Open Access Article
Open Access ArticleAn innovative biotransformation to produce resveratrol by Bacillus safensis†
Xiaoyan Hu ab,
Yexue Liuab,
Dengke Liab,
Wei Fengab,
Hanmeng Niab,
Shan Caoc,
Fuping Lu*abd and
Yu Li
ab,
Yexue Liuab,
Dengke Liab,
Wei Fengab,
Hanmeng Niab,
Shan Caoc,
Fuping Lu*abd and
Yu Li *abd
*abd
aKey Laboratory of Industrial Fermentation Microbiology, Ministry of Education, Tianjin University of Science & Technology, Tianjin 300457, China. E-mail: liyu@tust.edu.cn; lfp@tust.edu.cn
bCollege of Biotechnology, Tianjin University of Science & Technology, Tianjin 300457, China
cCollege of Leather Chemistry and Engineering, Qilu University of Technology, Shandong, China
dTianjin Engineering Research Center of Microbial Metabolism and Fermentation Process Control, China
First published on 17th May 2019
Abstract
Resveratrol is considered as a potential food supplement, cosmetic ingredient and nutraceutical. In this study, resveratrol was produced by biotransformation successfully. In detail, a β-glucosidase producing strain was isolated and identified as Bacillus safensis, and it could convert polydatin to resveratrol efficiently and rapidly. Further research showed that the conversion rate to resveratrol reached 93.1% in 8 h at 37 °C. The production of resveratrol was confirmed by HPLC, LC-MS and 1H-NMR to identify its structure and it was verified to possess antibacterial properties especially against Escherchia coli. To illustrate the resveratrol transformation mechanism, several glucosidases from B. safensis CGMCC 13129 were expressed and analyzed. The results showed that BGL4 and BGL5 had higher transformation activity compared with other tested glucosidases. This research provides a novel approach to produce resveratrol, and would promote the application of resveratrol in health-promoting pharmaceutical and food products.
1. Introduction
Resveratrol (trans-3,5,4′-trihydroxystilbene), a polyphenolic compound of the stilbene family, has been found in many plants including grapes, peanuts, blueberries, cassia seed and knotweed.1,2 It has attracted a lot of attention owing to its beneficial properties,3 such as defense against pathogen infection, antitumor, anti-inflammatory, antidiabetic, antithrombotic, and antiaging.4,5 In recent years, resveratrol has been applied as an over-the-counter nutritional supplement in health products, energy drinks and cosmetics products.6 With increasing emphasis on resveratrol, the demand is expected to constantly increase in the future.Due to its low yield in plants and high production cost, the amount of resveratrol available is clearly limited.7 At present, resveratrol is mainly produced by plant extraction, chemical synthesis and biotransformation.8 In skin and flesh of grape, the resveratrol contents were only 0.5 mg kg−1 and 0.25 mg kg−1, respectively.9 The resveratrol yield extracted from resveratrol-producing plants is usually unideal and confined by the long growth cycle of plants.10 Keylor reported a chemical synthesis to produce resveratrol. The yield was 5.1% and there were 13 linear synthetic steps in the total syntheses.11 However, the chemical synthesis is usually limited by the use of large amount of organic solvents, which not only increase the cost but also be considered as a potential threat to the environment. Besides, chemical synthesis is also hindered by poor water solubility and limited chemical stability of resveratrol.12
Owing to cost-effectiveness and environmental friendly, the most widely research on resveratrol producing is biotransformation.13 Che developed an enzymatic system for producing resveratrol from glucose and the yield was 224.4 μg L−1.14 Zhou proved polydatin in P. cuspidatum was able to be converted to resveratrol by polydatin-β-D-glucosidase.15 While β-glucosidase has the ability to hydrolyze glycosidic bonds to release glucose and glucosyl residues.16 This peculiarity allows β-glucosidase to specifically transform polydatin to resveratrol. Furtherly, microbial production possesses more advantages including rapid growth, easy of cultivation and high-level production.17 Until now, researches of microbial transformation mainly focus on grey mold for producing resveratrol.18,19 However, compared to mold, using bacterial for biotransformation may provide more excellent properties such as shorter production cycles, low cost, high conversion efficiency and easy control of reaction conditions.20 Therefore, bacteria should be considered as a potential candidate for resveratrol production.
