implemented in ImageJ software. All images were converted to grayscale, and the edges were enhanced and sharpened by the Sobel method. After the threshold was set to obtain a binary image, the edges of wound area were detected with ImageJ's wand tool. The enclosed area was calculated and expressed as a percentage of initial scratch wound area.
000 cells/500 μL in a 4-well, 35 mm dish (Cellvis, Sunnyvale, CA). In this experiment, the cells were seeded at the higher density to become fully confluent on day 1. On day 1, after cytokine treatment, the cell layer was scratched with a 200 μL pipette tip and the culture medium was replaced with medium containing EVE or EVE-loaded liposomes at a concentration equivalent to 1 μM EVE. Live cell imaging was performed every hour for 3 h at 10× magnification using TD (transmitted image) scanning by a confocal microscope (Nikon A1RMP/Ti-E/PFS, Nikon Instruments Inc., Melville, NY, USA) equipped with NIS-elements AR 4.13.00 software. The scratch wound area was determined using Fiji package implemented in ImageJ software. All images were converted to grayscale, and the edges were enhanced and sharpened by the Sobel method. After the threshold was set to obtain a binary image, the edges of wound area were detected with ImageJ's wand tool. The enclosed area was calculated and expressed as a percentage of initial scratch wound area.
Inhibition of capillary tube formation
Prior to cell seeding, 450 μL of Matrigel (Corning, Bedford, MA, USA) was added to each well in a 2-well chamber slide glass (Matsunami glass Industry, Osaka, Japan) and incubated at 37 °C for 30 min. HUVECs were seeded at a density of 240![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells/0.8 mL and incubated at 37 °C. Thirty minutes later, 0.8 mL of culture medium supplemented with 200 ng mL−1 TNF-α and 20 ng mL−1 IL-1β, and 10% Matrigel was added to the chamber well. Five hours later, the EVE or EVE-loaded liposomes at a concentration equivalent to 1 μM EVE were added and incubated for 5 h. The morphology of the cell culture was investigated under a microscope (Biozero, BZ-8100, Keyence Corporation, Osaka, Japan).
000 cells/0.8 mL and incubated at 37 °C. Thirty minutes later, 0.8 mL of culture medium supplemented with 200 ng mL−1 TNF-α and 20 ng mL−1 IL-1β, and 10% Matrigel was added to the chamber well. Five hours later, the EVE or EVE-loaded liposomes at a concentration equivalent to 1 μM EVE were added and incubated for 5 h. The morphology of the cell culture was investigated under a microscope (Biozero, BZ-8100, Keyence Corporation, Osaka, Japan).
Inhibition of mTOR signaling pathway
HUVECs were plated at a density of 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells/2 mL in a 6-well plate. On day 1, after cytokine treatment, the cells were treated with EVE or EVE-loaded liposomes at a concentration equivalent to 1 μM EVE and incubated at 37 °C for 4 h. To stimulate mTOR signaling, 50 ng mL−1 human epidermal growth factor (hEGF, Cell Signaling Technology, Denver, MA, USA) was added 15 min before the end of incubation, and the cells were washed with ice cold PBS. Eighty microliters of RIPA buffer with Halt phosphatase and protease inhibitor (Thermo Fisher Scientific, Waltham, MA, USA) was added to lyse the cells and the cells lysate was collected with a cell scraper. After centrifugation at 18
000 cells/2 mL in a 6-well plate. On day 1, after cytokine treatment, the cells were treated with EVE or EVE-loaded liposomes at a concentration equivalent to 1 μM EVE and incubated at 37 °C for 4 h. To stimulate mTOR signaling, 50 ng mL−1 human epidermal growth factor (hEGF, Cell Signaling Technology, Denver, MA, USA) was added 15 min before the end of incubation, and the cells were washed with ice cold PBS. Eighty microliters of RIPA buffer with Halt phosphatase and protease inhibitor (Thermo Fisher Scientific, Waltham, MA, USA) was added to lyse the cells and the cells lysate was collected with a cell scraper. After centrifugation at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g at 4 °C for 20 min, the supernatant was aliquoted and heat-treated at 95 °C for 5 min. The protein concentration was measured using the Pierce BCA protein assay kit. A 15 μg protein aliquot of each sample was subjected to SDS-PAGE on a 12.5% Precast gel (SuperSep Ace, Wako Pure Chemical Industry, Osaka, Japan) and transferred to a PVDF membrane (Immobilon, Merck Millipore, County Cork, Ireland). The membrane was blocked using Tris-buffered saline containing 3% bovine serum albumin (BSA, Sigma Life Science, St. Louis, MO, USA) at 25 °C for 1 h and stained with Phospho-p70 S6 kinase (Thr389) antibody (Cell Signaling Technology, Denver, MA, USA) at a concentration of 1
000 × g at 4 °C for 20 min, the supernatant was aliquoted and heat-treated at 95 °C for 5 min. The protein concentration was measured using the Pierce BCA protein assay kit. A 15 μg protein aliquot of each sample was subjected to SDS-PAGE on a 12.5% Precast gel (SuperSep Ace, Wako Pure Chemical Industry, Osaka, Japan) and transferred to a PVDF membrane (Immobilon, Merck Millipore, County Cork, Ireland). The membrane was blocked using Tris-buffered saline containing 3% bovine serum albumin (BSA, Sigma Life Science, St. Louis, MO, USA) at 25 °C for 1 h and stained with Phospho-p70 S6 kinase (Thr389) antibody (Cell Signaling Technology, Denver, MA, USA) at a concentration of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 at 4 °C for 14 h as the primary antibody, followed by treatment with horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G at a concentration of 1
1000 at 4 °C for 14 h as the primary antibody, followed by treatment with horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G at a concentration of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at 25 °C for 1 h (Abcam, Cambridge, U.K.) as the secondary antibody. A β-actin antibody (Santa Cruz Biotechnology, Dallas, TX, USA) was used as loading control at a concentration of 1
000 at 25 °C for 1 h (Abcam, Cambridge, U.K.) as the secondary antibody. A β-actin antibody (Santa Cruz Biotechnology, Dallas, TX, USA) was used as loading control at a concentration of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at 25 °C for 1 h. All antibodies were diluted in Tris-buffered saline containing 1% BSA. The protein was detected using chemiluminescent HRP substrate (Merck, Billerica, MA) and imaged using ImageQuant LAS 4000 (GE Life Sciences, Buckinghamshire, UK).
