Open Access Article
Open Access ArticleGut microbiota: a new angle for traditional herbal medicine research
Longfei Lin†
 a,
Liyu Luo†c,
Ming Zhongd,
Tanggui Xied,
Yuling Liu
a,
Liyu Luo†c,
Ming Zhongd,
Tanggui Xied,
Yuling Liu a,
Hui Li*a and
Jian Ni*b
a,
Hui Li*a and
Jian Ni*b
aInstitute Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, China
bSchool of Chinese Material Medica, Beijing University of Chinese Medicine, Beijing, China
cSchool of Pharmaceutical Science and Technology, Tianjin University, Tianjin, China
dGuangxi Key Laboratory of Traditional Chinese Medicine Quality Standards, Guangxi Institute of Chinese Medicine and Pharmaceutical Science, Nanning 530022, China
First published on 7th June 2019
Abstract
Traditional Herbal Medicine (THM) has been used for thousands of years, and is popular worldwide due to its effectiveness in a variety of diseases. THM has also formed the basis of the discovery of modern drugs like artemisinin and paclitaxel. However, at present, studies that focus on development in the field of THM are stagnant because currently, the effective ingredients in the herbal formulations and the ambiguity of the underlying mechanisms of action are unknown. In this review, we have investigated the studies available that focused on the efficacy, active ingredients and bioavailability of THM, and the function of gut microbiota in THM-mediated treatment of disease. We hypothesized that most THMs treat diseases via three mechanisms: (1) metabolizing into active metabolites by the action of gut microbiota, (2) regulation of gut microbiota balance, and (3) regulating the fermentation products of the gut microbes. Therefore, focusing on these aspects can help elucidate the pharmacodynamic constituents of THM preparations, and their therapeutic mechanisms of action.
1. The therapeutic efficacy and spectrum of THM
In China, herbs have been used for thousands of years to treat various disorders, and form the basis of a theoretical and practical system for the treatment and prevention of diseases. Traditional Herbal Medicine (THM) is a fully institutionalized part of Chinese health care that is extensively used along with Western medicine. An estimated 40% of health care in China is based on THM, and 75% of health centers have a THM department.1,2 According to the China Food and Drug Administration (CFDA), 8760 pharmaceutical manufacturers currently market THM in China, and thereby use a total of 59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027 types of herbs. THM is also widely used in other countries, and by 20% to 25% of the population in Europe.3 THM is categorized in European countries along with Asiatic medicines, and mainly includes depurative plants used for the treatment of dermatological illnesses or hay baths for cancer treatment.4 European countries spend almost $5 billion on over-the-counter herbal medicines; Germany accounted for 40% of the expenses.5 In the United States (US), alternative medicine, including herbs among others, has also gained popularity in routine health care.6 In retail pharmacy in the US, the sale of herbal medicines has grown by approximately 20% annually, with nearly 20% of the population using herbal preparations.7,8 THM has also been widely used to treat diseases, such as malaria, diabetes mellitus, and hypertensive in Africa,9,10 as well as in an Indian traditional medicinal system, such as Ayurveda, which is a comprehensive scientific medicinal system, including Charaka Samhita, Susruta Samhita, Madhava Nidana, et al.11
027 types of herbs. THM is also widely used in other countries, and by 20% to 25% of the population in Europe.3 THM is categorized in European countries along with Asiatic medicines, and mainly includes depurative plants used for the treatment of dermatological illnesses or hay baths for cancer treatment.4 European countries spend almost $5 billion on over-the-counter herbal medicines; Germany accounted for 40% of the expenses.5 In the United States (US), alternative medicine, including herbs among others, has also gained popularity in routine health care.6 In retail pharmacy in the US, the sale of herbal medicines has grown by approximately 20% annually, with nearly 20% of the population using herbal preparations.7,8 THM has also been widely used to treat diseases, such as malaria, diabetes mellitus, and hypertensive in Africa,9,10 as well as in an Indian traditional medicinal system, such as Ayurveda, which is a comprehensive scientific medicinal system, including Charaka Samhita, Susruta Samhita, Madhava Nidana, et al.11
In general, the therapeutic efficiency of THM is not supported by clinical evidence. Due to the increasing usage and popularity of THM, several studies have been undertaken in recent years to clinically evaluate THM.12 In 1986, it was showed that ginseng could improve certain psychomotor functions in healthy subjects.13 A randomized, double-blind, placebo-controlled clinical study was performed to indicate that daily consumption of Lycium barbarum for 14 days could improve neurological performance and gastrointestinal function.14 The biminne formulation is composed of 11 traditional Chinese herbs, and after 1 year of treatment was demonstrated to be safe and effective in relieving the symptoms of perennial allergic rhinitis.15 A Chinese herbal formulation improved the symptoms of irritable bowel syndrome in some patients, as proven by a randomized controlled trial in which 116 patients were enrolled.16 The efficiency of this formula was also verified by another randomized placebo-controlled trial.17 “Ba Wei Di Huang Wan’’ is a Japanese concoction of eight herbs that has successfully been used as a remedy for the elderly who present with cognitive decline and disability, and was shown to improve cognitive and physical functioning in a randomized trial in patients with dementia.18 Many studies have been performed that focus on the clinical efficacy of THM, and all of them cannot be enumerated here.
Taken together, THM is used worldwide, especially in China, and is used as an alternative medicine in the US and Europe. Although some THM preparations have shown clinical evidence, the treatment mechanism is still unclear.
2. Active ingredients or inactive ingredients?
There are hundreds of ingredients in a THM preparation, which can be classified as either active or inactive. The World Health Organization (WHO) defines active ingredients of THM as those “with therapeutic activity.” For herbal medicines with known active ingredients, their preparation should be standardized to allow for a defined amount of these ingredients, if adequate analytical methods are available. In any case it is not possible to identify active ingredients, the complete preparation may be considered as an active ingredient.19 Inactive ingredients are those without detectable physiological activity and clinical effect. Pharmaceutically active ingredients, either one or more compounds, are used to evaluate THM, including Chinese herb medicine, Chinese patent medicine, and Traditional Chinese Medicine (TCM), regarding their therapeutic activity.20 Although few active ingredients do not accurately represent the quality of TCM, the CFDA uses a single or multiple components as the quality control indicator(s) for most TCM formulations. Several techniques, including chemical fingerprinting, gas chromatography-flame ionization detection (GC-FID), and high-performance liquid chromatography-mass spectrometry, (HPLC-MS)21,22 can be used to analyze TCM composition, and help in quality control. However, due to the complex composition of THM not every component is isolated, therefore the biological activity cannot be studied. The Chinese Pharmacopoeia part one 2015 edition includes information of 2598 species used in Chinese herb medicine and Chinese patent medicine, with information on less than 1% quality indicator components (Table 1). The question arises as to whether these few indicators are fully representative of the efficacy of the herbs, and if perhaps unknown components may in fact represent the inactive ingredients. For example, the active ingredient of one of the listed herbs, Cacumen platycladi, which is used for treating epistaxis, hematemesis, hemptysis, hematochezia, abnormal uterine bleeding, hair loss, and premature greying, is quercetin. Quercetin is a flavonoid that inhibits the ferrous ion-dependent lipid peroxidation of lecithin liposomes,23 and shows antioxidant and anti-cancer effects by inhibiting protein kinases.24,25 However, to our knowledge, no association has been reported between quercetin and epistaxis, hematemesis, hemoptysis, and hematochezia. Furthermore, the recommended dose of C. platycladi is 12 g, with an active dose of quercetin 12 mg (0.1%), which may or may not be the clinically effective dose.| Traditional herbal medicine | Dosage per day | Quality indicator component | Content limit ≥ |
|---|---|---|---|
| Radix ginseng | 3–9 g | Ginsenoside Rb1 | 0.18% |
| Ginsenoside Rg1, Ginsenoside Re | Sum 0.27% | ||
| Schisandra chinensis fructus | 2–6 g | Schisandrin | 0.40% |
| Plantaginis semen | 9–15 g | Geniposide acid | 0.40% |
| Verbascoside | 0.30% | ||
| Radix scrophulariae | 9–15 g | Harpagide, harpagoside | Sum 0.45% |
| Cassiae semen | 9–15 g | Chrysophanol | 0.12% |
| Aurantio-obtusin | 0.080% | ||
| Radix saposhnikovia | 5–10 g | Prim-o-glucosylcimifugin, 5-o-methyl Weiss Amie alcohol glycoside | Sum 0.24% |
| Forsythiae fructus | 6–15 g | Forsythin | 0.15% |
| Forsythoside A | 0.25% | ||
| Lonicerae japonicae caulis | 9–30 g | Chlorogenic acid | 0.070% |
| Loganin | 0.10% | ||
| Cacumen platycladi | 6–12 g | Quercetin | 0.10% |
| Radix rubiae | 6–10 g | Rubimaillin | 0.20% |
| Purpurin | 0.080% | ||
| Curcumae longae rhizoma | 3–10 g | Curcumin | 0.90% |
| Radix astragali | 9–30 g | Astragaloside IV | 0.040% |
| Genistein glucose | 0.020% |
Patent herbal medicine is produced after extraction, concentration (refinement), and drying of THM, with higher daily dosage requirement compared to Western medicine. Ganmao Qingre granules, also described in the Chinese Pharmacopoeia, are used to treat colds, fever, cough, and pharyngalgia. Its dosage is described as “oral or boiled in water, 12 g twice a day”, and its active ingredient is puerarin, which is present at only one thousandth (∼10 mg) of the final formulation (Table 2). The pharmacological properties of puerarin include vasodilation, cardio-protection, neuroprotection, anti-cancer, anti-inflammation, analgesia, bone formation, inhibition of alcohol intake, inhibition of oxidative stress and apoptosis, and attenuation of insulin resistance.26,27 Several studies on puerarin have indicated that alleviating colds is not its main pharmacological effect. Taken together, most of the components in a patent medicine are still unknown, and therefore the main therapeutic substances and their pharmacological effects remain unclear.
| Name | Indication | Specification | Dosage | Quality indicator component | Content limit ≥ |
|---|---|---|---|---|---|
| Ganmao Qingre granules | Colds, fever, cough, and pharyngalgia | 12 g per pouch | Once pouch and twice a day | Puerarin | 10 mg per pouch |
| Huoxiang Zhengqi water | Heatstroke, gastrointestinal cold | 10 mL per bottle | 5–10 mL per time and twice a day | Hesperidin | 0.18 mg mL |
| Niuhuang Shangqing tablets | Headache, dizziness, sore throats, aphtha, swollen gums, constipation | 0.265 g per tablets | 4 Tablets per time, two times a day | Baicalin | 1.1 mg per tablet |
| Jasminoidin | 0.45 mg per tablet | ||||
| Weikang Ling capsules | Acute and chronic gastritis, duodenal ulcer, gastrohelcosis, gastrorrhagia | 0.4 g per capsule | 4 Capsules and three times a day | Paeoniflorin | 1.0 mg per capsule |
| Hugan tablets | Chronic hepatitis and early cirrhosis, reduced aminotransferase | 0.36 g per tablet | 4 Tablets per time, 3 times a day | Schisandrin | 0.28 mg per tablet |
| Lidan Paishi granules | Cholecystitis, gallstone | 3 g per pouch | Removing urinary calculus: two pouchs per time, twice a day; diminish inflammation: one pouch per time, twice a day | Emodin | 3.0 mg per pouch |
| Fufang Danshen tablets | Coronary disease, angina pectoris | (1) 0.32 g per tablet | 3 Tablets (1, 2) or 1 tablet (3) per time, 3 times a day | Ginsenoside Rg1, Rb1, Re, and notoginsenoside R1 | (1, 2): Sum 6.0 mg per tablet |
| (2) Sugar-coated tablet | (3): Sum 18.0 mg per tablet | ||||
| (3) 0.8 g per tablet |
3. Low bioavailability of THM
THM formulations are complex, often composed of hundreds or even thousands of ingredients, such as flavonoids, alkaloids, saponins, anthraquinones, tannins, lignin, and terpenoids. The active ingredients in THM have low absorption and bioavailability via the oral route, which is significant since the oral route is the usual mode of THM administration. The main reasons for their low bioavailability include the following:28 (1) low levels of the active ingredient due to the relatively crude extraction and purification process. (2) Poor solubility, lipid solubility, permeability, and gastrointestinal stability, and improper molecular size of the active substances. For example, water soluble phenolics are unable to cross the lipid membranes of intestinal cells,29 and molecules larger than 0.4 nm in diameter have difficulty in passing through the aqueous channels in the intestinal epithelium.29 (3) Drug–drug interactions (DDIs) of individual THM components, which are essential for its therapeutic effects.30 These components may be dual or multi substrates of drug efflux transporters, such as P-glycoprotein (P-gp), multidrug resistance associated protein (MRP), cytochrome P450 (CYP450) and others.31–33 Thus, these specificity multi substrates of P-gp, MRP and CYP450 might be results of the low bioavailability of THM.Given its successful use worldwide, evaluation of the pharmacokinetics and bioavailability of THM is critical for assessing their safety and efficacy.34 Sophisticated clinical pharmacokinetic and bioavailability evaluations in recent years (Table 3.)35–42 indicated that most THM are not absorbed via the circulation. With increasing evidence implicating gut microbiota in human health and disease, it is highly likely that the therapeutic effects of THM are related to the gut microbiota.