The aim of this study was to produce resveratrol by bacteria transformation successfully. In this study, a bacteria strain which produced β-glucosidases was screened, characterized and named as B. safensis CGMCC 13129. Polydatin could be directly converted to resveratrol through enzyme produced from CGMCC 13129. The conversion activity was also analyzed in this study, and after optimized the biotransformation conditions, the conversion rate reached 93.1%. To explore the mechanism why this strain has high biotransformation activity, different glucosidases from this strain were cloned and analyzed, and two glucosidases encoded by bgl4 and bgl5 were found the more important enzymes involved in this biotransformation. This was the first report of resveratrol biotransformation by bacteria. And this research would provide significant support in developing resveratrol application though improving resveratrol yield by bacteria transformation.
2. Materials and methods
2.1 Materials, reagents and culture medium
B. safensis CGMCC 13129 was screened from planting polygonum cuspidatum soil collected from Suizhou City, Hubei Province, and isolated by dilution plate technology. E. coli JM109, E. coli BL21 (DE3) and the plasmid pET28a were all preserved in our laboratory. The screening medium contained 1% (w/v) tryptone, 0.5% (w/v) yeast extract, 1% NaCl (w/v), 0.3% geniposide (w/v, 50% purity) and 2% agar (w/v). The seed medium consisted of 1% tryptone (w/v), 0.5% yeast extract (w/v) and 1% NaCl (w/v). The transformation medium was composed of 0.5% beef extract (w/v), 1% tryptone (w/v), 0.5% NaCl (w/v) and polygonum cuspidatum (50% purity). Methanol (chromatographically purity), acetonitrile (chromatographically purity), geniposide (50%), semi-finished herbs of polygonum cuspidatum (50%), the reference substance of resveratrol (98%) were purchased from Nanjing Dilger Medical Technology Co. Ltd (Nanjing, China). All other chemicals and reagents were analytical grade and purchased from commercial sources (Sinopharm Chemical Reagent Co. Ltd.).2.2 Strain screening
2.3 Classification of strain
2.4 Biotransformation process analysis
2.5 Product identification and analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 59.82
59.82![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.18, v/v/v). The HPLC-MS was also carried on using Agilent Venusil XBP column with the same column temperature and detection wavelength as HPLC analysis. The mobile phase was composed by acetonitrile, water and formic acid (40
0.18, v/v/v). The HPLC-MS was also carried on using Agilent Venusil XBP column with the same column temperature and detection wavelength as HPLC analysis. The mobile phase was composed by acetonitrile, water and formic acid (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 59.94
59.94![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.06, v/v/v). The 1H spectra were recorded in acetone-d6 (C3D6O) and in acetone-d6 with appropriate amount of D2O on a Bruker AV-500 spectrometer at working frequencies 400 MHz. Chemical shifts were corresponded with ppm (δ) values and coupling constants (J).31