000 at 25 °C for 1 h. All antibodies were diluted in Tris-buffered saline containing 1% BSA. The protein was detected using chemiluminescent HRP substrate (Merck, Billerica, MA) and imaged using ImageQuant LAS 4000 (GE Life Sciences, Buckinghamshire, UK).
Intracellular distribution
Cy5.5-labeled 3′-CE sLeX mimic liposomes were prepared according to the hydration method reported previously.24 Briefly, DSPC, cholesterol, 3′-CE sLeX mimic-linked DSPE-PEG, and DSPE-PEG-Cy5.5 were mixed in chloroform-methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at the molar ratio of 55
1) at the molar ratio of 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39
39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and then dried under reduced pressure using a rotary evaporator and vacuum desiccator. The lipid film was swollen and was suspended in PBS at 65 °C. The suspension was sonicated in a bath-type sonicator at 65 °C for 10 min and with a tip-type sonicator for 3 min, and then extruded through a 100 nm pore membrane equipped in an extruder (Avanti Mini-Extruder, Avanti Polar Lipids, Alabaster, AL, USA) maintained at 65 °C. The liposome solution was purified using a PD-10 gel filtration column. To evaluate the intracellular distribution of 3′-CE sLeX mimic liposomes, HUVECs were plated at a density of 40
1 and then dried under reduced pressure using a rotary evaporator and vacuum desiccator. The lipid film was swollen and was suspended in PBS at 65 °C. The suspension was sonicated in a bath-type sonicator at 65 °C for 10 min and with a tip-type sonicator for 3 min, and then extruded through a 100 nm pore membrane equipped in an extruder (Avanti Mini-Extruder, Avanti Polar Lipids, Alabaster, AL, USA) maintained at 65 °C. The liposome solution was purified using a PD-10 gel filtration column. To evaluate the intracellular distribution of 3′-CE sLeX mimic liposomes, HUVECs were plated at a density of 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells/250 μL in an 8-well chamber slide. On day 1, after cytokine treatment of the cells, 3′-CE sLeX mimic liposomes were added and incubated for 1–4 h. At 2 h before the end of each incubation period, LysoTracker Green DND-26 (Thermo Fisher Scientific, Waltham, MA) was added to yield a final concentration of 75 nM. At the end of incubation, the medium was replaced with fresh culture medium and the cells were observed using a confocal microscope. A Golgi-ID green assay kit (Enzo Life Sciences, Farmingdale, NY, USA) was also used to stain the Golgi bodies. The Golgi Green Detection Reagent at concentration of 0.25 nmol mL−1 was co-incubated with 3′-CE sLeX mimic liposomes in the last 30 min. The cells were washed once with assay solution and once with fresh medium before observation under a confocal microscope (Nikon A1RMP/Ti-E/PFS, Nikon Instruments Inc., Melville, NY, USA) equipped with NIS-elements AR 4.13.00 software. The excitation wavelength was set at 488.5 and 637.3 nm, and the emission wavelengths were scanned in the range of 500–550 nm and 663–738 nm for FITC and Cy5.5, respectively.
000 cells/250 μL in an 8-well chamber slide. On day 1, after cytokine treatment of the cells, 3′-CE sLeX mimic liposomes were added and incubated for 1–4 h. At 2 h before the end of each incubation period, LysoTracker Green DND-26 (Thermo Fisher Scientific, Waltham, MA) was added to yield a final concentration of 75 nM. At the end of incubation, the medium was replaced with fresh culture medium and the cells were observed using a confocal microscope. A Golgi-ID green assay kit (Enzo Life Sciences, Farmingdale, NY, USA) was also used to stain the Golgi bodies. The Golgi Green Detection Reagent at concentration of 0.25 nmol mL−1 was co-incubated with 3′-CE sLeX mimic liposomes in the last 30 min. The cells were washed once with assay solution and once with fresh medium before observation under a confocal microscope (Nikon A1RMP/Ti-E/PFS, Nikon Instruments Inc., Melville, NY, USA) equipped with NIS-elements AR 4.13.00 software. The excitation wavelength was set at 488.5 and 637.3 nm, and the emission wavelengths were scanned in the range of 500–550 nm and 663–738 nm for FITC and Cy5.5, respectively.