| Medicinal | Dosage | Samples | Target compound | Contents in extract or dosage | Tmax (h) | Cmax (ng mL−1) | T1/2 (h) | AUC (ng h mL−1) |
|---|---|---|---|---|---|---|---|---|
| a “Tmax”: peak time of component concentration in vivo; “Cmax”: maximum concentration; “T1/2”: eliminate half-time; “AUC”: area under the curve. | ||||||||
| Rhei undulati rhizoma | 100 mg kg−1 | 12 Healthy volunteers (9 men/3 women | Rhein | 3.3759 ± 0.335 | 9746.689 ± 1011.72 | |||
| Rhubarb | 50 mg kg−1 | Eight male volunteers | Rhein | 12.308 mg g−1 | 59.69 ± 28.63 min | 2.79 ± 1.02 μg mL | 199.66 ± 63.50 min | 18.64 ± 5.67 μg |
| Milk thistle (Silybum marianum) | 600 mg milk thistle extracts | 3 Healthy volunteers | Silychristin | 22.8% | 3.7 ± 0.6 h | 37 ± 7.8 | 7.9 ± 1.7 | 401 ± 93 |
| Silydianin | 8.93% | 2.8 ± 1.3 | 21 ± 3.1 | 3.3 ± 0.6 | 140 ± 47 | |||
| Silybin A | 22.0% | 1.0 ± 0.4 | 23 ± 7.9 | 1.3 ± 0.5 | 63 ± 38 | |||
| Silybin B | 34.0% | 1.3 ± 0.3 | 131 ± 28 | 3.4 ± 0.8 | 647 ± 234 | |||
| Isosilybin A | 8.59% | 1.7 ± 0.3 | 113 ± 30 | 4.1 ± 1.3 | 799 ± 403 | |||
| Isosilybin B | 3.67% | 1.5 ± 0.0 | 59 ± 16 | 4.0 ± 1.1 | 383 ± 195 | |||
| Artichoke leaf extracts | 2.4 g | 14 Healthy volunteers | Caffeic acid | 107.0 mg | 0.83 ± 0.3 | 6.51 ± 1.89 | 3.08 ± 1.53 | 23.02 ± 7.15 |
| 0.625 g | 14 Healthy volunteers | Caffeic acid | 153.8 mg | 0.94 ± 0.47 | 8.04 ± 2.64 | 2.69 ± 1.80 | 26.32 ± 10.69 | |
| Geriaforce™ tincture | 90 drops, 2.73 mL | 6 Healthy volunteers | Bilobalide | 646.93 μg | 1.09 ± 0.46 | 6.17 ± 5.67 | 2.33 ± 1.04 | 26.53 ± 26 |
| Ginkgolide A | 298.14 μg | 0.92 ± 0.38 | 3.81 ± 0.96 | 2.31 ± 0.66 | 12.42 ± 5.27 | |||
| Ginkgolide B | 147.45 μg | 0.88 ± 0.38 | 1.45 ± 0.98 | 2.34 ± 0.81 | 6.17 ± 3.77 | |||
| Ginkgo fresh plant extract tablets | 4 Tablets | 6 Healthy volunteers | Bilobalide | 1974.96 μg | 0.88 ± 0.3 | 11.6 ± 3.77 | 2.52 ± 1.7 | 41.56 ± 31.94 |
| Ginkgolide A | 881.52 μg | 0.80 ± 0.34 | 7.27 ± 3.83 | 2.66 ± 2.05 | 29.75 ± 29.64 | |||
| Ginkgolide B | 524.56 μg | 2.35 ± 2.27 | 4.61 ± 2.87 | 4.01 ± 2.58 | 43.48 ± 27.64 | |||
| EGb 761™ | 3 Tablets | 6 Healthy volunteers | Bilobalide | 3672.39 μg | 1.06 ± 0.4 | 29.87 ± 18.42 | 2.08 ± 0.23 | 100.57 ± 38.91 |
| Ginkgolide A | 1571.37 μg | 0.93 ± 0.43 | 16.97 ± 5.79 | 3.6 ± 2.96 | 80.83 ± 57.47 | |||
| Ginkgolide B | 836.46 μg | 2.0 ± 1.63 | 10.9 ± 5.59 | 3.94 ± 1.42 | 71.73 ± 26.98 | |||
| Cardiotonic pills | 30 Pills | 6 Male healthy volunteers | Tanshinol | 160 μg per pill | 1.33 ± 0.52 | 19.3 ± 5.1 ng | 1.16 ± 0.42 | 48.5 ± 3.4 |
| 6 Female healthy volunteers | Tanshinol | 160 μg per pill | 1.17 ± 0.68 | 31.3 ± 10.6 | 1.27 ± 0.21 | 77.6 ± 31.6 | ||
| Safflower oral solution | Single dose | 12 Healthy volunteers | Hydroxysafflor yellow A | 140 mg | 1 h | 73.5 ± 8.7 | 2.6–3.5 | |
| American ginseng powder | 10 g | 6 Healthy volunteers | Ginsenoside Rb1 | 31.7 mg g−1 | 4 h | 19.90 ± 5.43 | 155.0 ± 19.5 | |
4. Gut microbiota in human health and disease
The gut microbiome is very important for human health and is often considered a “hidden organ”. Recent estimates have suggested that the microbes residing in or on human body slightly exceed the number of human cells, which is approximately 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.43–45 The total number of genes encoded by the gut bacteria is at least 150 times that of the human genome.46,47 The ensemble of genetic material retrieved from the gut known as the “microbial group”, is called the “gut meta-genome”.48 The human body can be considered a “super-organism” made of both eukaryotic cells and symbiotic microbes.49 Under normal circumstances, human tissues and bacteria interact and maintain a functional balance, in addition to forming biological, chemical, mechanical, and immune barriers against invading pathogens.50 The gut meta-genome and the host genome influence pathophysiological conditions and the drug metabolism.51,52 In 2007, the National Microbiology Program was launched by the National Institutes of Health (NIH) for human health assessment and surveillance, and early diagnosis and treatment of chronic diseases.46,48 Previous studies have shown that the gut microbiota closely interact with the gastrointestinal tract, liver, skin, and central nervous system among other organs, and aid in physiological functions, such as digestion and absorption of nutrients, neurodevelopment and transmission, fat metabolism, and immune responses.53–56 Therefore, any disturbance in the gut microbiota may lead to various disorders, including metabolic diseases (obesity, diabetes), cardiovascular disease (hypertension), and mental disorders (depression, anxiety, cognitive decline, Parkinson's Disease).57–62
1.43–45 The total number of genes encoded by the gut bacteria is at least 150 times that of the human genome.46,47 The ensemble of genetic material retrieved from the gut known as the “microbial group”, is called the “gut meta-genome”.48 The human body can be considered a “super-organism” made of both eukaryotic cells and symbiotic microbes.49 Under normal circumstances, human tissues and bacteria interact and maintain a functional balance, in addition to forming biological, chemical, mechanical, and immune barriers against invading pathogens.50 The gut meta-genome and the host genome influence pathophysiological conditions and the drug metabolism.51,52 In 2007, the National Microbiology Program was launched by the National Institutes of Health (NIH) for human health assessment and surveillance, and early diagnosis and treatment of chronic diseases.46,48 Previous studies have shown that the gut microbiota closely interact with the gastrointestinal tract, liver, skin, and central nervous system among other organs, and aid in physiological functions, such as digestion and absorption of nutrients, neurodevelopment and transmission, fat metabolism, and immune responses.53–56 Therefore, any disturbance in the gut microbiota may lead to various disorders, including metabolic diseases (obesity, diabetes), cardiovascular disease (hypertension), and mental disorders (depression, anxiety, cognitive decline, Parkinson's Disease).57–62
The gut microbiota is thought to be comprised of more than 1000 various microbial species. Due to various reasons, the microbial composition can alter throughout their lifespan, therefore, its composition is not static and changes to its structure are dynamic, and overall gut enterotypes may vary from person to person.46,63 The Meta-genomics of Human Intestinal Tract (MetaHIT)64 study identified nine bacteriophyta in the human intestine, including Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Fusobacteria, Verrucomicrobia, Cyanobacteria, Spirochaetes, and VadinBE97.65–67 Based on their relationship with the host, gut microbiota can be divided into the following three categories: (1) physiological bacteria that are beneficial to the host, including Bifidobacterium and Lactobacillus, (2) conditional pathogen or neutral bacteria, including Enterococcus and Escherichia coli, and (3) pathogenic bacteria, including Proteus, Pseudomonas, and Staphylococcus aureus.68 Gut microbiota disorders can lead to metabolic disorders of carbohydrates, lipids, and other substances in the host, and thereby enhance inflammatory reactions and oxidative stress in the body, resulting in promoting the occurrence or deterioration of cardiovascular diseases.69–71 Gut microbiota can participate in bidirectional regulation of intestinal and central nervous system (CNS) through neurotransmitter, endocrine, immune and metabolic pathways. When the gut microbiota is disordered, it can affect the development of CNS diseases, including cerebral ischemia, Parkinson's disease, Alzheimer's disease, disseminated sclerosis, hepatic encephalopathy, and psychogenia.72–74 The intestinal epithelial system is the natural barrier of intestinal exogenous pathogens. In addition, gut microbiota disorders can increase intestinal permeability, and promote intestinal pathogens and intestinal toxins to enter the liver through the portal system, then will cause a series of pathophysiological reactions. The mechanism of action involved alterations in gut epithelial permeability, choline metabolism, endogenous alcohol production, release of inflammatory cytokines, regulation of hepatic Toll-like receptor (TLR), and bile acid metabolism.75,76 In addition, gut microbiota is also associated with a variety of diseases, such as diabetes, obesity, rheumatoid arthritis, inflammatory bowel disease (Fig. 1 and Table 4).