0.06, v/v/v). The 1H spectra were recorded in acetone-d6 (C3D6O) and in acetone-d6 with appropriate amount of D2O on a Bruker AV-500 spectrometer at working frequencies 400 MHz. Chemical shifts were corresponded with ppm (δ) values and coupling constants (J).312.6 Optimization of biotransformation
To enhance the conversion rate of B. safensis CGMCC 13129, different parameters were selected for optimization, including temperatures (24 °C, 29 °C, 33 °C, 37 °C, 41 °C), pH levels (pH 5, 6, 7, 8, 9), substrate concentration of 0.10% (w/v), 0.15% (w/v), 0.20% (w/v), 0.25% (w/v), 0.30% (w/v) and conversion time (6 h, 8 h, 10 h, 12 h and 14 h).3. Results and discussion
3.1 Strains screening and identification
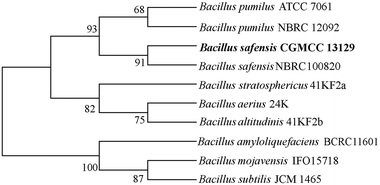
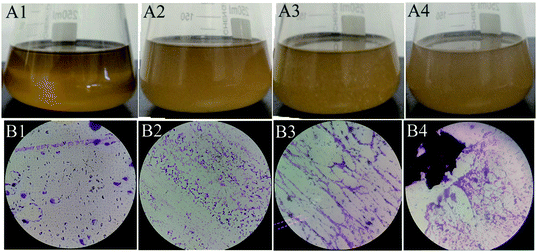
From the soil samples, 32 bacteria strains which produced β-glucosidase were isolated. According to the ratio of hydrolytic zone diameter to colony diameter (H/C), 12 strains with higher β-glucosidases activity were further screened out for the conversion experiment. A strain which had the highest β-glucosidase production ability was isolated, identified and analyzed. As shown in Fig. 1(A), due to genipin generated from geniposide hydrolysis by β-glucosidase could form blue color,33 the blue color around the colony represented the ability of producing β-glucosidase. The strain exhibited the morphological characteristics such as Gram-positive, rod-shaped, spore-forming (Fig. 1(B–D)). Based on the typical features, it was suggested that this strain could be described as Bacillus. In order to identify bacterial species of the screened strains, the partial 16S rDNA gene was amplified using 27F and 1492R primer. The sequences produced were compared with sequences from NCBI using BLAST method, and showed the similarity as Bacillus safensis. The phylogenetic tree showed the closest sequences in Fig. 2. Based on homology research,34 it indicated that the strain was similar to Bacillus safensis in the evolutionary relationship and named as Bacillus safensis CGMCC 13129. The similarity of DNA sequences between B. safensis CGMCC 13129 and B. safensis NBRC 100820 was 91%. Thus it revealed that the screened strain was identified as Bacillus safensis. So far, the strain B. safensis CGMCC 13129 was the first bacteria with ability to producing-resveratrol.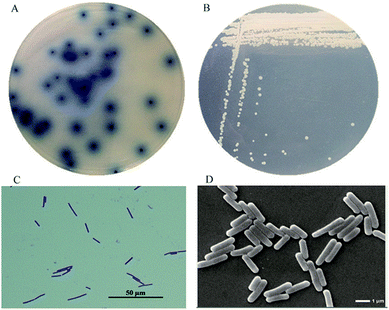 | ||
| Fig. 1 Morphological characteristics of CGMCC 13129. (A) Hydrolysis circle shape (B) colony morphology (C) Gram-staining analysis (D) SEM analysis. | ||
3.2 Analysis of resveratrol-producing process
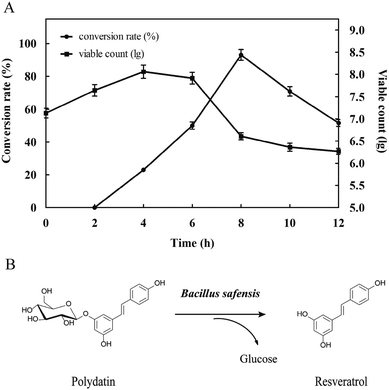 | ||
| Fig. 4 Analysis of resveratrol conversion. (A) The change of conversion rate and viable count with time (B) the conversion process. | ||
3.3 Identification of products
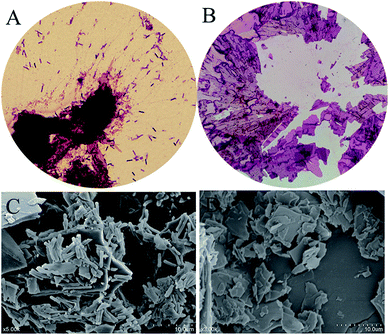
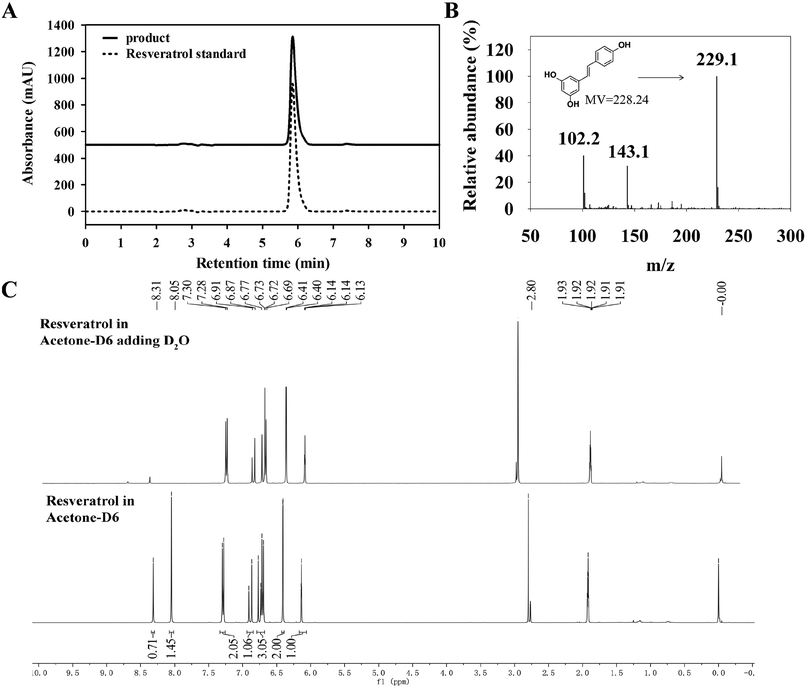