Statistical analysis
Statistical significance was evaluated by one-way or two-way ANOVA followed by the Tukey–Kramer's post hoc test. The type of ANOVAs adopted was indicated in each figure caption. A value of p < 0.05 was considered statistically significant.Results and discussion
Preparation and characterization of EVE-loaded liposomes
The ethanol injection method has been used to prepare EVE- or rapamycin-loaded liposomes.32,41,42 The EVE-loaded liposomes prepared in this study were comparable in size and drug-to-lipid molar ratio to those in past reports.32,43 Table 1 summarizes characteristics of the prepared liposomes. The average diameter was approximately 80 nm for both EVE/PEG- and EVE/3′-CE sLeX mimic liposomes. Their polydispersity indices were 0.11 and 0.14, indicating that both liposomes have an acceptably uniform size distribution. The EVE/3′-CE sLeX mimic liposome was highly negative with a zeta potential of −26.7 mV, whereas EVE/PEG-liposomes was slightly negative indicating a weaker surface charge. Total lipid recovery of both liposomes was more than 80% confirming no significant loss during preparation. On the other hand, the amount of EVE recovered was 17.97 and 19.90% for EVE/PEG-liposomes and EVE/3′-CE sLeX mimic liposomes, respectively. The low recoveries were associated with recrystallization of overloaded EVE during the ethanol-injection preparation. The drug-to-lipid molar ratio for both liposomes was comparable with the liposomes reported previously.32 The fluorescent labelled 3′-CE sLeX mimic liposomes were prepared by the same method as reported previously.24 Regardless of film hydration or ethanol injection methods, 3′-CE sLeX mimic liposomes prepared possessed basically the same characteristics.| Liposomes | Average diameter (nm) | Polydispersity index | Zeta potential (mV) | Drug recovery (%) | Total lipid recovery (%) | Drug-to-lipid molar ratio |
|---|---|---|---|---|---|---|
| a Results are expressed as mean ± SD of three samples. | ||||||
| EVE/PEG | 80.90 ± 8.17 | 0.11 ± 0.06 | −11.62 ± 2.09 | 17.97 ± 2.43 | 84.01 ± 6.04 | 0.015 ± 0.003 |
| EVE/3′-CE sLeX mimic | 80.28 ± 11.93 | 0.14 ± 0.06 | −26.70 ± 2.66 | 19.90 ± 5.48 | 85.68 ± 4.32 | 0.016 ± 0.001 |
| Cy5.5-labeled 3′-CE sLeX mimic | 87.58 ± 7.00 | 0.08 ± 0.02 | −25.21 ± 2.95 | — | 91.62 ± 2.90 | — |
Cellular toxicity
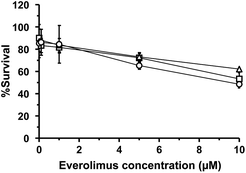
WST-8 assay was carried out to evaluate the cellular toxicity of plain and liposomal EVE and determine the dose used for cellular uptake and anti-angiogenic studies. Fig. 2 shows percent viability of cytokine-treated HUVECs following treatment at different concentrations of the drugs for 3 h. Regardless of treatment with plain or liposomal EVE, the cell viability was more than 80% at concentrations of up to 1 μM. Therefore, 1 μM EVE was chosen for further studies to ensure minimal cell death during the experiment. The designated concentration was consistent with past studies30,34 that had investigated an inhibitory effect of EVE on angiogenesis-related endothelial functions in HUVECs.Cellular uptake
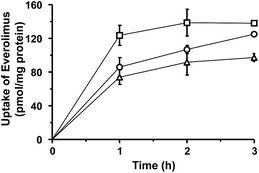
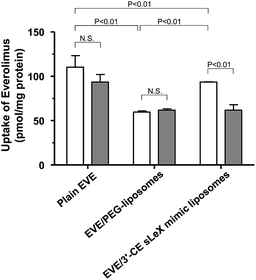
The uptake of plain or liposomal EVE in HUVECs treated with proinflammatory cytokines was investigated. As shown in Fig. 3, cellular uptake of EVE was the highest with plain EVE, followed by EVE/3′-CE sLeX mimic liposomes, and then EVE/PEG-liposomes. When plain EVE was administered, the uptake appeared to reach a plateau within almost 1 h. The relatively rapid equilibrium achieved with plain EVE is associated with the mechanism of simple diffusion across the plasma membrane in accordance with the lipophilic nature of EVE. In contrast, the uptake of EVE/3′-CE sLeX mimic liposomes increased steadily over 3 h and almost caught up with plain EVE. We have previously reported that fluorescein-labeled 3′-CE sLeX mimic liposomes have been taken up by E-selectin-mediated endocytotic processes.24 The specialized process involving cytotic membrane transport could be slower than simple diffusion of small lipophilic molecules. The uptake of EVE/3′-CE sLeX mimic liposomes was significantly higher than that of EVE/PEG-liposomes, but the difference between the two liposome formulations was not so large as that seen with fluorescein-labeled liposomes in the previous study. Leakage of EVE from liposomes could not be ruled out (see ESI†). Although a dialysis release experiment might be hampered by adsorption onto membranes, it appeared that the release of EVE from the liposome formulations was biphasic with an initial burst followed by a slow release. The convex-shaped profile for the uptake of EVE/PEG-liposomes similar to that of plain EVE (Fig. 3) might result from the uptake of EVE liberated during the burst phase. The initial-burst release appeared more remarkable with EVE/3′-CE sLeX mimic liposomes (Fig. S1†), but nevertheless the uptake of EVE from the liposomes lasted more than 1 h (Fig. 3). It suggests that drug molecules remaining in 3′-CE sLeX mimic liposomes might be taken up efficiently via the receptor-mediated endocytosis pathway.To clarify the involvement of the E-selectin-mediated uptake process, the effect of anti-E-selectin antibody on cellular uptake was investigated. Although sLeX is a ligand of E-selectin, P-selectin, and L-selectin44,45 only E-selectin blockade was tested, because our previous study24 concluded that cellular association of 3′-CE sLeX mimic liposomes be not affected by anti-P-selectin and anti-L-selectin antibodies. Fig. 4 shows that the uptake of EVE/3′-CE sLeX mimic liposomes was significantly inhibited in the presence of anti-E-selectin antibody, whereas it was not observed with either EVE/PEG-liposomes or plain EVE. Interestingly, the uptake of EVE/3′-CE sLeX mimic liposomes under antibody blockade was comparable to that of EVE/PEG-liposomes, implying that the release of EVE from liposome formulations, if any, was not different between the two. Therefore, the improved uptake of EVE/3′-CE sLeX mimic liposomes would be primarily due to an E-selectin-mediated uptake process.