| Body system | Disease | Related gut microbiota | References |
|---|---|---|---|
| Metabolic system | Hepatocirrhosis | Veillonella atypica, Streptococcus salivarius | 77 |
| Liver cancer | Helicobacter hepaticus | 78 | |
| Type 2 diabetes | Akkermansia muciniphila | 79 | |
| NAFLD | Lachnospiraceae bacterium 609, Barnesiella intestinihominis | 80 | |
| Obesity | Enterobacter cloacae B29 | 81 | |
| Alimentary system | Diarrhea | Escherichia/Shigella, Granulicatella | 82 |
| Irritable bowel syndrome | Veillonella, Lactobacillus, Prevotella, Bacteroides | 83 and 84 | |
| Inflammatory bowel disease | Roseburia, Enterobacteriaceae, Ruminococcaceae, Campylobacter, Salmonella | 83 and 85 | |
| Colorectal cancer | Bacteroides fragilis, Streptococcus gallolyticus | 86 and 87 | |
| Paradentitis | Porphyromonas gingivalis | 88 and 89 | |
| Immune system | Rheumatic arthritis | Prevotella copri, Lactobacillus, Tropheryma whipplei | 90 and 91 |
| Systemic lupus erythematosus | Enterobacteriaceae, Firmicutes/Bacteroidetes | 92 and 93 | |
| Respiratory system | Asthma | Aspergillus fumigatus | 94 |
| Chronic bronchitis | Clostridium perfringens, Enterobacter, Enterococcus | 95 | |
| Circulatory system | Atherosclerosis | Collinsella, Eubacterium, Roseburia | 96 |
| Hyperlipemia | Lactobacillus | 97 | |
| Hypertension | Bacteroides acidifaciens | 98 | |
| Nervous system | Depression | Spore-forming bacteria | 99 |
| Anxiety neurosis | Campylobacter jejuni, Citrobacter rodentium | 100 and 101 | |
| Autism | Bacteroides fragilis | 102 | |
| Parkinson's disease | Enterobacteriaceae, Escherichia coli | 103 | |
| Multiple sclerosis | Methanobrevibacter, Akkermansia | 104 | |
| Alcohol dependence | Ruminococcus, Faecalibacterium, Subdoligranulum, Oscillibacter, Anaerofilum, Dorea, Blautia | 105 |
5. Therapeutic effects of THM through gut microbiota
In the circulation after oral administration, the concentration of active THM components is usually low. This opens the possibility of THM acting via intestinal absorption, with considerable influence on the gut microbiota.5.1 Metabolic effect of gut microbiota on THM
β-D-glucosidase mainly plays a role in cleavage of β-glycoside bonds (β-1,1; β-1,2; β-1,3; β-1,6), and due to its poor specificity, the C–O glycoside bond, C–S bond, C–N bond, and C–F bond can also be cleaved by β-D-glucosidase. β-D-glucosidase is involved in the glycolysis pathway and is highly expressed in Bifidobacteria and Bacteroides thetaiotaomicron.106,107 β-D-glucuronidase, an important phase II metabolic enzyme, cleaved the β-1,4-glucuronide bond, and its activity was only observed in Firmicutes, such as Lactobacillus, Enterococcus, Clostridium paraputrificum, B. fragilis, and B. uniformis.108 β-Xylosidase is an exonuclease that mainly hydrolyzes xyloside, which could be hydrolyzed to xylooligosaccharide from the non-reducing end in an exo-cutting manner, and the hydrolyzed product is xylose. β-Xylosidase exists in Bifidobacterium, Clostridium stercorarium, and Thermoanaerobacterium saccharolyticum.109–111 β-Galactosidase can catalyze and hydrolyze lactose to produce glucose and galactose, and catalyzes the galactosyl transfer reaction to form galactooligosaccharides. The activity of β-galactosidase is closely related to the composition of the gut microbiota, and the higher the activity of β-galactosidase when probiotics were dominantly present in intestinal flora.112 β-Galactosidase is mainly produced by probiotics, such as Bifidobacterium, Lactobacillus acidophilus, and Streptococcus thermophiles.112,113 Nitroreductase is an NAD(P)H-dependent flavoenzyme, which can reduce the nitrogroup into amine or amino, and catalyze the reduction reaction of nitroaromatic compounds into aromatic amine. The metabolites can be used as a nitrogen source for bacterial growth. The activity of nitroreductase can be found in various intestinal bacteria, such as Lactobacillus, Bifidobacterium, and Escherichia coli.114–116
In addition to the above-mentioned enzymes, many other metabolic enzymes are present in the gut, including α-L-rhamnosidase, azoreductase, and mannosidase. Most of these are produced by intestinal probiotics, such as Bifidobacterium, and produce α/β-galactosidase, α/β-glucosidase, β-fructofuranosidase, mannosidase, D-xylanase, D-xylose enzyme.113 Bifidobacterium has been shown to adjust intestinal dysfunction, improve human immunity, inhibit pathogen growth, and have anti-inflammatory, and anti-aging effects.117,118 Bacteroides thetaiotaomicron is one of the largest bacteria in the gut that can express β-glucosidase, β-galactosidase, β-fructofuranosidase, β-mannosidase, α-mannanase, and arabinase.119
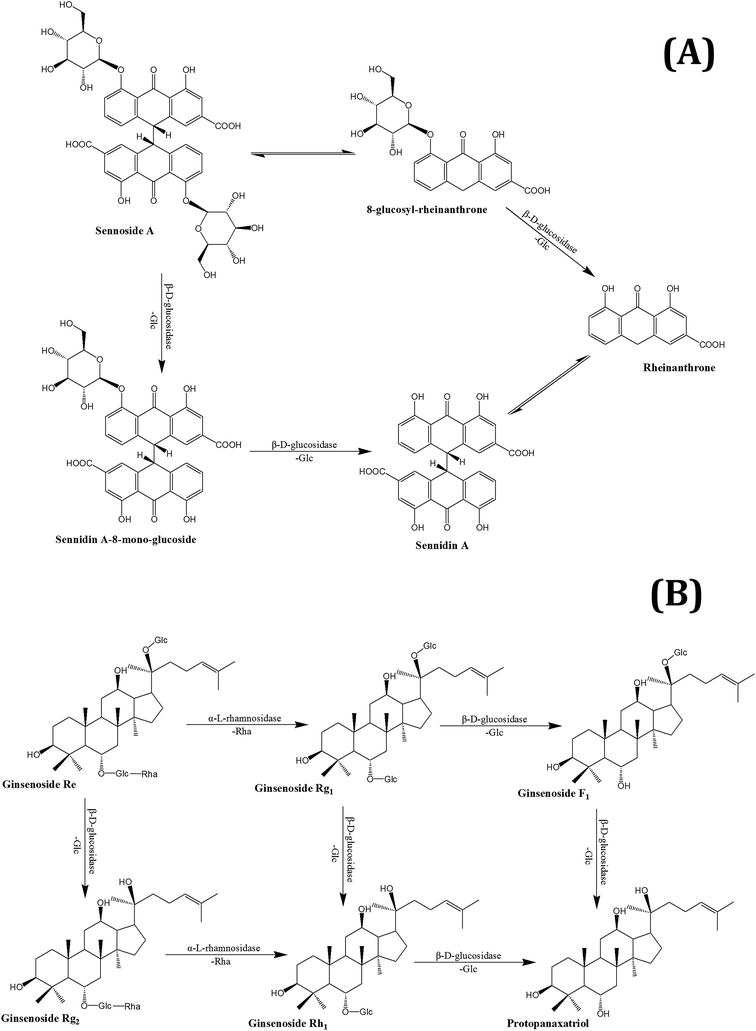
Chemical constituents in THM are degraded and metabolized in the gut by the above-mentioned enzymes. Rutin has two metabolic pathways, which mainly work through Bifidobacterium in the gut; one was first hydrolyzed into isoquercetin by the enzyme α-L-rhamnosidase, and further hydrolyzed to quercetin by β-D-glucosidase. The other was converted to leucocyanidin under the action of α-L-rhamnosidase and β-D-glucosidase.120,121 Under the action of α-L-rhamnosidase and β-D-glucosidase that are expressed by gut microbiota, ginsenoside Re can be converted into Rh1, F1, and a small amount of protopanaxadiol. There are also two metabolic pathways of sennoside in the intestinal flora. The first involves the hydrolysis of senna into senna glycoside by β-D-glucosidase, which is then reduced to form rheinanthrone, the other involves reduction of sennoside to 8-glucosyl-rheinanthrone, which is then hydrolyzed by β-D-glucosidase to rheinanthrone.122 Geniposide is hydrolyzed by β-D-glucuronidase of intestinal bacteria to genipin.123 Paeoniflorin produced deglucose glucosides, and 7S- and 7R-paeonimetabolines I and II under the action of β-D-glucosidase, β-D-glucoesterase and other enzymes, which are produced by Bacteroides fragilis, Peptostreptococcus, and Lactobacillus brevis.124,125 The metabolic pathways of several THM compounds in the gut are shown in Fig. 1.
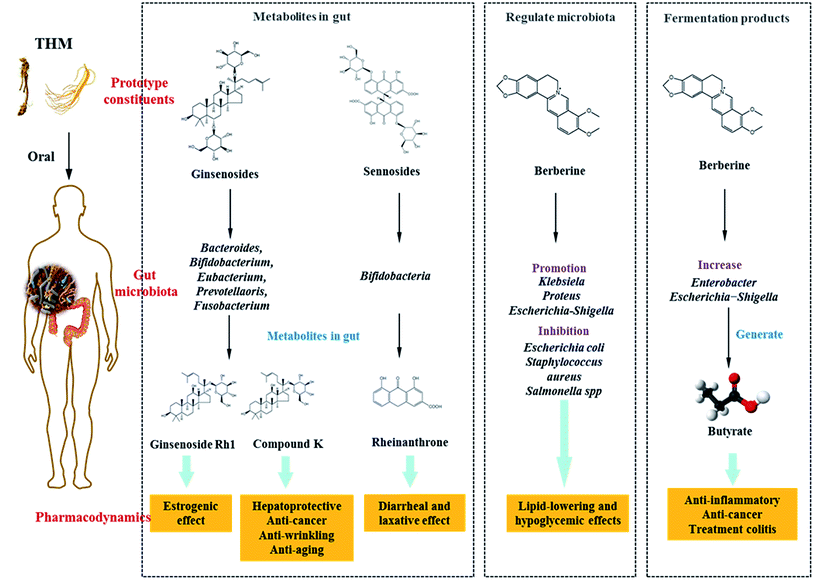
Saponins: Saponins have antipyretic, sedative, anti-cancer and other pharmacological effects, and are commonly present in ginseng, Panax notoginseng and Bupleurum. Ginsenosides are the main active constituents of P. ginseng, and are metabolized to compound K (20-O-(b-D-glucopyranosyl)-20(S)-protopanaxadiol) by gut microbiota before absorption into the circulation. Compound K has hepato-protective, anti-cancer, anti-wrinkling, and anti-aging properties, and is more potent than the parental ginsenosides.128–130 Ginsenosides can be metabolized by Bacteroides JY-6, Bifidobacterium K506, Eubacterium A-44, Prevotella oris, and Fusobacterium K-60.131,132 Ginsenoside Re can be converted into Rh1, which exerts a strong estrogenic effect. Ginsenoside Rh1 increased the proliferation of MCF-7 cells 2.1-fold at a concentration of 1 mM, however levels of ginsenoside Re were not significantly increased when compared with the blank control group.133 Furthermore, Ardipusilloside-I is a triterpenoid saponin isolated from Ardisia pusilla DC, and has potent anti-tumor activity. Ardipusilloside-I is de-glycosylated into four compounds by the action of bacterial enzymes, two of them have the same anti-tumor capacity as Ardipusilloside-I.134 Glycyrrhizin is difficult to be absorbed in the intestine. It interacts with intestinal bacteria to produce glycyrrhetic acid, which was absorbed by the body and showed stronger anti-hepatotoxicity activity when compared with glycyrrhizin.135
Flavonoids: Flavonoids are the active ingredients of various THM formulations. Baicalin, a flavone glycoside, can be de-glycosylated into baicalein or methylated into aglycon orohylin A by gut microbes. Baicalein and orohylin A have been shown to improve antihistamine-induced pruritus responses in mice when compared to baicalin, and compared to baicalin inhibited the contraction of guinea pig ileum induced by histamine (with 50% inhibitory concentration of 0.28 mmol L−1).136 Poncirin is the active constituent of Poncirus trifoliate, and is metabolized by gut microbiota to ponciretin both in vitro and in vivo. The anti-inflammatory effect of ponciretin is superior to that of poncirin, and can attenuate colitis by inhibiting lipopolysaccharide (LPS) binding to TLR4 on macrophages, and restore the Th17/Treg balance.137 The anti-inflammatory and anti-tumor effects of lancemaside A (also has β-glycosidic bonds and α-rhamnoside bond in the structure), the active constituent of Codonopsis pilosula, might be due to echinocystic acid, which is a metabolite produced by the action of gut microbiota.138 Codonolaside II, echinocystic acid 28-O-b-D-xylopyranosyl-(1,4)-a-L-rhamnopyranosyl-(1,2)-a-L-arabinopyranosyl ester, and echinocystic acid 28-O-a-L-rhamnopyranosyl-(1,2)-a-L-arabinopyranosyl ester are other products that were produced by glycosidic bond hydrolysis.139 In a previous study, it was shown that echinocystic acid potently inhibited acetylcholinesterase activity, and significantly reversed scopolamine-induced memory and learning deficits on passive avoidance tasks. Echinocystic acid more potently shortened the escape latencies prolonged by treatment with scopolamine when compared to lancemaside A in a Morris water maze task.140
Sennoside: Sennoside is the main active ingredient in rhubarb, and has poor diarrhea effect due to it poor intestinal absorption. Sennoside glycoside (rheinanthrone), which is produced by hydrolysis of sennoside by the Bifidobacteria secreted β-D-glucosidase, has diarrheal effects.122 Rheinanthrone reflects the laxative effect by stimulating the activation of macrophages and reducing the expression of colonic aquaporin-3.141 Similarly, barbaloin can be hydrolyzed to aloe-rhein anthrone by the action of Eubacterium spp. BAR, which has a pronounced diarrheal effect.142
Others: Rhapontin is metabolized into rhapontigenin by intestinal bacteria, which has stronger antithrombotic and anti-allergic effects when compared to its parental compound.143 Geniposide is hydrolyzed into genipin by intestinal bacteria, which have cholagogue and anti-hepatitis effects, however an intravenous injection of geniposide does not have a cholagogue effect.123
Gut microbiota not only increase the pharmacological activity of active THM ingredients, but also reduce the toxicity of certain ingredients. For example, aconitine is decarboxylated, methylated, hydroxylated, and esterified by intestinal bacteria into mono-, di-, and lipid alkaloids, and metabolites, which have significantly less toxicity but the same pharmacological effect when compared to the parental compound.144 In addition, some active THM ingredients, including ginsenoside Rb1, crocin, amygdalin, geniposide, puerarin, ginsenoside Re, poncirin, hesperidin, and baicalin are metabolized by the gut microbiota to compound K, crocetin, benzaldehyde, genipin, daidzein, ginsenoside Rh1, ponciretin, hesperetin, and baicalein, which have a more potent anti-tumor effect compared to the parental compounds.145 The pharmacodynamics of part of the prototype compounds in THM and their metabolites in the gut are presented in Table 5.