To further confirm resveratrol structure produced by B. safensis CGMCC 13129, subsequent analysis was performed with LC-MS. The compound exhibited ion peaks at m/z 102.1, 143.1 and 229.1 (Fig. 7(B)). The main peak at m/z 229.1 represented the compound was resveratrol, which was formed by acquiring a molecule from the deprotonated species [M + H]+.39 Based on these results, it indicated that resveratrol was successfully generated by bacterial biotransformation with CGMCC 13129 strain, and extraction with methanol was effective for resveratrol production.
As known, cis–trans isomers had significant differences on chemical properties, physiological activities and pharmacological effects. It was found that resveratrol derived from natural plants was described with a mixture of cis & trans structures.40 The trans-resveratrol was considered as a biologically active isomer41 and contributed to a variety of health benefits such as the antioxidant, antiaging, anti-inflammatory and anti-tumor biological properties.42 In order to investigate the structure of resveratrol produced by B. safensis CGMCC 13129, it was unambiguously confirmed by 1H-NMR analysis (Fig. 7(C)). The 1H-NMR spectra data for resveratrol was analyzed as follows: 1H-NMR (400 Hz, C3D6O): δ 6.27 (1H, d, J = 2.1 Hz), 6.54 (2H, d, J = 2.1 Hz), 6.84 (2H, d, J = 8.3 Hz), 6.89 (1H, d, J = 16.3 Hz), 7.02 (1H, d, J = 16.3 Hz) and 7.42 (2H, d, J = 8.3 Hz). Therefore, the compound was identified as trans-resveratrol and our results were similar to other studies.22,43
3.4 Optimization of conversion conditions
A successful conversion process depended on suitable conditions.45 In order to optimize conversion process, factors such as temperature, pH, substrate concentration and reaction time were determined. The results showed that the highest conversion rate appeared at 37 °C, while lower temperature resulted in the decreasing of conversion rate due to slower rate of cell growth (Fig. 8(A)). It was obvious that pH 7 was the optimum pH and the conversion rate was 89.7%. Both alkaline and acidic conditions resulted in a lower conversion rate of resveratrol (Fig. 8(B)). It was explained that enzyme from B. safensis CGMCC 13129, which hydrolyzed polydatin to form resveratrol, mainly catalyzed by neutral enzyme and the optimum temperature of this enzyme should be 37 °C. Meantime, the highest conversion rate of 93.1% was obtained with 0.15% substrate concentration (Fig. 8(C)). The conversion obviously decreased with increasing substrate concentration after the substrate concentration was over 0.15%. It was explained that excessive substrate was negative on the growth of this bacteria. From Fig. 8(D), the conversion rate reached the highest level at 92.5% through the first 8 hours, and then declined over time. With the transformation time increasing after 8 h, the resveratrol was accumulated and adsorption between bacteria and resveratrol occurred. It was not conducive for resveratrol producing. Besides, resveratrol slightly degraded over time, which also resulted in the decrease of conversion rate. This phenomenon was also reported in Trela and Liazid's presentations.46,47 Based on these results, the resveratrol was extracted and