Anti-angiogenic effect
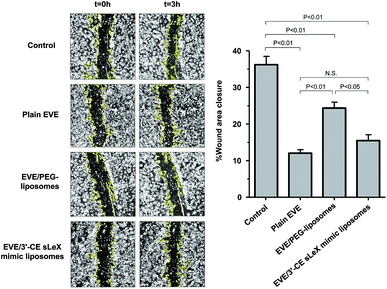
The main endothelial functions involved in angiogenesis are cell migration and capillary tube formation. Cell migration plays an important role in the initial steps of angiogenesis, starting with basement membrane break down and followed by mitosis and migration in response to angiogenic factors including VEGF. After migration, endothelial cells initiate tube formation and develop new vessels to complete angiogenesis.46,47 To investigate the anti-angiogenic effect of plain and liposomal EVE, a scratch assay and capillary tube formation were conducted. Moreover, the regulation of the mTOR cascade was confirmed by the degree of Thr389 phosphorylation of pS6 kinase (Ser235/236).Migration of cytokine-treated HUVECs in the presence of EVE or EVE-loaded liposomes was evaluated in a scratch assay. Fig. 5 shows the percentage of wound closure within 3 h incubation with EVE or EVE-loaded liposomes. The control cells, i.e., HUVECs treated with TNF-α and IL-1β, showed 36.2% wound closure within 3 h. However, treatment with plain EVE and EVE/3′-CE sLeX mimic liposomes suppressed the wound closure by more than half (12% and 15.5%, respectively). The suppressive effect of EVE/PEG-liposomes was moderate, resulting in 24.3% wound closure. These trends regarding suppression of cell migration were in agreement with cellular uptake of EVE from each formulation. Damiano et al. also found that 1 μM EVE was effective for suppression of HUVEC migration.30 According to their results, the degree of suppression was ∼35% at 24 h following the onset of treatment and almost zero at 48 h. However, the longer time of incubation might have reduced the estimate of the drug effect because of the limitation of available area for cell migration. Indeed, real-time monitoring of HUVEC migration has indicated that the index of migration increased linearly up to 3 h and the rate of migration slowed down thereafter.34 In this experiment, the incubation time of 3 h gave the degree of suppression by 1 μM plain EVE of ∼67%. By improving the detection sensitivity to the drug effect, we could more easily evaluate whether the influence of liposomal formulations was accelerative or suppressive.
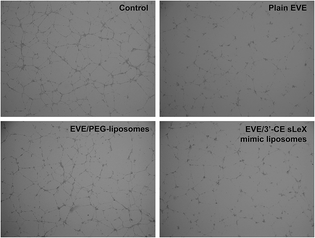
Tube formation assay is another popular in vitro method to evaluate angiogenesis. It utilizes the nature of endothelial progenitor cells such as HUVECs that form tubes when placed on a 3D extracellular matrix. One micromolar EVE has been known to inhibit tube formation of HUVECs in several reports.30,34 Fig. 6 shows the effect of plain EVE and EVE-loaded liposomes on tube formation. Formation of tubes and networks from cytokine-treated HUVECs was significantly inhibited by plain EVE and EVE/3′-CE sLeX mimic liposomes after 5 h of exposure, whereas the EVE/PEG-liposomes did not significantly interrupt the formation. Morphology of the tube formation in this experiment and other studies30,34 are consistent, control cells formed completed ring, whereas everolimus-treated cells showed disrupted-branches of tube formation.
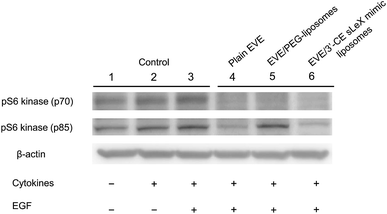
Western blot analysis was performed to confirm phosphorylation of pS6 kinase at Thr389 amino acid residues, which is a downstream target of EVE in the mTOR signaling pathway.48 Phosphorylation of pS6 kinase was induced by EGF according to the method described by Damiano V., et al.,30 in addition to TNF-α and IL-1β.49–51 Although Damiano, et al.30 incubated HUVECs with EVE for 24 h, we shortened the time of incubation to 4 h to minimize a pharmacological effect due to EVE leaked from the liposomes. As shown in Fig. 7, treatment with plain EVE and EVE/3′-CE sLeX mimic liposomes remarkably suppressed phosphorylation of pS6 kinase to less than the endogenous level. The four-hour incubation appeared to be enough for EVE to modulate the kinase activity.