| Prototype compounds in THM | Metabolites in gut | The efficacy of metabolites superior to prototypes |
|---|---|---|
| Ginsenosides/ginsenoside Re | Compound K/ginsenoside Rh1 | Hepato-protective, anti-cancer, anti-wrinkling, and anti-aging properties/estrogenic effect |
| Glycyrrhizin | Glycyrrhetic acid | Anti-hepatotoxicity |
| Lancemaside A | Echinocystic acid | Anti-inflammatory, anti-tumor and anti-Alzheimer's disease |
| Baicalin | Baicalein, orohylin A | Anti-histamine-induced pruritus response |
| Poncirin | Ponciretin | Anti-inflammatory |
| Sennosides | Rheinanthrone | Diarrheal and laxative effect |
| Barbaloin | Aloe-rheinanthrone | Diarrheal effect |
| Rhapontin | Rhapontigenin | Antithrombotic and anti-allergic effects |
| Geniposide | Genipin | Cholagogue effects |
O-glycoside bond compounds: Oxyglucosides are one of the most common compounds present in THM, and include oxyglucosides, glucosinolates, and azaglycosides, which are characterized by glycosidic bonds. Glycosides are highly susceptible to hydrolysis, and glycosidic bond cleavage results in the formation of aglycones. Water-soluble psoralen and isopsoralen glycosides can be hydrolyzed into psoralen and isopsoralen by the gut microbiota.146 Hattori et al. showed that paeoniflorin is primarily metabolized into paeonian I, followed by paeoniins II, III, and IV by Peptostreptococcus anaerobius and Lactobacillus.124,125,147 Arctiin is the major lignan glycoside of Fructus arctii, and is hydrolyzed by Blautia spp. AUH-JLD56 into 3′-desmethylarctigenin.148 Loganin is an important constituent of Fructus corni, and is metabolized by intestinal bacteria into log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and log
1 and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, which have immuno-modulatory, anti-shock, and anti-arrhythmia functions.149 Astilbin, the active ingredient of Smilax glabra Roxb, exhibits a variety of pharmacological activities, including anti-inflammation, anti-cancer properties. It is metabolized by intestinal bacteria into two main bioactive metabolites that are identified as eriodictyol and quercetin.150 Myricitrin is an important botanical flavonol glycoside with significant anti-inflammatory and antioxidant properties. It is de-glycosylated and de-hydroxylated into quercetin-3-O-rhamnoside and quercetin, respectively.151 Ziyuglycoside I is one of the major active ingredients in Sanguisorba officinali with free radical scavenging and elastase inhibitory activities. The gut microbiota degrade ziyuglycoside I into M0-Glu-Ara+O, M0-Ara, M0-Glu-COOH, M0-Glu, M0-Glu-Ara+O and M0-Ara+H2O.152 Rutin, the active ingredient of Flos sophorae, is metabolized by Eubacterium ramulus into quercetin. Reduction and hydrogenation of quercetin into alphitonin, followed by oxidation cleaved generated 3,4-dihydroxyphenylacetic acid and phloroglucinol.153,154 Moreover, studies have shown that α-L-rhamnosidase, a rhamno-glycoside metabolizing enzyme that is secreted by Bifidobacterium dentium, and can hydrolyze rutin, poncirin, naringin, and ginsenoside Re. It is more effective in hydrolyzing (1 → 6) bonds than (1 → 2) bonds of rhamnoglycosides.155
2, which have immuno-modulatory, anti-shock, and anti-arrhythmia functions.149 Astilbin, the active ingredient of Smilax glabra Roxb, exhibits a variety of pharmacological activities, including anti-inflammation, anti-cancer properties. It is metabolized by intestinal bacteria into two main bioactive metabolites that are identified as eriodictyol and quercetin.150 Myricitrin is an important botanical flavonol glycoside with significant anti-inflammatory and antioxidant properties. It is de-glycosylated and de-hydroxylated into quercetin-3-O-rhamnoside and quercetin, respectively.151 Ziyuglycoside I is one of the major active ingredients in Sanguisorba officinali with free radical scavenging and elastase inhibitory activities. The gut microbiota degrade ziyuglycoside I into M0-Glu-Ara+O, M0-Ara, M0-Glu-COOH, M0-Glu, M0-Glu-Ara+O and M0-Ara+H2O.152 Rutin, the active ingredient of Flos sophorae, is metabolized by Eubacterium ramulus into quercetin. Reduction and hydrogenation of quercetin into alphitonin, followed by oxidation cleaved generated 3,4-dihydroxyphenylacetic acid and phloroglucinol.153,154 Moreover, studies have shown that α-L-rhamnosidase, a rhamno-glycoside metabolizing enzyme that is secreted by Bifidobacterium dentium, and can hydrolyze rutin, poncirin, naringin, and ginsenoside Re. It is more effective in hydrolyzing (1 → 6) bonds than (1 → 2) bonds of rhamnoglycosides.155
C-glycoside bond compounds: The C-glycoside bonds of pueraria isoflavones (active ingredient of Kudzu root) are difficult to hydrolyze. The CG19-1 strain of Corynebacterium isolated from human intestine specifically hydrolyzes the C-glycoside bonds of puerarin, and other aromatic compounds.156
Others: Procyanidins are common polyphenols present in many THM, and exhibit antibacterial, antioxidant, antiviral and anti-inflammatory properties. A-type and B-type procyanidins are metabolized to benzoic acid, 2-phenylacetic acid, and 3-phenylpropionic acid, respectively, by gut microbiota.157 Ellagitannin, a polyphenol isolated from Polygonum capitatum with potential antitumor activity, is metabolized into thirteen metabolites by the intestinal bacteria in vitro.158 In addition, Eubacterium ramulus SY8519 dehydrodaidze into O-demethylandrostatin.159
Taken together, although the activities of the above-mentioned metabolites have not yet been confirmed, gut microbiota can certainly alter the composition of THM in the gut.
5.2 THM regulate gut microbiota balance
Under normal circumstances, the intestinal microflora maintains a symbiotic or antagonistic relationship with the host, and constitutes a micro-ecological balance in the human body. If this balance is disturbed during pathophysiological conditions, dysbiosis may occur. Therefore, the physiological relationship between normal flora and host is transformed into a pathological relationship. Intestinal dysbiosis is largely the result of the overuse of antibiotics, and is associated with various diseases of the digestive system, allergies, and immune disorders among others.In previous studies, it has been shown that several THM and its active compounds have prebiotic-like effects that promote the proliferation of intestinal bacteria. Berberine, the active ingredient of Rhizoma coptidis, prevents high-fat diet-induced obesity and diabetes. Berberine also exerts a significant antimicrobial activity by inhibiting the assembly function of FtsZ and halting bacterial division. Berberine improves gut microbiota composition in rats, promotes the growth of probiotics and production of short-chain fatty acids, lowers the intestinal pH and the accumulation of exogenous antigens, increases the levels of lipopolysaccharide-binding protein and leptin, reduces serum adiponectin, and exerts lipid-lowering and hypoglycemic effects.160,161 Furthermore, Berberine significantly enriches the genera of Klebsiella, Proteus, Escherichia, and Shigella of the Enterobacteriaceae family in gut microbiota, and inhibits the growth of Lactobacillus in vitro.162,163 Astragali Radix is a THM, which is known for its immunogenic effect. Its active ingredient is calycosin-7-O-glucoside, which is hydrolyzed to aglycone calycosin by gut microbiota in vitro. Calycosin has a higher permeability when compared with calycosin-7-O-glucoside, although the parental compound stimulates the growth of Lactobacillus and Bifidobacterium.164
Extracts of THM or its active compounds also have regulatory effects on the gut microbiota. THM polysaccharides can restore the homeostasis of gut microbiota and promote the absorption of small molecules. Ginseng polysaccharides in the Du-Shen-Tang formulation have shown to improve intestinal metabolism and absorption of certain ginsenosides and restore perturbed gut microbiota, and enhanced the growth of Lactobacillus spp. and Bacteroides spp.165 Saponins and polysaccharides of Polygonatum kingianum prevented the increase in fasting blood glucose levels and enhanced the amount of fasting insulin, which may improve intestinal micro-ecology by decreasing the abundance of Bacteroidetes and Proteobacteria, thereby increasing that of Firmicutes.166 Ganoderma lucidum extracts added to a high-fat mouse diet not only reduced the ratio of Firmicutes/Bacteroidetes and endotoxin-producing bacteria induced by the high-fat diet, but also maintained the integrity of the intestinal barrier, and reduced endotoxemia.167,168 In addition, extracts of ginseng and Semen coicis have shown to improve intestinal dysbiosis and symptoms of colonic inflammation, as demonstrated in an ulcerative colitis model induced by trinitrobenzene sulfonic acid. Furthermore, these extracts also promoted the growth of probiotics including Lactobacillus and Bifidobacterium, and inhibited the growth of pathogens, such as E. coli, S. aureus and Salmonella spp., which was verified in vitro.169 Chinese herbs and herbal extracts, such as polysaccharides from the seed of Plantago asiatica L. and Dendrobium candidum could treat diabetes mellitus by regulating the gut microbiota.170 Furthermore, Ulmus rubra and triphala significantly increased the relative abundance of butyrate-producing bacteria, and Glycyrrhiza glabra was increased in propionate-producing species.171
5.3 Gut microbiota fermentation products are influenced by THM
The major gut fermentation products in healthy adults involve gases and organic acids, in particular the three short-chain fatty acids (SCFAs) acetate, propionate, and butyrate, which have anti-inflammatory and anti-cancer effects.172 Acetate is the main product of colonic fermentation, most of which is produced by the fermentation of undigested carbohydrates in the intestinum tenue by colonic anaerobic bacteria.173 Propionate is the main product of Bacteroides fermentation, whereas butyrate is the main metabolite of Firmicutes, which can be absorbed and utilized by colonic epithelial cells, and are the preferred source of energy for both the colon and cecum.174 Bacteria that produce SCFAs include Bacteroides, Clostridium, Bifidobacterium, Eubacterium, Streptococcus, and Peptostreptococcus.Immune regulation and anti-inflammatory: SCFAs participated in intestinal immune regulation by affecting the release of inflammatory factors, immunochemokines, and inhibition of epidemic effector cell proliferation, and play an important role in the intestinal defense against pathogenic bacteria.175 Recent data have shown that SCFAs also regulate the size and function of the colonic Treg cell pool, and protect against colitis in a Ffar2-dependent manner.176 SCFAs could inhibit histone deacetylase, reduce NF-κB activity, and inhibit IL-8 and TNF-α, which are released from neutrophils and macrophages, thereby reducing the inflammatory response in the intestine.177 Butyrate could also inhibit intestinal inflammation though inhibiting the IFN-γ/STAT1 signaling pathway, tyrosine and serine phosphorylation, nuclear translocation, and DNA binding activity.178 A total of 30 mmol L−1 acetate, propionate and butyrate reduced the release of TNF-α without affecting the release of IL-κ protein, thereby indicating that SCFAs have a good therapeutic effect on colitis.179