the conversion rate reached more than 90%. While in Chen's report, the polydatin was transformed to resveratrol by piceid-β-D-glucosidase from Aspergillus oryzae sp. 100 with the conversion rate of 2 g h−1 U−1 under 60 °C, pH 5.0, 4 h.48 The biotransformation method of producing resveratrol with co-immobilized Aspergillus niger and yeast was investigated in Jin's study and the conversion rate reached about 96.7% under 30 °C, pH 6.5, 2 days.49 The conversion rate reached 85.21% under 31 °C pH 6.0 and 60 h with the method of Aspergillus oryzae negative pressure cavitation bioreactor.50 In this study, the time of conversion to resveratrol was shortened to 8 h successfully and it provided a more convenient and simple procedure, so our research provided a promising and competitive biological material for the production of resveratrol (Fig. 9).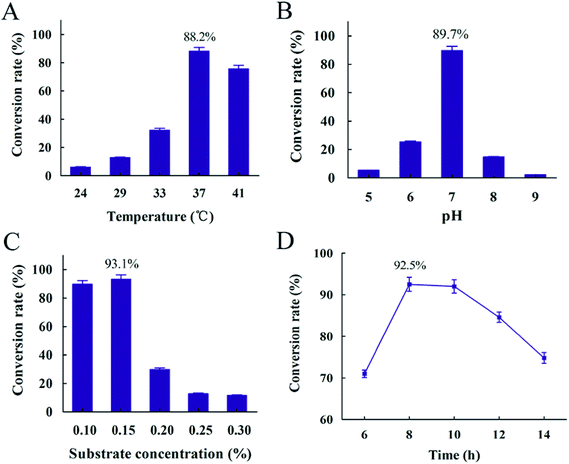 | ||
| Fig. 9 Conversion rate influenced by different conversion conditions (A) temperature (B) pH (C) substrate concentration (D) time. | ||
4. Conclusions
This research presented that bacteria could be applied to produce resveratrol by biotransformation. In this paper, a strain which produced β-glucosidase was screened from the soil planted polygonum cuspidatum yearly. It was identified as Bacillus safensis and named as B. safensis CGMCC 13129. Based on its enzyme-specific activities and biotransformation, the resveratrol was produced efficiently. The biotransformation conditions, including temperature, pH, substrate concentration and time, were optimized. The results showed that the optimal conversion time was 8 h (37 °C, pH = 7), and the conversation rate reached 93.1% with methanol extraction. By 1H-NMR analysis, it demonstrated that B. safensis CGMCC 13129 had the ability to produce trans-resveratrol efficiently and rapidly. Our study provided innovative biological producing process for potential resveratrol application.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by The National Key Research and Development Program of China (2017YFB0308401), National Natural Science Foundation of China (Grant No. 31871741) and Public Service Platform for Selection and Fermentation Technology of Industrial Microorganisms (17PTGCCX00190).References
- Y. Z. Mei, R. X. Liu, D. P. Wang, X. Wang and C. C. Dai, Biotechnol. Lett., 2015, 37, 9–18 CrossRef CAS PubMed.
- A. Y. Berman, R. A. Motechin, M. Y. Wiesenfeld and M. K. Holz, NPJ Precis. Oncol., 2017, 1, 35 CrossRef PubMed.
- I. Ahuja, R. Kissen and A. M. Bones, Trends Plant Sci., 2012, 17, 73–90 CrossRef CAS PubMed.
- M. Li, K. R. Kildegaard, Y. Chen, A. Rodriguez, I. Borodina and J. Nielsen, Metab. Eng., 2015, 32, 1 CrossRef PubMed.
- L. Huang, S. Zhang, J. Zhou and X. Li, RSC Adv., 2019, 9, 2572–2580 RSC.
- J. Christenson, S. J. Whitby, D. Mellor, J. Thomas, A. McKune, P. D. Roach and N. Naumovski, Metab. Syndr. Relat. Disord., 2016, 14, 323–333 CrossRef CAS PubMed.
- S. Jin, M. Luo, W. Wang, C. J. Zhao, C. B. Gu, C. Y. Li, Y. G. Zu, Y. J. Fu and Y. Guan, Bioresour. Technol., 2013, 136, 766–770 CrossRef CAS PubMed.