EVE/PEG-liposomes suppressed phosphorylation of pS6 kinase (p70) but not that of pS6 kinase (p85). pS6 kinase (p85) is a splicing variant of pS6 kinase (p70), having 23 extra amino acids at the amino-terminus. Since the additional peptide sequence encodes a nuclear localizing signal, pS6 kinase (p85) is exclusively localized in the nuclei.49,52,53
The difference in subcellular localization or protein structure between the two S6 kinases might relate to the differential suppression of phosphorylation observed with EVE/PEG-liposomes, which could be emphasized only when the suppressive effect was mild. Nevertheless, a high correlation between cellular uptake, cell migration, tube formation, and phosphorylation of pS6 kinases implied that not only plain EVE but liposomal EVE are likely to suppress angiogenesis via mTOR signaling pathway.
Intracellular distribution
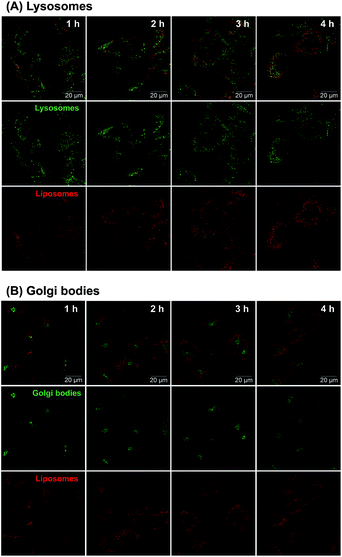
Considering the uncertain observation that E-selectin-targeted liposomes are transported intracellularly via clathrin-independent pathways followed by transport to the Golgi bodies and endoplasmic reticulum54 or the endosome/lysosome pathway followed by release in the cytoplasm,55,56 intracellular disposition behavior of 3′-CE sLeX mimic liposomes was also investigated.Confocal fluorescence microscopy was employed to clarify subcellular distribution of fluorescence-labeled 3′-CE sLeX mimic liposomes. As shown in Fig. 8, red fluorescence signal associated with 3′-CE sLeX mimic liposomes increased over time. The images indicating such gradual uptake of 3′-CE sLeX mimic liposomes corresponded to the result of quantification of EVE by LC-MS/MS (Fig. 3), as well as our previous observation.24 The previous study was not conclusive in terms of subcellular localization of the liposomes because of low resolution and no subcellular staining, but Fig. 8 clearly indicates that the liposomes appear to be transported preferentially to lysosomes than to the Golgi bodies.
Taking all findings together, it was suggested that EVE/3′-CE sLeX mimic liposomes are taken up and disassembled in an endosome/lysosome pathway, leading to the release of EVE in the cytoplasm. Our result that sLeX-liposomes are not directly transported to the nucleus are in agreement with the conclusion of Stahn et al.57
Conclusion
The present study demonstrated that EVE/3′-CE sLeX mimic liposomes were intracellularly taken up by E-selectin and prompted an anti-angiogenic effect of EVE involved in the mTOR signaling pathway. However, the effectiveness of 3′-CE sLeX mimic liposomes was limited in case of the delivery of EVE, presumably due to moderate retention of the drug. The drug delivery potential of 3′-CE sLeX mimic liposomes and the formulations of EVE need to be further investigated and optimized.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported in part by JSPS KAKENHI under grant numbers JP16H01861(MH) and 18H03531(FY) and the Research on Development of New Drugs from Japan Agency for Medical Research and Development, AMED under grant number JP19ak0101081 (FY).References
- T. Li, G. Kang, T. Wang and H. Huang, Tumor angiogenesis and anti-angiogenic gene therapy for cancer, Oncol. Lett., 2018, 16, 687–702 Search PubMed.
- A. Dutour and M. Rigaud, Tumor endothelial cells are targets for selective therapies: in vitro and in vivo models to evaluate antiangiogenic strategies, Anticancer Res., 2005, 25, 3799–3808 CAS.
- R. S. Samant and L. A. Shevde, Recent advances in anti-angiogenic therapy of cancer, Oncotarget, 2011, 2, 122–134 CrossRef PubMed.
- G. Bergers and D. Hanahan, Modes of resistance to anti-angiogenic therapy, Nat. Rev. Cancer, 2008, 8, 592–603 CrossRef CAS PubMed.
- S. Ma, S. Pradeep, W. Hu, D. Zhang, R. Coleman and A. Sood, The role of tumor microenvironment in resistance to anti-angiogenic therapy, F1000Research, 2018, 326(7), 1–19 Search PubMed.
- A. M. E. Abdalla, L. Xiao, M. W. Ullah, M. Yu, C. Ouyang and G. Yang, Current challenges of cancer anti-angiogenic therapy and the promise of nanotherapeutics, Theranostics, 2018, 8(2), 533–548 CrossRef CAS PubMed.
- F. Tan, X. H. Mo, J. Zhao, H. Liang, Z. J. Chen and X. L. Wang, A novel delivery vector for targeted delivery of the antiangiogenic drug paclitaxel to angiogenic blood vessels: TLTYTWS-conjugated PEG–PLA nanoparticles, J. Nanopart. Res., 2017, 19, 51 CrossRef.
- E. A. Murphy, B. K. Majeti, L. A. Barnes, M. Makale, S. M. Weis, K. Lutu-Fuga, W. Wrasidlo and D. A. Cheresh, Nanoparticle-mediated drug delivery to tumor vasculature suppresses metastasis, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 9343–9348 CrossRef CAS PubMed.
- S. Zhu, W. Kisiel, Y. J. Lu, L. C. Petersen, J. M. Ndungu, T. W. Moore, E. T. Parker, A. Sun, D. C. Liotta, B. F. El-Rayes, D. J. Brat, J. P. Snyder and M. Shoji, Tumor angiogenesis therapy using targeted delivery of paclitaxel to the vasculature of breast cancer metastases, J. Drug Delivery, 2014, 865732 Search PubMed.