Anti-tumor: physiological doses of acetate, propionate, and butyrate inhibited the growth and proliferation of rectal tumor cells, and induced tumor cell differentiation, and apoptosis.180 SLC5A8, which is predominantly expressed in the colon, is a Na(+)-coupled transporter for short-chain fatty acids that inhibited tumor growth by regulating the bacterial fermentation product butyrate.181 SCFAs also protected the colonic mucosa from tumorigenesis by regulating colon cell phenotype, DNA synthesis and methylation, and c-myc levels.182,183
Energy metabolism: in the context of energy metabolism, SCFAs activate the GPR43, thereby suppressing insulin signaling in adipocytes, which inhibits fat accumulation and promotes the metabolism of unincorporated lipids and glucose in other tissues.184 SCFAs may also be the key factors involved in improving the pathophysiology of diabetes.185
Others: SCFAs function in maintaining the balance of water and electrolytes in the gut. Perfusion of SCFAs can significantly reduce the secretion of water in the colon, and subsequently, butyrate significantly reduced the secretion of Na+, K+, and Cl−, while propionate reduced the secretion of HCO3−, and acetate inhibited the secretion of Na+ and Cl−.186 Intravenous SCFAs effectively maintained the height, width, crypto depth and mucosal thickness of intestinal villi, increased the proliferation capacity of intestinal mucosal cells, reduce intestinal mucosal atrophy caused by triphosphopyridine nucleotides, maintained the morphology of intestinal mucosa and maintained the integrity of intestinal epithelial cells.187 SCFAs also promoted glucagon-like peptide-2 secretion in intestinal cells, which is an intestinal epithelial-specific growth factor that promotes intestinal mucosal growth and repairs intestinal epithelial damage.188
In recent studies, it was confirmed that THM affected the fermentation products of gut microbiota, for example berberine lowered blood lipid and glucose levels via multi-target mechanisms, and affected the SCFAs. Oral administration of berberine modified intestinal bacterial composition by increasing the abundance of butyrate-producing bacteria, which then entered the circulation and reduced blood lipid and glucose levels.189 Polysaccharides from seeds of Plantago asiatica L. increased SCFAs and fecal moisture along with lowering the pH in the colon, which might be beneficial for colon health.190 Polysaccharides from Polygonatum kingianum decreased total SCFAs, acetate, propionate, and butyrate in high-fat diet-induced type 2 diabetic rat gut.166 Moreover, the leaves of Passiflora edulis decreased acetate and butyrate levels in normal male Wistar rat gut.191 Polysaccharides from Chrysanthemum morifolium, Xiexin Tang, polysaccharides S-3 from Sijunzi decoction increased acetate, propionate and butyrate levels in rat gut.192
6. Perspectives
THM is the foundation of modern medicine, and many successful THM-based pharmaceutical drugs have been formulated. Prominent examples include morphine, which was isolated from opium in 1806, strychnine (1818), ephedrine (1887), paclitaxel (1971), and more recently artemisinin, which was the subject of the 2015 Nobel Prize in Physiology or Medicine. These drugs have made significant contributions to human health, and have indicated the importance of THM in modern medical research and development. However, to date very few active ingredients of THM have been identified by pharmacodynamical and biological assays either in vitro or in vivo. Furthermore, the content of these substances in THM preparations is very low, and most of them have poor bioavailability. Nevertheless, thousands of years of observing the practical benefits of THM and modern clinical research has proven the efficacy of THM in several diseases. Further research and development is still required to elucidate the underlying mechanism of action.Disturbances in gut microbiota composition are related to the occurrence, development, and treatment of various diseases. Several studies have shown that active components of THM or its extracts could interact with gut microbiota, thereby resulting in positive effects (Fig. 2). These findings suggested that the role of THM in treating diseases may be related to gut microbiota, and there may be three likely modes of interaction: (1) metabolism of THM by microbes into active and even more potent metabolites, (2) regulation of gut microbiota by THM components, and (3) promoting the synthesis of therapeutic fermentation products, such as SCFAs by the gut microbiota. In addition, gut microbiota can increase the toxicity of several THM, such as amygdalin, hydrolyze this to HCN under the anaerobic Bacteroidetes phylum in the gut and cause toxicity.193
In summary, both historic and modern science validate the efficacy of THM, however research and development in this field has staggered due to the complexity of ingredients and the unknown mechanism of action. Thus, in recent years, research on gut microbiota has brought a new perspective into the study of THM.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was financially supported by the Fundamental Research Funds for the Central public welfare research institutes (project no. ZXKT17013 and ZZ11-092) and Young Elite Scientists Sponsorship Program by CACM (2018QNRC2-C01)..References
- T. Hesketh and W. X. Zhu, BMJ, 1997, 315, 115–117 CrossRef CAS PubMed.
- J. Xu and Y. Yang, Health Policy, 2009, 90, 133–139 CrossRef PubMed.
- P. Fisher and A. Ward, BMJ, 1994, 309, 107–111 CrossRef CAS PubMed.
- F. Firenzuoli and L. Gori, Evid. Based Complement Altern. Med., 2007, 4, 37–40 CrossRef PubMed.
- P. A. De Smet, N. Engl. J. Med., 2005, 352, 1176–1178 CrossRef CAS PubMed.
- D. M. Eisenberg, R. C. Kessler, C. Foster, F. E. Norlock, D. R. Calkins and T. L. Delbanco, N. Engl. J. Med., 1993, 328, 246–252 CrossRef CAS PubMed.
- R. C. Kessler, R. B. Davis, D. F. Foster, M. I. Van Rompay, E. E. Walters, S. A. Wilkey, T. J. Kaptchuk and D. M. Eisenberg, Ann. Intern. Med., 2001, 135, 262–268 CrossRef CAS.
- C. H. Wu, C. C. Wang and J. Kennedy, Clin. Ther., 2011, 33, 1749–1758 CrossRef PubMed.
- O. S. Olorunnisola, A. Adetutu, E. A. Balogun and A. J. Afolayan, J. Ethnopharmacol., 2013, 150, 71–78 CrossRef CAS PubMed.
- A. C. Liwa, L. R. Smart, A. Frumkin, H. A. Epstein, D. W. Fitzgerald and R. N. Peck, Curr. Hypertens. Rep., 2014, 16, 437 CrossRef PubMed.
- S. Sen and R. Chakraborty, J. Tradit. Complement. Med., 2017, 7, 234–244 CrossRef PubMed.
- J. L. Tang and T. W. Wong, Hong Kong Med. J., 1998, 4, 208–210 CAS.
- L. D'Angelo, R. Grimaldi, M. Caravaggi, M. Marcoli, E. Perucca, S. Lecchini, G. M. Frigo and A. Crema, J. Ethnopharmacol., 1986, 16, 15–22 CrossRef.
- H. Amagase and D. M. Nance, J. Altern. Complement. Med., 2008, 14, 403–412 CrossRef PubMed.
- G. Hu, R. S. Walls, D. Bass, B. Ramon, D. Grayson, M. Jones and V. Gebski, Ann. Allergy, Asthma, Immunol., 2002, 88, 478–487 CrossRef.
- A. Bensoussan, N. J. Talley, M. Hing, R. Menzies, A. Guo and M. Ngu, JAMA, 1998, 280, 1585–1589 CrossRef CAS PubMed.
- W. K. Leung, J. C. Wu, S. M. Liang, L. S. Chan, F. K. Chan, H. Xie, S. S. Fung, A. J. Hui, V. W. Wong, C. T. Che and J. J. Sung, Am. J. Gastroenterol., 2006, 101, 1574–1580 CrossRef PubMed.
- K. Iwasaki, S. Kobayashi, Y. Chimura, M. Taguchi, K. Inoue, S. Cho, T. Akiba, H. Arai, J. C. Cyong and H. Sasaki, J. Am. Geriatr. Soc., 2004, 52, 1518–1521 CrossRef PubMed.
- World Health Organization, General guidelines for methodologies on research and evaluation of traditional medicine, World Health Organization, Geneva, 2000 Search PubMed.
- S. Li, Q. Han, C. Qiao, J. Song, C. Lung Cheng and H. Xu, Chin. Med., 2008, 3, 7 CrossRef PubMed.
- X. M. Liang, Y. Jin, Y. P. Wang, G. W. Jin, Q. Fu and Y. S. Xiao, J. Chromatogr. A, 2009, 1216, 2033–2044 CrossRef CAS PubMed.
- Y.-Z. Liang, P. Xie and K. Chan, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2004, 812, 53–70 CrossRef CAS.
- I. B. Afanas'ev, A. I. Dorozhko, A. V. Brodskii, V. A. Kostyuk and A. I. Potapovitch, Biochem. Pharmacol., 1989, 38, 1763–1769 CrossRef.
- G. L. Russo, M. Russo, C. Spagnuolo, I. Tedesco, S. Bilotto, R. Iannitti and R. Palumbo, Cancer Treat. Res., 2014, 159, 185–205 CAS.
- D. W. Lamson and M. S. Brignall, Alternative Medicine Review, 2000, 5, 196–208 CAS.
- S. Y. Wei, Y. Chen and X. Y. Xu, Chin. J. Nat. Med., 2014, 12, 407–414 CAS.
- Y. X. Zhou, H. Zhang and C. Peng, Phytother. Res., 2014, 28, 961–975 CrossRef CAS PubMed.
- K. Kesarwani and R. Gupta, Asian Pac. J. Trop. Biomed., 2013, 3, 253–266 CrossRef CAS.
- F. Veiga, C. Fernandes and F. Teixeira, Int. J. Pharm., 2000, 202, 165–171 CrossRef CAS PubMed.
- T. H. Xue and R. Roy, Science, 2003, 300, 740–741 CrossRef CAS PubMed.
- D. Pal, D. Kwatra, M. Minocha, D. K. Paturi, B. Budda and A. K. Mitra, Life Sci., 2011, 88, 959–971 CrossRef CAS PubMed.
- X. K. Huo, Q. Liu, C. Y. Wang, Q. Meng, H. J. Sun, J. Y. Peng, X. C. Ma and K. X. Liu, Eur. J. Pharm. Sci., 2013, 50, 420–428 CrossRef CAS PubMed.
- C. Mohn, H. G. Hacker, R. A. Hilger, M. Gutschow and U. Jaehde, Pharmazie, 2013, 68, 622–627 CAS.
- P. Goldman, Ann. Intern. Med., 2001, 135, 594–600 CrossRef CAS PubMed.
- J. H. Lee, J. M. Kim and C. Kim, J. Ethnopharmacol., 2003, 84, 5–9 CrossRef CAS PubMed.
- W. Zhu, X. M. Wang, L. Zhang, X. Y. Li and B. X. Wang, Am. J. Chin. Med., 2005, 33, 839–850 CrossRef CAS PubMed.
- Z. Wen, T. E. Dumas, S. J. Schrieber, R. L. Hawke, M. W. Fried and P. C. Smith, Drug Metab. Dispos., 2008, 36, 65–72 CrossRef CAS PubMed.
- S. M. Wittemer, M. Ploch, T. Windeck, S. C. Muller, B. Drewelow, H. Derendorf and M. Veit, Phytomedicine, 2005, 12, 28–38 CrossRef CAS PubMed.
- K. Woelkart, E. Feizlmayr, P. Dittrich, E. Beubler, F. Pinl, A. Suter and R. Bauer, Phytother. Res., 2010, 24, 445–450 CrossRef CAS PubMed.