- A. Martínez-Márquez, J. A. Morante-Carriel, K. Ramírez-Estrada, R. M. Cusidó, J. Palazon and R. Bru-Martínez, J. Plant Biotechnol., 2016, 14, 1813–1825 CrossRef PubMed.
- M. T. Barcia, P. B. Pertuzatti, D. Rodrigues, S. Gómez-Alonso, I. Hermosín-Gutiérrez and H. T. Godoy, Food Res. Int., 2014, 62, 500–513 CrossRef CAS.
- L. Z. Kang, X. L. Zeng, Z. W. Ye, J. F. Lin and L. Q. Guo, Food Chem., 2014, 164, 211–218 CrossRef CAS PubMed.
- M. H. Keylor, B. S. Matsuura, M. Griesser, J. R. Chauvin, R. A. Harding, M. S. Kirillova, X. Zhu, O. J. Fischer, D. A. Pratt and C. R. Stephenson, Science, 2016, 354, 1260 CrossRef CAS PubMed.
- L. Hai, D. He, X. He, K. Wang, X. Yang, J. Liu, H. Cheng, X. Huang and J. Shangguan, J. Mater. Chem. B, 2017, 5, 29 Search PubMed.
- S. Jin, B. Yang, Y. Cheng, J. Tan, H. Kuang, Y. Fu, X. Bai, H. Xie, Y. Gao and C. Lv, Food Bioprod. Process., 2017, 102, 177–185 CrossRef CAS.
- J. Che, J. Shi, Z. Gao and Y. Zhang, J. Microbiol. Biotechnol., 2016, 26, 1348 CrossRef CAS PubMed.
- L. Zhou, S. Li, T. Zhang, W. Mu and B. Jiang, J. Sci. Food Agric., 2016, 96, 2588–2595 CrossRef CAS PubMed.
- J. R. K. Cairns and A. Esen, Cell. Mol. Life Sci., 2010, 67, 3389–3405 CrossRef PubMed.
- J. Wu, P. Liu, Y. Fan, H. Bao, G. Du, J. Zhou and J. Chen, J. Biotechnol., 2013, 167, 404–411 CrossRef CAS PubMed.
- B. Houillé, N. Papon, L. Boudesocque, E. Bourdeaud, S. Besseau, V. Courdavault, C. Enguehardgueiffier, G. Delanoue, L. Guérin and J. P. Bouchara, J. Nat. Prod., 2014, 77, 1658–1662 CrossRef PubMed.
- M. Salgado, S. Rodríguez-Rojo, F. M. Alves-Santos and M. J. Cocero, J. Food Eng., 2015, 165, 13–21 CrossRef CAS.
- F. Lai, Y. E. Miao, Y. Huang, Y. Zhang and T. Liu, ACS Appl. Mater. Interfaces, 2015, 8, 3558–3566 CrossRef PubMed.
- S. Fujikawa, Y. Fukui, K. Koga and J. i. Kumada, J. Ferment. Technol., 1987, 65, 419–424 CrossRef CAS.
- A. Todaro, R. Palmeri, R. N. Barbagallo, P. G. Pifferi and G. Spagna, Food Chem., 2008, 107, 1570–1575 CrossRef CAS.
- K. Minamida, M. Tanaka, A. Abe, T. Sone, F. Tomita, H. Hara and K. Asano, J. Biosci. Bioeng., 2006, 102, 247–250 CrossRef CAS PubMed.
- J. Hong, W. Guan, G. Jin, H. Zhao, X. Jiang and J. Dai, Microbiol. Res., 2015, 170, 69–77 CrossRef CAS PubMed.
- J. Sambrook, E. F. Fritsch and T. Maniatis, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, 1989 Search PubMed.
- E. W. Sayers, T. Barrett, D. A. Benson, E. Bolton, S. H. Bryant, K. Canese, V. Chetvernin, D. M. Church, M. DiCuccio and S. Federhen, Nucleic Acids Res., 2012, 40, D13–D25 CrossRef CAS PubMed.
- S. F. Altschul, T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller and D. J. Lipman, Nucleic Acids Res., 1997, 25, 3389–3402 CrossRef CAS PubMed.
- N. Saitou and M. Nei, Mol. Biol. Evol., 1987, 4, 406 CAS.