- V. Bhaskar, D. A. Law, E. Ibsen, D. Breinberg, K. M. Cass, R. B. DuBridge, F. Evangelista, S. M. Henshall, P. Hevezi, J. C. Miller, M. Pong, R. Powers, P. Senter, D. Stockett, R. L. Sutherland, U. von Freeden-Jeffry, D. Willhite, R. Murray, D. E. Afar and V. Ramakrishnan, E-selectin up-regulation allows for targeted drug delivery in prostate cancer, Cancer Res., 2003, 63, 6387–6394 CAS.
- S. R. Barthel, J. D. Gavino, L. Descheny and C. J. Dimitroff, Targeting selectins and selectin ligands in inflammation and cancer, Expert Opin. Ther. Targets, 2007, 11, 1473–1491 CrossRef CAS PubMed.
- H. Kang, L. Josephson, A. Petrovsky, R. Weissleder and A. Bogdanov Jr, Magnetic resonance imaging of inducible E-selectin expression in human endothelial cell culture, Bioconjugate Chem., 2002, 13, 122–127 CrossRef CAS PubMed.
- A. L. Harris, Antiangiogenesis for cancer therapy, Lancet, 1997, 349, 13–15 CrossRef CAS.
- S. Hiratsuka, S. Goel, W. S. Kamoun, Y. Maru, D. Fukumura, D. G. Duda and R. K. Jaina, Endothelial focal adhesion kinase mediates cancer cell homing to discrete regions of the lungs via E-selectin up-regulation, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(9), 3725–3730 CrossRef CAS PubMed.
- A. M. Khatib, L. Fallavollita, E. V. Wancewicz, B. P. Monia and P. Brodt, Inhibition of hepatic endothelial E-selectin expression by C-raf antisense oligonucleotides blocks colorectal carcinoma liver metastasis, Cancer Res., 2002, 62, 5393–5398 CAS.
- Y. Shamay, L. Raviv, M. Golan, E. Voronov, R. N. Apte and A. David, Inhibition of primary and metastatic tumors in mice by E-selectin-targeted polymer–drug conjugates, J. Controlled Release, 2015, 217, 102–112 CrossRef CAS PubMed.
- N. Hashida, N. Ohguro, N. Yamazaki, Y. Arakawa, E. Oiki, H. Mashimo, N. Kurokawa and Y. Tano, High-efficacy site-directed drug delivery system using sialyl-Lewis X conjugated liposome, Exp. Eye Res., 2008, 86(1), 138–149 CrossRef CAS PubMed.
- E. L. Vodovozova, E. V. Moiseeva, G. K. Grechko, G. P. Gayenko, N. E. Nifant'ev, N. V. Bovin and J. G. Molotkovsky, Antitumour activity of cytotoxic liposomes equipped with selectin ligand SiaLe(X), in a mouse mammary adenocarcinoma model, Eur. J. Cancer, 2000, 36(7), 942–949 CrossRef CAS PubMed.
- M. Hirai, H. Minematsu, Y. Hiramatsu, H. Kitagawa, T. Otani, S. Iwashita, T. Kudoh, L. Chen, Y. Li, M. Okada, D. S. Salomon, K. Igarashi, M. Chikuma and M. Seno, Novel and simple loading procedure of cisplatin into liposomes and targeting tumor endothelial cells, Int. J. Pharm., 2010, 391(1–2), 274–283 CrossRef CAS PubMed.
- E. Jubeli, L. Moine, J. V. Gauduchon and G. Barratt, E-selectin as a target for drug delivery and molecular imaging, J. Controlled Release, 2012, 158, 194–206 CrossRef CAS PubMed.
- C. B. Pattillo, B. Venegas, F. J. Donelson, L. Del Valle, L. C. Knight, P. L. Chong and M. F. Kiani, Radiation-guided targeting of combretastatin encapsulated immunoliposomes to mammary tumors, Pharm. Res., 2009, 26(5), 1093–1100 CrossRef CAS PubMed.
- Y. Shamay, D. Paulin D, G. Ashkenasy and A. David, E-selectin binding peptide-polymer-drug conjugates and their selective cytotoxicity against vascular endothelial cells, Biomaterials, 2009, 30(32), 6460–6468 CrossRef CAS PubMed.
- A. P. Mann, A. Somasunderam, R. Nieves-Alicea, X. Li, A. Hu, A. K. Sood, M. Ferrari, D. G. Gorenstein and T. Tanaka, Identification of thioaptamer ligand against E-selectin: potential application for inflamed vasculature targeting, PLoS One, 2010, 5(9), e13050 CrossRef PubMed.
- C. Chantarasrivong, A. Ueki, R. Ohyama, J. Unga, S. Nakamura, I. Nakanishi, Y. Higuchi, S. Kawakami, H. Ando, A. Imamura, H. Ishida, F. Yamashita, M. Kiso and M. Hashida, Synthesis and functional characterization of novel sialyl lewisx mimic-decorated liposomes for e-selectin-mediated targeting to inflamed endothelial cells, Mol. Pharm., 2017, 14, 1528–1537 CrossRef CAS PubMed.