- T. Lu, J. Yang, X. Gao, P. Chen, F. Du, Y. Sun, F. Wang, F. Xu, H. Shang, Y. Huang, Y. Wang, R. Wan, C. Liu, B. Zhang and C. Li, Drug Metab. Dispos., 2008, 36, 1578–1586 CrossRef CAS PubMed.
- A. Wen, J. Yang, Y. Jia, Z. Yang, Y. Tian, Y. Wu, Z. Wang and Z. He, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2008, 876, 41–46 CrossRef CAS PubMed.
- C. Z. Wang, K. E. Kim, G. J. Du, L. W. Qi, X. D. Wen, P. Li, B. A. Bauer, M. B. Bissonnette, M. W. Musch, E. B. Chang and C. S. Yuan, Am. J. Chin. Med., 2011, 39, 1161–1171 CrossRef CAS PubMed.
- R. Sender, S. Fuchs and R. Milo, Cell, 2016, 164, 337–340 CrossRef CAS PubMed.
- R. Sender, S. Fuchs and R. Milo, PLoS Biol., 2016, 14, e1002533 CrossRef PubMed.
- T. S. Postler and S. Ghosh, Cell Metab., 2017, 26, 110–130 CrossRef CAS PubMed.
- J. Qin, R. Li, J. Raes, M. Arumugam, K. S. Burgdorf, C. Manichanh, T. Nielsen, N. Pons, F. Levenez, T. Yamada, D. R. Mende, J. Li, J. Xu, S. Li, D. Li, J. Cao, B. Wang, H. Liang, H. Zheng, Y. Xie, J. Tap, P. Lepage, M. Bertalan, J. M. Batto, T. Hansen, D. Le Paslier, A. Linneberg, H. B. Nielsen, E. Pelletier, P. Renault, T. Sicheritz-Ponten, K. Turner, H. Zhu, C. Yu, S. Li, M. Jian, Y. Zhou, Y. Li, X. Zhang, S. Li, N. Qin, H. Yang, J. Wang, S. Brunak, J. Dore, F. Guarner, K. Kristiansen, O. Pedersen, J. Parkhill, J. Weissenbach, H. I. T. C. Meta, P. Bork, S. D. Ehrlich and J. Wang, Nature, 2010, 464, 59–65 CrossRef CAS PubMed.
- S. R. Gill, M. Pop, R. T. Deboy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett and K. E. Nelson, Science, 2006, 312, 1355–1359 CrossRef CAS PubMed.
- C. Human, Microbiome Project, Nature, 2012, 486, 215–221 CrossRef PubMed.
- J. Lederberg, Science, 2000, 288, 287–293 CrossRef CAS PubMed.
- N. Arpaia, C. Campbell, X. Fan, S. Dikiy, J. van der Veeken, P. deRoos, H. Liu, J. R. Cross, K. Pfeffer, P. J. Coffer and A. Y. Rudensky, Nature, 2013, 504, 451–455 CrossRef CAS PubMed.
- W. Jia, H. Li, L. Zhao and J. K. Nicholson, Nat. Rev. Drug Discovery, 2008, 7, 123–129 CrossRef CAS PubMed.
- M. G. Rooks and W. S. Garrett, Nat. Rev. Immunol., 2016, 16, 341–352 CrossRef CAS PubMed.
- P. Bercik, S. M. Collins and E. F. Verdu, Neurogastroenterol. Motil., 2012, 24, 405–413 CrossRef CAS PubMed.
- A. L. Kau, P. P. Ahern, N. W. Griffin, A. L. Goodman and J. I. Gordon, Nature, 2011, 474, 327–336 CrossRef CAS PubMed.
- G. Sharon, T. R. Sampson, D. H. Geschwind and S. K. Mazmanian, Cell, 2016, 167, 915–932 CrossRef CAS PubMed.
- D. Kelly, S. Conway and R. Aminov, Trends Immunol., 2005, 26, 326–333 CrossRef CAS PubMed.
- M. Bajzer and R. J. Seeley, Nature, 2006, 444, 1009–1010 CrossRef CAS PubMed.
- L. M. Bustos Fernandez, J. S. Lasa and F. Man, J. Clin. Gastroenterol., 2014, 48, 657–666 CrossRef PubMed.
- J. L. Pluznick, R. J. Protzko, H. Gevorgyan, Z. Peterlin, A. Sipos, J. Han, I. Brunet, L. X. Wan, F. Rey, T. Wang, S. J. Firestein, M. Yanagisawa, J. I. Gordon, A. Eichmann, J. Peti-Peterdi and M. J. Caplan, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 4410–4415 CrossRef CAS PubMed.
- H. S. Ejtahed, J. Mohtadi-Nia, A. Homayouni-Rad, M. Niafar, M. Asghari-Jafarabadi and V. Mofid, Nutrition, 2012, 28, 539–543 CrossRef CAS PubMed.
- T. R. Sampson, J. W. Debelius, T. Thron, S. Janssen, G. G. Shastri, Z. E. Ilhan, C. Challis, C. E. Schretter, S. Rocha, V. Gradinaru, M. F. Chesselet, A. Keshavarzian, K. M. Shannon, R. Krajmalnik-Brown, P. Wittung-Stafshede, R. Knight and S. K. Mazmanian, Cell, 2016, 167, 1469–1480 CrossRef CAS PubMed.
- J. Qin, Y. Li, Z. Cai, S. Li, J. Zhu, F. Zhang, S. Liang, W. Zhang, Y. Guan, D. Shen, Y. Peng, D. Zhang, Z. Jie, W. Wu, Y. Qin, W. Xue, J. Li, L. Han, D. Lu, P. Wu, Y. Dai, X. Sun, Z. Li, A. Tang, S. Zhong, X. Li, W. Chen, R. Xu, M. Wang, Q. Feng, M. Gong, J. Yu, Y. Zhang, M. Zhang, T. Hansen, G. Sanchez, J. Raes, G. Falony, S. Okuda, M. Almeida, E. LeChatelier, P. Renault, N. Pons, J. M. Batto, Z. Zhang, H. Chen, R. Yang, W. Zheng, S. Li, H. Yang, J. Wang, S. D. Ehrlich, R. Nielsen, O. Pedersen, K. Kristiansen and J. Wang, Nature, 2012, 490, 55–60 CrossRef CAS PubMed.
- E. S. Bliss and E. Whiteside, Front. Physiol., 2018, 9, 900 CrossRef PubMed.
- M. Arumugam, J. Raes, E. Pelletier, D. Le Paslier, T. Yamada, D. R. Mende, G. R. Fernandes, J. Tap, T. Bruls, J. M. Batto, M. Bertalan, N. Borruel, F. Casellas, L. Fernandez, L. Gautier, T. Hansen, M. Hattori, T. Hayashi, M. Kleerebezem, K. Kurokawa, M. Leclerc, F. Levenez, C. Manichanh, H. B. Nielsen, T. Nielsen, N. Pons, J. Poulain, J. Qin, T. Sicheritz-Ponten, S. Tims, D. Torrents, E. Ugarte, E. G. Zoetendal, J. Wang, F. Guarner, O. Pedersen, W. M. de Vos, S. Brunak, J. Dore, H. I. T. C. Meta, M. Antolin, F. Artiguenave, H. M. Blottiere, M. Almeida, C. Brechot, C. Cara, C. Chervaux, A. Cultrone, C. Delorme, G. Denariaz, R. Dervyn, K. U. Foerstner, C. Friss, M. van de Guchte, E. Guedon, F. Haimet, W. Huber, J. van Hylckama-Vlieg, A. Jamet, C. Juste, G. Kaci, J. Knol, O. Lakhdari, S. Layec, K. Le Roux, E. Maguin, A. Merieux, R. Melo Minardi, C. M'Rini, J. Muller, R. Oozeer, J. Parkhill, P. Renault, M. Rescigno, N. Sanchez, S. Sunagawa, A. Torrejon, K. Turner, G. Vandemeulebrouck, E. Varela, Y. Winogradsky, G. Zeller, J. Weissenbach, S. D. Ehrlich and P. Bork, Nature, 2011, 473, 174–180 CrossRef CAS PubMed.
- P. B. Eckburg, E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson and D. A. Relman, Science, 2005, 308, 1635–1638 CrossRef PubMed.
- F. Backhed, R. E. Ley, J. L. Sonnenburg, D. A. Peterson and J. I. Gordon, Science, 2005, 307, 1915–1920 CrossRef PubMed.
- R. E. Ley, D. A. Peterson and J. I. Gordon, Cell, 2006, 124, 837–848 CrossRef CAS PubMed.
- F. Backhed, J. K. Manchester, C. F. Semenkovich and J. I. Gordon, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 979–984 CrossRef CAS PubMed.
- G. Rogler and G. Rosano, Eur. Heart J., 2014, 35, 426–430 CrossRef PubMed.
- A. F. Ahmad, N. C. Ward and G. Dwivedi, Curr. Opin. Cardiol., 2018, 34, 1 Search PubMed.
- Y. Nagatomo and W. H. Tang, J. Card. Fail., 2015, 21, 973–980 CrossRef CAS PubMed.
- J. F. Cryan and S. M. O'Mahony, Neurogastroenterol. Motil., 2011, 23, 187–192 CrossRef CAS PubMed.
- Y. Wang and L. H. Kasper, Brain Behav. Immun., 2014, 38, 1–12 CrossRef CAS PubMed.
- E. A. Mayer, R. Knight, S. K. Mazmanian, J. F. Cryan and K. Tillisch, J. Neurosci., 2014, 34, 15490–15496 CrossRef PubMed.
- H. Chu, B. Williams and B. Schnabl, Liver Res., 2018, 2, 43–51 CrossRef PubMed.
- L. Bird, Nat. Rev. Immunol., 2012, 12, 153 CrossRef CAS PubMed.
- N. Qin, F. Yang, A. Li, E. Prifti, Y. Chen, L. Shao, J. Guo, E. Le Chatelier, J. Yao, L. Wu, J. Zhou, S. Ni, L. Liu, N. Pons, J. M. Batto, S. P. Kennedy, P. Leonard, C. Yuan, W. Ding, Y. Chen, X. Hu, B. Zheng, G. Qian, W. Xu, S. D. Ehrlich, S. Zheng and L. Li, Nature, 2014, 513, 59–64 CrossRef CAS PubMed.
- J. G. Fox, Y. Feng, E. J. Theve, A. R. Raczynski, J. L. Fiala, A. L. Doernte, M. Williams, J. L. McFaline, J. M. Essigmann, D. B. Schauer, S. R. Tannenbaum, P. C. Dedon, S. A. Weinman, S. M. Lemon, R. C. Fry and A. B. Rogers, Gut, 2010, 59, 88–97 CrossRef CAS PubMed.
- A. Everard, C. Belzer, L. Geurts, J. P. Ouwerkerk, C. Druart, L. B. Bindels, Y. Guiot, M. Derrien, G. G. Muccioli, N. M. Delzenne, W. M. de Vos and P. D. Cani, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 9066–9071 CrossRef CAS PubMed.
- T. Le Roy, M. Llopis, P. Lepage, A. Bruneau, S. Rabot, C. Bevilacqua, P. Martin, C. Philippe, F. Walker, A. Bado, G. Perlemuter, A. M. Cassard-Doulcier and P. Gerard, Gut, 2013, 62, 1787–1794 CrossRef CAS PubMed.
- N. Fei and L. Zhao, ISME J., 2013, 7, 880–884 CrossRef CAS PubMed.
- M. Pop, A. W. Walker, J. Paulson, B. Lindsay, M. Antonio, M. A. Hossain, J. Oundo, B. Tamboura, V. Mai, I. Astrovskaya, H. Corrada Bravo, R. Rance, M. Stares, M. M. Levine, S. Panchalingam, K. Kotloff, U. N. Ikumapayi, C. Ebruke, M. Adeyemi, D. Ahmed, F. Ahmed, M. T. Alam, R. Amin, S. Siddiqui, J. B. Ochieng, E. Ouma, J. Juma, E. Mailu, R. Omore, J. G. Morris, R. F. Breiman, D. Saha, J. Parkhill, J. P. Nataro and O. C. Stine, Genome Biol., 2014, 15, R76 CrossRef PubMed.