- S. Kumar, G. Stecher and K. Tamura, Mol. Biol. Evol., 2016, 33, 1870–1874 CrossRef CAS PubMed.
- D. G. Wang, W. Y. Liu and G. T. Chen, J. Pharm. Anal., 2013, 3, 241–247 CrossRef CAS PubMed.
- H. E. Gottlieb, V. Kotlyar and A. Nudelman, J. Org. Chem., 1997, 62, 7512–7515 CrossRef CAS PubMed.
- G. Zhang, M. Hu, L. He, P. Fu, L. Wang and J. Zhou, Food Bioprod. Process., 2013, 91, 158–168 CrossRef CAS.
- A. S. Bellé, C. R. Hackenhaar, L. S. Spolidoro, E. Rodrigues, M. P. Klein and P. F. Hertz, Food Chem., 2018, 246, 266–274 CrossRef PubMed.
- A. Levican, S. Rubio-Arcos, A. Martinez-Murcia, L. Collado and M. J. Figueras, Syst. Appl. Microbiol., 2015, 38, 30–35 CrossRef CAS PubMed.
- B. Y. M. S and B. VS, Crit. Rev. Biotechnol., 2002, 22, 375–407 CrossRef PubMed.
- P. González-Pombo, L. Fariña, F. Carrau, F. Batista-Viera and B. M. Brena, Process Biochem., 2011, 46, 385–389 CrossRef.
- L. Jaitz, K. Siegl, R. Eder, G. Rak, L. Abranko, G. Koellensperger and S. Hann, Food Chem., 2010, 122, 366–372 CrossRef CAS.
- S. Y. Shin, N. S. Han, Y. C. Park, M. D. Kim and J. H. Seo, Enzyme Microb. Technol., 2011, 48, 48–53 CrossRef CAS PubMed.
- Y. J. Jeong, S. G. Woo, C. H. An, H. J. Jeong, Y. S. Hong, Y. M. Kim, Y. B. Ryu, M. C. Rho, W. S. Lee and C. Y. Kim, Mol. Cells, 2015, 38, 318 CrossRef CAS PubMed.
- M. H. Keylor, B. S. Matsuura and C. R. Stephenson, Chem. Rev., 2015, 115, 8976–9027 CrossRef CAS PubMed.
- S. W. Tsang, H. Zhang, Z. Lin, H. Mu and Z. X. Bian, Biomed. Pharmacother., 2015, 71, 91–97 CrossRef CAS PubMed.
- A. Ahmad and R. Ahmad, Chem. Biol. Interact., 2014, 221, 1 CrossRef CAS PubMed.
- C. Caddeo, M. Manconi, M. C. Cardia, O. Diez-Sales, A. M. Fadda and C. Sinico, Int. J. Pharm., 2015, 484, 138–145 CrossRef CAS PubMed.
- L. Paulo, S. Ferreira, E. Gallardo, J. A. Queiroz and F. Domingues, World J. Microbiol. Biotechnol., 2010, 26, 1533–1538 CrossRef CAS.
- H. P. Kuo, R. Wang, Y. S. Lin, J. T. Lai, Y. C. Lo and S. T. Huang, Bioresour. Technol., 2017, 243, 986–993 CrossRef CAS PubMed.
- A. Liazid, M. Palma, J. Brigui and C. G. Barroso, J. Chromatogr. A, 2007, 1140, 29–34 CrossRef CAS PubMed.
- B. C. Trela and A. L. Waterhouse, J. Agric. Food Chem., 1996, 44, 1253–1257 CrossRef CAS.
- M. Chen, D. Li, Z. Gao and C. Zhang, Bioproc. Biosyst. Eng., 2014, 37, 1–6 CrossRef.
- S. Jin, M. Luo, W. Wang, C. J. Zhao, C. B. Gu, C. Y. Li, Y. G. Zu, Y. J. Fu and Y. Guan, Bioresour. Technol., 2013, 136, 766–770 CrossRef CAS PubMed.
- J. Shuang, B. Yang, Y. Cheng, J. Tan, H. Kuang, Y. Fu, X. Bai, H. Xie, G. Yuan and L. Chen, Food Bioprod. Process., 2016, 102, 177–185 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra01338e |
| This journal is © The Royal Society of Chemistry 2019 |