- H. A. Lane, J. M. Wood, P. M. McSheehy, P. R. Allegrini, A. Boulay, J. Brueggen, A. Littlewood-Evans, S. M. Maira, G. Martiny-Baron, C. R. Schnell, P. Sini and T. O'Reilly, mTOR Inhibitor RAD001 (everolimus) has antiangiogenic/vascular properties distinct from a VEGFR tyrosine kinase inhibitor, Clin. Cancer Res., 2009, 15(5), 1612–1622 CrossRef CAS PubMed.
- L. H. Meng and X. S. Zheng, Toward rapamycin analog (rapalog)-based precision cancer therapy, Acta Pharmacol. Sin., 2015, 36, 1163–1169 CrossRef CAS PubMed.
- P. J. Houghton, Everolimus, Clin. Cancer Res., 2010, 16(5), 1368–1372 CrossRef CAS PubMed.
- S. Faes, T. Santoro, N. Demartines and O. Dormond, Evolving significance and future relevance of anti-angiogenic activity of mTOR inhibitors in cancer therapy, Cancers, 2017, 9, 152 CrossRef PubMed.
- A. Albini, F. Tosetti, V. W. Li, D. M. Noonan and W. W. Li, Cancer prevention by targeting angiogenesis, Nat. Rev. Clin. Oncol., 2012, 1–12 Search PubMed.
- V. Damiano, R. Rosa, L. Formisano, L. Nappi, T. Gelardi, R. Marciano, I. Cozzolino, G. Troncone, S. Agrawal, B. M. Veneziani, S. De Placido, R. Bianco and G. Tortora, Toll-like receptor 9 agonist IMO cooperates with everolimus in renal cell carcinoma by interfering with tumour growth and angiogenesis, Br. J. Cancer, 2013, 108, 1616–1623 CrossRef CAS PubMed.
- Y. Pignochino, C. Dell'Aglio, M. Basiricò, F. Capozzi, M. Soster, S. Marchiò, S. Bruno, L. Gammaitoni, D. Sangiolo, E. Torchiaro, L. D'Ambrosio, F. Fagioli, S. Ferrari, M. Alberghini, P. Picci, M. Aglietta and G. Grignani, The Combination of Sorafenib and Everolimus Abrogates mTORC1 and mTORC2 Upregulation in Osteosarcoma Preclinical Models, Clin. Cancer Res., 2013, 19(8), 2117–2131 CrossRef CAS PubMed.
- Y. Iwase and Y. Maitani, Preparation and in vivo evaluation of liposomal everolimus for lung carcinoma and thyroid carcinoma, Biol. Pharm. Bull., 2012, 35(6), 975–979 CrossRef CAS.
- M. Kasper, D. Gabriel, M. Möller, D. Bauer, L. Wildschütz, H. Courthion, M. R. R. Böhm, M. Busch, K. Loser, S. Thanos, R. Gurny and A. Heiligenhaus, Novel everolimus-loaded nanocarriers for topical treatment of murine experimental autoimmune uveoretinitis (EAU), Exp. Eye Res., 2018, 168, 49–56 CrossRef CAS PubMed.
- G. P. Mishra, B. S. Doddapaneni, D. Nguyen and A. W. Alani, Antiangiogenic effect of docetaxel and everolimus as individual and dual-drug-loaded micellar nanocarriers, Pharm. Res., 2014, 3, 660–669 CrossRef PubMed.
- B. Pezeshkian, C. Donnelly, K. Tamburo, T. Geddes and G. J. Madlambayan, Leukemia Mediated Endothelial Cell Activation Modulates Leukemia Cell Susceptibility to Chemotherapy through a Positive Feedback Loop Mechanism, PLoS One, 2013, 8(4), e60823 CrossRef CAS PubMed.
- C. Eichbaum, A. S. Meyer, E. Bischofs, A. Steinborn, T. Bruckner, P. Brodt, C. Sohn and M. H. R. Eichbaum, Breast cancer cell-derived cytokines, macrophages and cell adhesion: implications for metastasis, Anticancer Res., 2011, 31, 3219–3228 CAS.
- T. Narita, N. K. Kimura, Y. Kasai, J. Hosono, T. Nakashio, N. Matsuura, M. Sato and R. Kannagi, Induction of E-selectin expression on vascular endothelium by digestive system cancer cells, J. Gastroenterol., 1996, 31, 299–301 CrossRef CAS PubMed.
- E. Bischofs, D. Lubs, F. Fritzsche, A. S. Meyer, T. Bruckner, C. Sohn and M. H. R. Eichbaum, In vitro blockade of adhesion of breast cancer cells to endothelial cells using anti-inflammatory drugs, Anticancer Res., 2012, 32(3), 767–771 CAS.
- H. Bachtarzi, M. Stevenson, V. Šubr, K. Ulbrich, L. W. Seymour and K. D. Fisher, Targeting adenovirus gene delivery to activated tumour-associated vasculature via endothelial selectins, J. Controlled Release, 2011, 150, 196–203 CrossRef CAS PubMed.
- K. C. S. Gasque, L. P. Al-Ahj, R. C. Oliveira and A. C. Magalhães, Cell Density and Solvent are Critical Parameters Affecting Formazan Evaluation in MTT Assay, Braz. Arch. Biol. Technol., 2014, 57(3), 381–385 CrossRef.
- Z. L. Miao, Y. J. Deng, H. Y. Du, X. B. Suo, X. Y. Wang, X. Wang, L. Wang, L. J. Cui and N. Duan, Preparation of a liposomal delivery system and its in vitro release of rapamycin, Exp. Ther. Med., 2015, 9(3), 941–946 CrossRef CAS PubMed.
- M. A. L. Alba, M. B. G. Guajardo, J. F. Fonzar, D. E. Brooks, G. A. G. Sánchez and M. J. B. Bernad, Preformulation studies of a liposomal formulation containing sirolimus for the treatment of dry eye disease, J. Ocul. Pharmacol. Ther., 2016, 32(1), 11–22 CrossRef PubMed.
- S. Gholizadeh, G. R. R. Visweswaran, G. Storm, W. E. Hennink, J. A. A. M. Kamps and R. J. Kok, E-selectin targeted immunoliposomes for rapamycin delivery to activated endothelial cells, Int. J. Pharm., 2018, 548, 759–770 CrossRef CAS PubMed.
- P. Malý, A. D. Thall, B. Petryniak, C. E. Rogers, P. L. Smith, R. M. Marks, R. J. Kelly, K. M. Gersten, G. Cheng, T. L. Saunders, S. A. Camper, R. T. Camphausen, F. X. Sullivan, Y. Isogai, O. Hindsgaul, U. H. von Andrian and J. B. Lowe, The α(1,3) Fucosyltransferase Fuc-TVII Controls Leukocyte Trafficking through an Essential Role in L-, E-, and P-selectin Ligand Biosynthesis, Cell, 1996, 86, 643–653 CrossRef.
- K. Neelu and E. T. Bert, Design and Synthesis of Sialyl Lewis x Mimics as E- and P-Selectin Inhibitors, Med. Res. Rev., 2002, 22, 566–601 CrossRef PubMed.
- A. Albini, R. Benelli, D. M. Noonan and C. Brigati, The “chemoinvasion assay”: a tool to study tumor and endothelial cell invasion of basement membranes, Int. J. Dev. Biol., 2004, 58, 563–571 CrossRef.
- Z. Tahergorabi and M. Khazaei, A review on angiogenesis and its assays, Iran. J. Basic Med. Sci., 2012, 15(6), 1110–1126 CAS.
- M. Matsuki, Y. Adachi, Y. Ozawa, T. Kimura, T. Hoshi, K. Okamoto, O. Tohyama, K. Mitsuhashi, A. Yamaguchi, J. Matsui and Y. Funahashi, Targeting of tumor growth and angiogenesis underlies the enhanced antitumor activity of lenvatinib in combination with everolimus, Cancer Sci., 2017, 108(4), 763–771 CrossRef CAS PubMed.
- L. W. Kraiss, A. S. Weyrich, N. M. Alto, D. A. Dixon, T. M. Ennis, V. Modur, T. M. McIntyre, S. M. Prescott and G. A. Zimmerman, Fluid flow activates a regulator of translation, p70/p85 S6 kinase, in human endothelial cells, Am. J. Physiol.: Heart Circ. Physiol., 2000, 278, H1537–H1544 CrossRef CAS PubMed.
- D. F. Lee, H. P. Kuo, C. T. Chen, J. M. Hsu, C. K. Chou, Y. Wei, H. L. Sun, L. Y. Li, B. Ping, W. C. Huang, X. He, J. Y. Hung, C. C. Lai, Q. Ding, J. L. Su, J. Y. Yang, A. A. Sahin, G. N. Hortobagyi, F. J. Tsai, C. H. Tsai and M. C. Hung, IKKb suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway, Cell, 2007, 130, 440–455 CrossRef CAS PubMed.
- J. Zhang, Z. Gao, J. Yin, M. J. Quon and J. Ye, S6K directly phosphorylates IRS-1 on Ser-270 to promote insulin resistance in response to TNF-α signaling through IKK2, J. Biol. Chem., 2008, 283, 35375–35382 CrossRef CAS PubMed.
- B. Magnuson, B. Ekim and D. C. Fingar, Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks, Biochem. J., 2012, 441, 1–21 CrossRef CAS PubMed.
- M. Pende, S. H. Um, V. Mieulet, M. Sticker, V. L. Goss, J. Mestan, M. Mueller, S. Fumagalli, S. C. Kozma and G. Thomas, S6K1-/-/S6K2-/- mice exhibit perinatal lethality and rapamycin-sensitive 5-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway, Mol. Cell. Biol., 2004, 24(8), 3112–3124 CrossRef CAS.
- A. Alekseeva, M. Kapkaeva, O. Shcheglovitova, I. Boldyrev, G. Pazynina, N. Bovin and E. Vodovozova, Interactions of antitumour sialyl Lewis X liposomes with vascular endothelial cells, Biochim. Biophys. Acta, 2015, 1848, 1099–1110 CrossRef CAS PubMed.
- D. D. Spragg, D. R. Alford, R. Greferath, C. E. Larsen, K. D. Lee, G. C. Gurtner, M. I. Cybulsky, P. F. Tosi, C. Nicolau and M. A. Gimbrone Jr, Immunotargeting of liposomes to activated vascular endothelial cells: a strategy for site-selective delivery in the cardiovascular system, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 8795–8800 CrossRef CAS PubMed.
- S. Kessner, A. Krause, U. Rothe and G. Bendas, Investigation of the cellular uptake of E-selectin-targeted immunoliposomes by activated human endothelial cells, Biochim. Biophys. Acta, 2001, 1514, 177–190 CrossRef CAS.
- S. Stahn, C. Grittner, R. Zeisig, U. Karsten, S. B. Felix and K. Wenzel, Sialyl Lewisx-liposomes as vehicles for site-directed, E-selectin-mediated drug transfer into activated endothelial cells, Cell. Mol. Life Sci., 2001, 58, 141–147 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra01943j |
| This journal is © The Royal Society of Chemistry 2019 |