- A. W. DuPont and H. L. DuPont, Nat. Rev. Gastroenterol. Hepatol., 2011, 8, 523–531 CrossRef PubMed.
- A. Swidsinski, J. Weber, V. Loening-Baucke, L. P. Hale and H. Lochs, J. Clin. Microbiol., 2005, 43, 3380–3389 CrossRef PubMed.
- X. C. Morgan, T. L. Tickle, H. Sokol, D. Gevers, K. L. Devaney, D. V. Ward, J. A. Reyes, S. A. Shah, N. LeLeiko, S. B. Snapper, A. Bousvaros, J. Korzenik, B. E. Sands, R. J. Xavier and C. Huttenhower, Genome Biol., 2012, 13, R79 CrossRef CAS PubMed.
- T. Wang, G. Cai, Y. Qiu, N. Fei, M. Zhang, X. Pang, W. Jia, S. Cai and L. Zhao, ISME J., 2012, 6, 320–329 CrossRef CAS PubMed.
- L. Aymeric, F. Donnadieu, C. Mulet, L. du Merle, G. Nigro, A. Saffarian, M. Berard, C. Poyart, S. Robine, B. Regnault, P. Trieu-Cuot, P. J. Sansonetti and S. Dramsi, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E283–E291 CrossRef CAS PubMed.
- G. Hajishengallis, R. P. Darveau and M. A. Curtis, Nat. Rev. Microbiol., 2012, 10, 717–725 CrossRef CAS PubMed.
- K. Honda, Cell Host Microbe, 2011, 10, 423–425 CrossRef CAS PubMed.
- J. U. Scher and S. B. Abramson, Nat. Rev. Rheumatol., 2011, 7, 569–578 CrossRef CAS PubMed.
- V. Moos and T. Schneider, Eur. J. Clin. Microbiol. Infect. Dis., 2011, 30, 1151–1158 CrossRef CAS PubMed.
- H. Z. Apperloo-Renkema, H. Bootsma, B. I. Mulder, C. G. Kallenberg and D. van der Waaij, Epidemiol. Infect., 1994, 112, 367–373 CrossRef CAS PubMed.
- A. Hevia, C. Milani, P. Lopez, A. Cuervo, S. Arboleya, S. Duranti, F. Turroni, S. Gonzalez, A. Suarez, M. Gueimonde, M. Ventura, B. Sanchez and A. Margolles, mBio, 2014, 5, e01548–01514 CrossRef CAS PubMed.
- M. R. Edwards, N. W. Bartlett, T. Hussell, P. Openshaw and S. L. Johnston, Nat. Rev. Microbiol., 2012, 10, 459–471 CrossRef CAS PubMed.
- X. L. Zheng, Y. Yang, B. J. Wang, J. Wang and H. Q. Tang, Chin. J. Integr. Med., 2017, 23, 196–200 CrossRef PubMed.
- F. H. Karlsson, F. Fak, I. Nookaew, V. Tremaroli, B. Fagerberg, D. Petranovic, F. Backhed and J. Nielsen, Nat. Commun., 2012, 3, 1245 CrossRef PubMed.
- W. C. Huang, Y. M. Chen, N. W. Kan, C. S. Ho, L. Wei, C. H. Chan, H. Y. Huang and C. C. Huang, Nutrients, 2015, 7, 3767–3782 CrossRef CAS PubMed.
- F. Z. Marques, E. Nelson, P. Y. Chu, D. Horlock, A. Fiedler, M. Ziemann, J. K. Tan, S. Kuruppu, N. W. Rajapakse, A. El-Osta, C. R. Mackay and D. M. Kaye, Circulation, 2017, 135, 964–977 CrossRef CAS PubMed.
- J. M. Yano, K. Yu, G. P. Donaldson, G. G. Shastri, P. Ann, L. Ma, C. R. Nagler, R. F. Ismagilov, S. K. Mazmanian and E. Y. Hsiao, Cell, 2015, 161, 264–276 CrossRef CAS PubMed.
- R. P. Gaykema, L. E. Goehler and M. Lyte, Brain Behav. Immun., 2004, 18, 238–245 CrossRef CAS PubMed.
- P. Bercik, E. F. Verdu, J. A. Foster, J. Macri, M. Potter, X. Huang, P. Malinowski, W. Jackson, P. Blennerhassett, K. A. Neufeld, J. Lu, W. I. Khan, I. Corthesy-Theulaz, C. Cherbut, G. E. Bergonzelli and S. M. Collins, Gastroenterology, 2010, 139, 2102–2112 CrossRef CAS PubMed.
- E. Y. Hsiao, S. W. McBride, S. Hsien, G. Sharon, E. R. Hyde, T. McCue, J. A. Codelli, J. Chow, S. E. Reisman, J. F. Petrosino, P. H. Patterson and S. K. Mazmanian, Cell, 2013, 155, 1451–1463 CrossRef CAS PubMed.
- F. Scheperjans, V. Aho, P. A. Pereira, K. Koskinen, L. Paulin, E. Pekkonen, E. Haapaniemi, S. Kaakkola, J. Eerola-Rautio, M. Pohja, E. Kinnunen, K. Murros and P. Auvinen, Mov. Disord., 2015, 30, 350–358 CrossRef PubMed.
- S. Jangi, R. Gandhi, L. M. Cox, N. Li, F. von Glehn, R. Yan, B. Patel, M. A. Mazzola, S. Liu, B. L. Glanz, S. Cook, S. Tankou, F. Stuart, K. Melo, P. Nejad, K. Smith, B. D. Topcuolu, J. Holden, P. Kivisakk, T. Chitnis, P. L. De Jager, F. J. Quintana, G. K. Gerber, L. Bry and H. L. Weiner, Nat. Commun., 2016, 7, 12015 CrossRef CAS PubMed.
- S. Leclercq, S. Matamoros, P. D. Cani, A. M. Neyrinck, F. Jamar, P. Starkel, K. Windey, V. Tremaroli, F. Backhed, K. Verbeke, P. de Timary and N. M. Delzenne, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E4485–E4493 CrossRef CAS.
- Y. Kim, J. A. Hollenbaugh, D. H. Kim and B. Kim, PLoS One, 2011, 6, e21781 CrossRef CAS PubMed.
- R. F. Thornton, E. C. Murphy, T. F. Kagawa, P. W. O'Toole and J. C. Cooney, BMC Microbiol., 2012, 12, 190 CrossRef CAS PubMed.
- M. Mroczynska and Z. Libudzisz, Pol. J. Microbiol., 2010, 59, 265–269 CAS.
- Y. J. Hyun, B. Kim and D. H. Kim, J. Microbiol. Biotechnol., 2012, 22, 535–540 CrossRef CAS.
- Suryani, T. Kimura, K. Sakka and K. Ohmiya, Biosci., Biotechnol., Biochem., 2004, 68, 609–614 CrossRef CAS.
- J. K. Yang, H. J. Yoon, H. J. Ahn, B. I. Lee, J. D. Pedelacq, E. C. Liong, J. Berendzen, M. Laivenieks, C. Vieille, G. J. Zeikus, D. J. Vocadlo, S. G. Withers and S. W. Suh, J. Mol. Biol., 2004, 335, 155–165 CrossRef CAS.
- P. Brigidi, B. Vitali, E. Swennen, G. Bazzocchi and D. Matteuzzi, Res. Microbiol., 2001, 152, 735–741 CrossRef CAS PubMed.
- M. Miwa, T. Horimoto, M. Kiyohara, T. Katayama, M. Kitaoka, H. Ashida and K. Yamamoto, Glycobiology, 2010, 20, 1402–1409 CrossRef CAS PubMed.
- V. Kadiyala, L. J. Nadeau and J. C. Spain, Appl. Environ. Microbiol., 2003, 69, 6520–6526 CrossRef CAS PubMed.
- A. de Moreno de LeBlanc and G. Perdigon, Biocell, 2005, 29, 15–24 CAS.
- S. Punj and G. H. John, Curr. Issues Mol. Biol., 2009, 11, 59–65 CAS.
- E. Isolauri, S. Rautava and S. Salminen, Gastroenterol. Clin. North Am., 2012, 41, 747–762 CrossRef PubMed.
- S. Rautava and E. Isolauri, Curr. Issues Mol. Biol., 2002, 3, 15–22 CAS.
- A. Cartmell, L. S. McKee, M. J. Pena, J. Larsbrink, H. Brumer, S. Kaneko, H. Ichinose, R. J. Lewis, A. Vikso-Nielsen, H. J. Gilbert and J. Marles-Wright, J. Biol. Chem., 2011, 286, 15483–15495 CrossRef CAS PubMed.
- A. Amaretti, S. Raimondi, A. Leonardi, A. Quartieri and M. Rossi, Nutrients, 2015, 7, 2788–2800 CrossRef CAS PubMed.
- P. F. Zhou, J. Z. Wang, X. X. Pang and H. C. Lv, J. Guangdong Pharm. Coll., 2011, 27, 582–586 CAS.
- M. Matsumoto, A. Ishige, Y. Yazawa, M. Kondo, K. Muramatsu and K. Watanabe, PLoS One, 2012, 7, e31700 CrossRef CAS.
- T. Akao, K. Kobashi and M. Aburada, Biol. Pharm. Bull., 1994, 17, 1573–1576 CrossRef CAS.
- T. Akao, Y. Z. Shu, Y. Matsuda, M. Hattori, T. Namba and K. Kobashi, Chem. Pharm. Bull., 1988, 36, 3043–3048 CrossRef CAS PubMed.
- Y. Z. Shu, M. Hattori, T. Akao, K. Kobashi, K. Kagei, K. Fukuyama, T. Tsukihara and T. Namba, Chem. Pharm. Bull., 1987, 35, 3726–3733 CrossRef CAS PubMed.
- P. J. Turnbaugh, C. Quince, J. J. Faith, A. C. McHardy, T. Yatsunenko, F. Niazi, J. Affourtit, M. Egholm, B. Henrissat, R. Knight and J. I. Gordon, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 7503–7508 CrossRef CAS PubMed.
- I. M. de Oliveira, J. A. Henriques and D. Bonatto, Biochem. Biophys. Res. Commun., 2007, 355, 919–925 CrossRef PubMed.
- K. A. Kim, I. H. Jung, S. H. Park, Y. T. Ahn, C. S. Huh and D. H. Kim, PLoS One, 2013, 8, e62409 CrossRef CAS PubMed.
- A. D. Kim, K. A. Kang, H. S. Kim, D. H. Kim, Y. H. Choi, S. J. Lee, H. S. Kim and J. W. Hyun, Cell Death Dis., 2013, 4, e750 CrossRef CAS PubMed.
- L. Chen, Y. Meng, Q. Sun, Z. Zhang, X. Guo, X. Sheng, G. Tai, H. Cheng and Y. Zhou, Cell Death Dis., 2016, 7, e2334 CrossRef CAS PubMed.
- E. A. Bae, M. J. Han, E. J. Kim and D. H. Kim, Arch. Pharmacal Res., 2004, 27, 61–67 CrossRef CAS.
- I. H. Jung, J. H. Lee, Y. J. Hyun and D. H. Kim, Biol. Pharm. Bull., 2012, 35, 573–581 CrossRef CAS.
- E. A. Bae, J. E. Shin and D. H. Kim, Biol. Pharm. Bull., 2005, 28, 1903–1908 CrossRef CAS.
- W. Y. Cao, Y. N. Wang, P. Y. Wang, W. Lei, B. Feng and X. J. Wang, Molecules, 2015, 20, 20569–20581 CrossRef CAS PubMed.
- M. Nose, M. Ito, K. Kamimura, M. Shimizu and Y. Ogihara, Planta Med., 1994, 60, 136–139 CrossRef CAS PubMed.
- H. T. Trinh, E. H. Joh, H. Y. Kwak, N. I. Baek and D. H. Kim, Acta Pharmacol. Sin., 2010, 31, 718–724 CrossRef CAS PubMed.
- G. D. Kang and D. H. Kim, J. Ethnopharmacol., 2016, 189, 175–185 CrossRef CAS PubMed.
- E. H. Joh and D. H. Kim, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2010, 878, 1875–1880 CrossRef CAS PubMed.
- N. Komoto, M. Ichikawa, S. Ohta, D. Nakano, T. Nishihama, M. Ushijima, Y. Kodera, M. Hayama, O. Shirota, S. Sekita and M. Kuroyanagi, J. Nat. Med., 2010, 64, 321–329 CrossRef CAS PubMed.
- I. H. Jung, S. E. Jang, E. H. Joh, J. Chung, M. J. Han and D. H. Kim, Phytomedicine, 2012, 20, 84–88 CrossRef CAS PubMed.
- R. Kon, N. Ikarashi, C. Nagoya, T. Takayama, Y. Kusunoki, M. Ishii, H. Ueda, W. Ochiai, Y. Machida, K. Sugita and K. Sugiyama, J. Ethnopharmacol., 2014, 152, 190–200 CrossRef CAS PubMed.
- T. Akao, Q. M. Che, K. Kobashi, M. Hattori and T. Namba, Biol. Pharm. Bull., 1996, 19, 136–138 CrossRef CAS.
- E. K. Park, M. K. Choo, H. K. Yoon and D. H. Kim, Arch. Pharmacal Res., 2002, 25, 528–533 CrossRef CAS.
- Y. Kawata, C. M. Ma, M. R. MeSelhy, N. Nakamura, H. Wang, M. Hattori, T. Namba, K. Satoh and Y. Kuraishi, J. Tradit. Med., 1999, 16, 15–23 CAS.
- Y. S. Kim, J. J. Kim, K. H. Cho, W. S. Jung, S. K. Moon, E. K. Park and D. H. Kim, J. Microbiol. Biotechnol., 2008, 18, 1109–1114 CAS.
- Y. F. Wang, Y. N. Liu, W. Xiong, D. M. Yan, Y. Zhu, X. M. Gao, Y. T. Xu and A. D. Qi, J. Ethnopharmacol., 2014, 151, 609–617 CrossRef CAS PubMed.
- M. Hattori, Y. Z. Shu, M. Shimizu, T. Hayashi, N. Morita, K. Kobashi, G. J. Xu and T. Namba, Chem. Pharm. Bull., 1985, 33, 3838–3846 CrossRef CAS PubMed.
- M. Y. Liu, M. Li, X. L. Wang, P. Liu, Q. H. Hao and X. M. Yu, J. Agric. Food Chem., 2013, 61, 12060–12065 CrossRef CAS PubMed.
- X. N. Li, C. H. Huo, Q. Wang, X. W. Zhang, X. N. Sheng and L. T. Zhang, Biomed. Chromatogr., 2008, 22, 367–373 CrossRef CAS PubMed.
- M. Zhao, J. Xu, D. W. Qian, J. M. Guo, S. Jiang, E. X. Shang and J. A. Duan, Biomed. Chromatogr., 2014, 28, 1024–1029 CrossRef CAS PubMed.
- L. Y. Du, M. Zhao, J. Xu, D. W. Qian, S. Jiang, E. X. Shang, J. M. Guo, P. Liu, S. L. Su, J. A. Duan and X. J. Leng, Expert Opin. Drug Metab. Toxicol., 2014, 10, 921–931 CrossRef CAS PubMed.
- G. J. Wang, H. X. Fu, W. Ye, X. Zheng, J. C. Xiao, D. Kang, T. Rao, Y. H. Shao, L. Xie and Y. Liang, J. Pharm. Biomed. Anal., 2016, 128, 191–200 CrossRef CAS PubMed.
- K. Keppler, E. M. Hein and H. U. Humpf, Mol. Nutr. Food Res., 2006, 50, 686–695 CrossRef CAS PubMed.
- S. Tranchimand, P. Brouant and G. Iacazio, Biodegradation, 2010, 21, 833–859 CrossRef CAS PubMed.
- S. H. Bang, Y. J. Hyun, J. Shim, S. W. Hong and D. H. Kim, J. Microbiol. Biotechnol., 2015, 25, 18–25 CrossRef CAS PubMed.
- A. Braune and M. Blaut, Environ. Microbiol., 2011, 13, 482–494 CrossRef CAS PubMed.
- K. Ou, P. Sarnoski, K. R. Schneider, K. Song, C. Khoo and L. Gu, Mol. Nutr. Food Res., 2014, 58, 2196–2205 CrossRef CAS PubMed.
- J. Fu, J. Y. Ma, X. F. Zhang, Y. Wang, R. Feng, Y. C. Chen, X. S. Tan, Y. Y. Zhang, Y. P. Sun, Y. Zhou, C. Ma, C. Y. He, Z. X. Zhao and X. W. Du, J. Pharm. Biomed. Anal., 2012, 71, 162–167 CrossRef CAS PubMed.
- S. Yokoyama, T. Niwa, T. Osawa and T. Suzuki, Arch. Microbiol., 2010, 192, 15–22 CrossRef CAS PubMed.
- X. Zhang, Y. Zhao, M. Zhang, X. Pang, J. Xu, C. Kang, M. Li, C. Zhang, Z. Zhang, Y. Zhang, X. Li, G. Ning and L. Zhao, PLoS One, 2012, 7, e42529 CrossRef CAS PubMed.
- J. Han, H. Lin and W. Huang, Med. Sci. Monit., 2011, 17, RA164–167 CrossRef CAS PubMed.
- W. Xie, D. Gu, J. Li, K. Cui and Y. Zhang, PLoS One, 2011, 6, e24520 CrossRef CAS PubMed.
- C. Y. Kang, X. Zhang, M. Zhao, M. Li, L. P. Zhao and Y. F. Zhao, Chin. J. Microecol., 2013, 25, 1117–1122 CAS.
- J. Q. Ruan, S. Li, Y. P. Li, W. J. Wu, S. M. Y. Lee and R. Yan, Drug Metab. Dispos., 2015, 43, 1601–1611 CrossRef CAS PubMed.
- S. S. Zhou, J. Xu, H. Zhu, J. Wu, J. D. Xu, R. Yan, X. Y. Li, H. H. Liu, S. M. Duan, Z. Wang, H. B. Chen, H. Shen and S. L. Li, Sci. Rep., 2016, 6, 22474 CrossRef CAS PubMed.
- H. Yan, J. Lu, Y. Wang, W. Gu, X. Yang and J. Yu, Phytomedicine, 2017, 26, 45–54 CrossRef CAS PubMed.
- C. J. Chang, C. S. Lin, C. C. Lu, J. Martel, Y. F. Ko, D. M. Ojcius, S. F. Tseng, T. R. Wu, Y. M. Chen, J. D. Young and H. C. Lai, Nat. Commun., 2017, 8, 16130 CrossRef CAS PubMed.
- C. J. Chang, C. S. Lin, C. C. Lu, J. Martel, Y. F. Ko, D. M. Ojcius, S. F. Tseng, T. R. Wu, Y. Y. Chen, J. D. Young and H. C. Lai, Nat. Commun., 2015, 6, 7489 CrossRef CAS PubMed.
- M. Guo, S. Ding, C. Zhao, X. Gu, X. He, K. Huang, Y. Luo, Z. Liang, H. Tian and W. Xu, J. Ethnopharmacol., 2015, 162, 7–13 CrossRef PubMed.
- B. Zhang, R. Yue, Y. Chen, M. Yang, X. Huang, J. Shui, Y. Peng and J. Chin, Evid. Based Complement Altern. Med., 2019, 2019, 2634898 Search PubMed.
- C. T. Peterson, V. Sharma, S. Uchitel, K. Denniston, D. Chopra, P. J. Mills and S. N. Peterson, J. Altern. Complement. Med., 2018, 24, 656–665 CrossRef PubMed.
- P. Louis, G. L. Hold and H. J. Flint, Nat. Rev. Microbiol., 2014, 12, 661–672 CrossRef CAS PubMed.
- O. Ezquerro, B. Pons and M. T. Tena, J. Chromatogr. A, 2002, 963, 381–392 CrossRef CAS PubMed.
- S. E. Pryde, S. H. Duncan, G. L. Hold, C. S. Stewart and H. J. Flint, FEMS Microbiol. Lett., 2002, 217, 133–139 CrossRef CAS PubMed.
- A. M. Kabat, N. Srinivasan and K. J. Maloy, Trends Immunol., 2014, 35, 507–517 CrossRef CAS PubMed.
- P. M. Smith, M. R. Howitt, N. Panikov, M. Michaud, C. A. Gallini, Y. M. Bohlooly, J. N. Glickman and W. S. Garrett, Science, 2013, 341, 569–573 CrossRef CAS PubMed.
- J. Tan, C. McKenzie, M. Potamitis, A. N. Thorburn, C. R. Mackay and L. Macia, Adv. Immunol., 2014, 121, 91–119 CAS.
- L. Klampfer, J. Huang, T. Sasazuki, S. Shirasawa and L. Augenlicht, Mol. Cancer Res., 2003, 1, 855–862 CAS.
- S. Tedelind, F. Westberg, M. Kjerrulf and A. Vidal, World J. Gastroenterol., 2007, 13, 2826–2832 CrossRef CAS PubMed.
- A. Hague, D. J. Elder, D. J. Hicks and C. Paraskeva, Int. J. Cancer, 1995, 60, 400–406 CrossRef CAS PubMed.
- M. Thangaraju, G. Cresci, S. Itagaki, J. Mellinger, D. D. Browning, F. G. Berger, P. D. Prasad and V. Ganapathy, J. Gastrointest. Surg., 2008, 12, 1773–1781 CrossRef PubMed ; discussion 1781–1772.
- N. J. Emenaker and M. D. Basson, Dig. Dis. Sci., 2001, 46, 96–105 CrossRef CAS PubMed.
- D. L. Worthley, V. L. Whitehall, R. K. Le Leu, N. Irahara, R. L. Buttenshaw, K. A. Mallitt, S. A. Greco, I. Ramsnes, J. Winter, Y. Hu, S. Ogino, G. P. Young and B. A. Leggett, Dig. Dis. Sci., 2011, 56, 387–396 CrossRef CAS PubMed.
- I. Kimura, K. Ozawa, D. Inoue, T. Imamura, K. Kimura, T. Maeda, K. Terasawa, D. Kashihara, K. Hirano, T. Tani, T. Takahashi, S. Miyauchi, G. Shioi, H. Inoue and G. Tsujimoto, Nat. Commun., 2013, 4, 1829 CrossRef PubMed.
- A. Puddu, R. Sanguineti, F. Montecucco and G. L. Viviani, Mediators Inflammation, 2014, 2014, 162021 Search PubMed.
- G. H. Rabbani, M. J. Albert, H. Rahman and A. K. Chowdhury, Dig. Dis. Sci., 1999, 44, 1547–1553 CrossRef CAS PubMed.
- D. R. Donohoe, N. Garge, X. Zhang, W. Sun, T. M. O'Connell, M. K. Bunger and S. J. Bultman, Cell Metab., 2011, 13, 517–526 CrossRef CAS PubMed.
- H. F. Mangian and K. A. Tappenden, JPEN, J. Parenter. Enteral Nutr., 2009, 33, 607–617 CrossRef CAS PubMed ; discussion 617.
- Y. Wang, J. W. Shou, X. Y. Li, Z. X. Zhao, J. Fu, C. Y. He, R. Feng, C. Ma, B. Y. Wen, F. Guo, X. Y. Yang, Y. X. Han, L. L. Wang, Q. Tong, X. F. You, Y. Lin, W. J. Kong, S. Y. Si and J. D. Jiang, Metabolism, 2017, 70, 72–84 CrossRef CAS PubMed.
- J. L. Hu, S. P. Nie, F. F. Min and M. Y. Xie, J. Agric. Food Chem., 2012, 60, 11525–11532 CrossRef CAS PubMed.
- B. Thomas, B. S. M. Vithiya, T. A. A. Prasad, S. B. Mohamed, C. M. Magdalane, K. Kaviyarasu and M. Maaza, J. Nanosci. Nanotechnol., 2019, 19, 2640–2648 CrossRef.
- W. Feng, H. Ao and C. Peng, Front. Pharmacol., 2018, 9, 1354 CrossRef CAS.
- V. Jaswal, J. Palanivelu and C. Ramalingam, Biochem. Biophys. Rep., 2018, 14, 125–132 Search PubMed.
Footnote |
| † These two authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |