Open Access Article
Open Access ArticleDiscovery of potential therapeutic targets for non-small cell lung cancer using high-throughput metabolomics analysis based on liquid chromatography coupled with tandem mass spectrometry†
Hong-dan Xua,
Wen Luob,
Yuanlong Linc,
Jiawen Zhangb,
Lijuan Zhangb,
Wei Zhang *b and
Shu-ming Huang*d
*b and
Shu-ming Huang*d
aCollege of Jiamusi, Heilongjiang University of Chinese Medicine, Jiamusi 154007, China
bDepartment of Respiratory and Critical Care, First Affiliated Hospital, Harbin Medical University, Harbin 150081, China. E-mail: weipoza@163.com; Fax: +86-451-85555787; Tel: +86-451-85555787
cInfectious Diseases Department, Fourth Affiliated Hospital, Harbin Medical University, Harbin, China
dInstitute of Chinese Medicine, Heilongjiang University of Medicine Chinese, Heping Road 24, Xiangfang District, Harbin 150040, China. E-mail: shmetabolites78@163.com; Tel: +86-451-87266816
First published on 8th April 2019
Abstract
Lung cancer is a severe health problem and threatens a patient's quality of life. The metabolites present in biological systems are expected to be key mediators and the changes in these metabolites play an important role in promoting health. Metabolomics can unravel the global metabolic changes and identify significant biological pathways involved in disease development. However, the role of metabolites in lung cancer is still largely unknown. In the present study, we developed a liquid chromatography coupled with tandem mass spectrometry method for biomarker discovery and identification of non-small cell lung cancer (NSCLC) from metabolomics data sets and aimed to investigate the metabolic profiles of NSCLC samples to identify potential disease biomarkers and to reveal the pathological mechanism. After cell metabolite extraction, the metabolic changes in NSCLC cells were characterized and targeted metabolite analysis was adopted to offer a novel opportunity to probe into the relationship between differentially regulated cell metabolites and NSCLC. Quantitative analysis of key enzymes in the disturbed pathways by proteomics was employed to verify metabolomic pathway changes. A total of 13 specific biomarkers were identified in NSCLC cells related with metabolic disturbance of NSCLC morbidity, which were involved in 4 vital pathways, namely glycine, serine and threonine metabolism, aminoacyl-tRNA biosynthesis, tyrosine metabolism and sphingolipid metabolism. The proteomics analysis illustrated the obvious fluctuation of the expression of the key enzymes in these pathways, including the downregulation of 3-phosphoglycerate dehydrogenase, phosphoserine phosphatase, tyrosinase and argininosuccinic acid catenase. NSCLC occurrence is mainly related to amino acid and fatty acid metabolic alteration. These findings highlight that the metabolome can provide information on the molecular profiles of cells, which can aid in investigating the metabolite changes to reveal the pathological mechanism.
Introduction
With the increase in the risk-factors for cancer, such as age, exposure to environmental carcinogens and unhealthy lifestyle, cancer has become a murderous killer that threatens life, in spite of decades of medical research, and is obviously characterized by cellular growth in an abnormal state.1–3 Although early diagnosis can improve the survival rates of patients, most individuals at risk are not effectively discovered until advanced stages due to the lack of sensitive early screening examinations, late-stage diagnosis and the metastatic behavior of tumors.4 Cancer patients are looking forward to emerging diagnostic techniques and methods. Lung cancer is the main cause of cancer-related death all over the world. Compared with other histological kinds of lung cancer, non-small cell lung cancer (NSCLC) has received less attention but accounts for approximately 85% to 90% of lung cancers.5,6 Prognostic measures for NSCLC are lacking on account of its higher metastatic potential and chemoresistance upon relapse. During the past few decades, the therapeutic schedule of NSCLC and the survival ratio of this illness have not distinctly improved. By combining radiotherapy and chemotherapy in the early stages, the effective rate of patient treatment is approximately 45–80%; however, almost all patients will relapse within half a year to one year.7 In addition, the genetic variability of NSCLC leads to limitations in the study of molecular targeted drugs, unlike in small cell lung cancer (SCLC).8Compared with normal cells, cancer cells exhibit the ability to alter their metabolic pathways to allow them to persistently survive in the pathological body and increase their energy requirements to support them to grow and reproduce.9 In recent years, metabolomic studies of cancer have become more and more popular, because the metabolome is a relatively more conserved trait than genomic or transcriptomic sequences.10 Cancer cells not only acquire energy from glycolysis in the presence of oxygen, but also use glutamine as a dominant source of energy.11,12 Now, a comprehensive understanding of the underlying metabolic alterations in NSCLC is urgently required to identify the distinct metabolic features of NSCLC and seek novel drug targets and biomarkers for prevention, diagnosis and therapy.
Metabolomics, a rapidly emerging field in the omics area, is focused on the qualitative and quantitative analysis of low-molecular weight metabolites in biofluids, tissues, and cells at a specified time in specific environmental conditions.13 It has become a complementary method to genomics and proteomics, and provides valuable information on small-molecule metabolites and the signaling pathways in various biological processes.14 Metabolomic approaches have been applied to lung cancer to identify cancer biomarkers and metabolic pathways associated with tumor progression.15,16 Using high-throughput techniques such as nuclear magnetic resonance, high-performance liquid chromatography/mass spectrometry and gas chromatography/MS, fundamental analysis is performed to build a chemical fingerprint of the samples and screen for biomarkers. Then, metabolomics increases the possibility of the validation of candidate biomarkers associated with cancer in validation studies as a more precise selection tool, which promotes researchers to apply metabolomics data to obtain the biological nature of diseases.17 Targeted metabolomics involves the quantitative measurement of specific metabolites such as amino acids, lipids, sugars and fatty acids, to detect specific metabolic pathways of diseases or to verify differentiated metabolites identified by non-targeted metabolic profiling. By comparison, non-targeted metabolomics refers to global coverage analysis of the metabolome in a hypothesis-generating condition.18,19 Because of the lack of access to primary tumor model material, cancer cell lines, which are simple and direct experimental models carrying highly relevant information about metabolic reprogramming in cells, are especially vital to NSCLC research.20 The aim of this study was to employ comparative metabolomics analysis in control and cancer cell groups to illuminate the pathological mechanism of non-small cell lung cancer by UPLC-MS and targeted metabolite analysis. The general flow chart of this study based on high-throughput metabolic analysis coupled with proteomics is shown in Fig. 1.
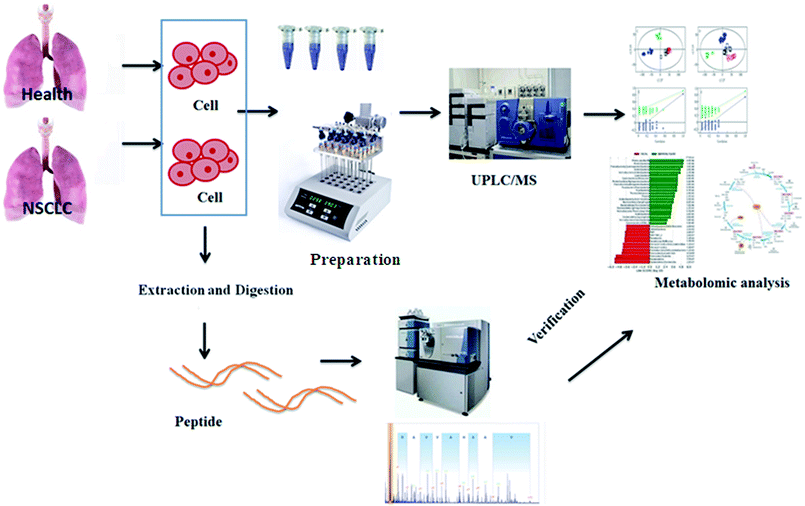 | ||
| Fig. 1 A general flow chart of this study based on high-throughput metabolic analysis coupled with proteomics. | ||
Experiments and methods
Reagents and materials
HPLC grade methanol and acetonitrile were purchased from Fischer Scientific Co. (Pittsburgh, PA, USA). Distilled water, used for preparing all of the aqueous solutions and mobile phase, was obtained from Watson's Food & Beverage Co. Ltd (Guangzhou, China). Formic acid, ammonium hydroxide and leucine enkephalin were obtained from Sigma-Aldrich (St. Louis, MO, USA). Cell culture reagents RPMI1640 and phosphate buffer saline (PBS) were obtained from Gibco Laboratories (Grand Island, USA). A Pierce™ Mass Spec Sample Prep Kit for Cultured Cells and a BCA Protein Assay Kit were purchased from Amresco Limited-Liability Company (Houston, USA). Standards of key enzymes in the study were purchased from Tianjin Chemical Reagent Co. (Tianjin, China).Cell culture
The cell lines, including normal human bronchial epithelial cells (HBEC) and human non-small cell lung cancer cells (NCI-H1975), were obtained from the JRDUN Cell Culture Collection (Shanghai, China). The cells lines were cultured in RPMI-1640 solution containing 10% fetal bovine serum, 110 U mL−1 penicillin, and 110 μg mL−1 streptomycin and maintained in an incubator at 37 °C with 5% CO2. The cells were collected when the cell density reached up to 80–90%, and then they were washed with PBS and digested with 0.3% trypsin containing 0.01% EDTA. After continuous cultivation, the logarithmic phase of the cells was applied for metabolomic analysis.Sample preparation
Each type of cell line was collected in the logarithmic phase. Eight replicate samples were prepared for the two kinds of cells, and the number of cells is approximately 1 million (SD#10%) for each replication. Firstly, the redundant culture medium was removed using a pipette and the cells were digested with 4 mL of 0.3% trypsin. The collected cells were then deposited into a 5 mL clean centrifuge tube and centrifuged for 10 min at 4 °C (800 × g), and the supernatant was removed. 500 μL of extraction buffer containing methanol and water (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v) was then added to the centrifuge tube. The mixture was centrifuged at 15
50, v/v) was then added to the centrifuge tube. The mixture was centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 min at 4 °C after ice incubation for 10 min. 200 μL of the supernatant was dried with nitrogen gas and stored at −80 °C in a hermetically sealed container until metabolomic analysis.
000 × g for 15 min at 4 °C after ice incubation for 10 min. 200 μL of the supernatant was dried with nitrogen gas and stored at −80 °C in a hermetically sealed container until metabolomic analysis.
UPLC-MS analysis
Global cell metabolite profiling was conducted using a high-resolution, accurate quadrupole-time-of-flight mass spectrometer (5500 Q-TRAP, AB SCIEX, USA) equipped with an ESI ion source that operates in positive ionization mode (ESI+) and negative ionization mode (ESI−) at 50–1000 m/z in full scan mode coupled to a UPLC UltiMate 3000 (Dionex Corporation, Sunnyvale, CA, USA). The chromatographic separation of the samples was performed by a 2.1 × 100 mm C18 reverse-phase column with a 1.8 μm particle size (Waters Corp., Milford, MA, USA) after they were redissolved in 100 μL of extraction buffer. The column was maintained at 35 °C; the injected sample volume was 5 μL. The flow rate was maintained at 0.4 mL min−1. The optimized mobile phase was as follows: 90% water + 10% acetonitrile + 15 μM ammonium hydroxide, pH 9.0 (A) and 100% acetonitrile (B). The UPLC gradient program was 0–2 min 100% A, 2–4 min linear from 100 to 70% A, 4–8 min linear from 70 to 30% A, 8–10 min linear from 30 to 100% A, and 10–12 min 100% A. Mass spectrometry (MS) was performed in multiple reaction monitoring mode at a mass range of 70–1000 m/z with positive and negative ionization. The measurement conditions were set as follows: capillary voltage, 4.0 kV; cone voltage, 45 kV; ion source temperature, 120 °C; and vaporizer temperature, 300 °C. During the course of the sequence of analyses, QC samples containing most of the components of the whole cell samples, including the NSCLC and control groups, were injected six times to ensure the stability and consistent performance of the analytical system. Nitrogen was utilized as the nebulizer gas and desolvation gas at a flow rate of 45 L h−1 and 600 L h−1, respectively.Method validation
The precision of the method was evaluated using one QC sample with six replicate injections. The repeatability of the method was investigated by analyzing six replicate solutions. The stability of the analytes was tested by analyzing the solution at 0, 2, 4, 6, 8, 12 and 24 h. The validation was expressed as the relative standard deviation (RSD).Data processing and metabolomics analysis
The obtained raw UPLC-MS data files were processed using Mass Hunter Qualitative Analysis Software (version B.03.01; Agilent Technologies). The process of data manipulation consisted of mass detection, chromatogram building, smoothing, chromatogram deconvolution, grouping of isotopic peaks, peak alignment with m/z tolerance of 10 ppm and retention time tolerance of 0.2 min, gap filling to fill in missing peaks, duplicate peak removal, and peak filtering. After data screening, at least 90% of the sample features were detected and preserved in the peak list. EZinfo 2.0 software (Waters Corporation, Manchester, UK) was used for multivariate analysis of the normalized metabolomics datasets. Unsupervised principal component analysis (PCA) and orthogonal projection to latent structure-discriminant analysis (OPLS-DA), as a model of mean-centering and unit-variance scaling application, ensure the quality of the multivariate models by calculating the cumulative values of the total Y explained variance (R2) and the Y predictable variation (Q2). The 7-fold cross-validation method for permutation tests with 100 iterations was conducted to reduce the risk of over-fitting. Differentially regulated cell metabolites that highlight the discrimination between the control and NSCLC group were obtained by the ions having a variable importance in projection (VIP) score value above 1 and a p-value less than 0.05 in Student's t-tests. To characterize each selected metabolite, a combination of the Mass Fragment software with MS/MS data and some vital databases such as LIPID MAPS, HMDB, MassBank, ChemSpider database and METLIN was used. In addition, MetaboAnalyst 4.0 was employed to analyze the construction, interaction, and pathways of the potential lung cancer biomarkers to attempt to reveal the pathological mechanism of non-small cell lung cancer.Enzyme validation by proteomics
By using a Pierce™ Mass Spec Sample Prep Kit for Cultured Cells and a BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA), proteins were extracted from the cells for quantitative analysis. After the proteins were degraded into polypeptides, they were resuspended in 0.1% (v/v) formic acid for LC-MS/MS analysis after being decomposed by trypsin as described in the Mass Spec Sample Prep Kit for Cultured Cells (Thermo Fisher Scientific). The separation of the peptides was achieved on a Thermo EASY-nLC 1000 nanoliquid chromatography system (Thermo Fisher Scientific) using a C 18 column (75 μm × 15 cm), and then a 6540 quadrupole time-of-flight (Q-TOF) mass detector (Agilent, Santa Clara, CA, USA) equipped with an electrospray ionization source was applied to perform the parallel reaction monitoring analysis. The LC mobile phases were set as follows: 100% water with 0.1% formic acid (mobile phase A) and 95% acetonitrile and 0.1% formic acid (mobile phase B). The gradient was as follows: 0–25 min, 0–30% B; 25–38 min, 30–40% B; 38–55 min, 40–65% B; 55–60 min, 100% B. The MS cycle embodies a full MS1 scan and the predetermined targeted MS2 scan with a resolution of 62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 40
000 and 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, respectively. The obtained data were imported into Skyline software (v3.6, MacCoss Lab, Seattle, WA, USA) for further analysis, such as selecting optimal charge number state and fragment ions of the polypeptide. Some critical parameters were set as follows: the peptide enzyme was set as trypsin (KR/P), the max missed cleavage was set as zero, 6–23 amino acid residues was set as the peptide length, the variable modification, including carbamidomethyl on Cys and oxidation on Met, precursor charges were set as 2 and 3, and 0.03 Da was set as the ion match tolerance.
000, respectively. The obtained data were imported into Skyline software (v3.6, MacCoss Lab, Seattle, WA, USA) for further analysis, such as selecting optimal charge number state and fragment ions of the polypeptide. Some critical parameters were set as follows: the peptide enzyme was set as trypsin (KR/P), the max missed cleavage was set as zero, 6–23 amino acid residues was set as the peptide length, the variable modification, including carbamidomethyl on Cys and oxidation on Met, precursor charges were set as 2 and 3, and 0.03 Da was set as the ion match tolerance.
Statistical analysis
Statistical analysis was performed using the SPSS version 19.0 software for Windows (IBM, Armonk, NY). One-way analysis of variance (ANOVA) was used to inspect the statistical significance of the values gained. When the p-value was less than 0.05, the result was deemed as statistically significant, and data are deemed as extremely statistically significant when the p-value is less than 0.01. In addition, all data are shown as the mean ± standard error of the mean (SEM).Results and discussion
Multivariate statistical analysis
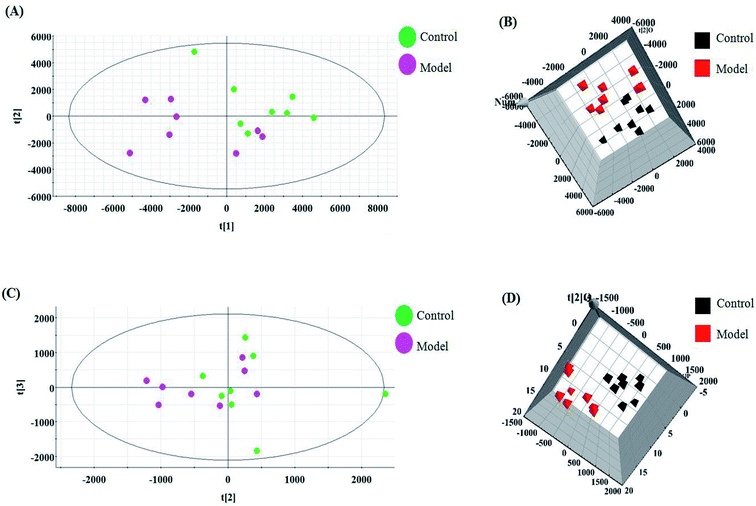
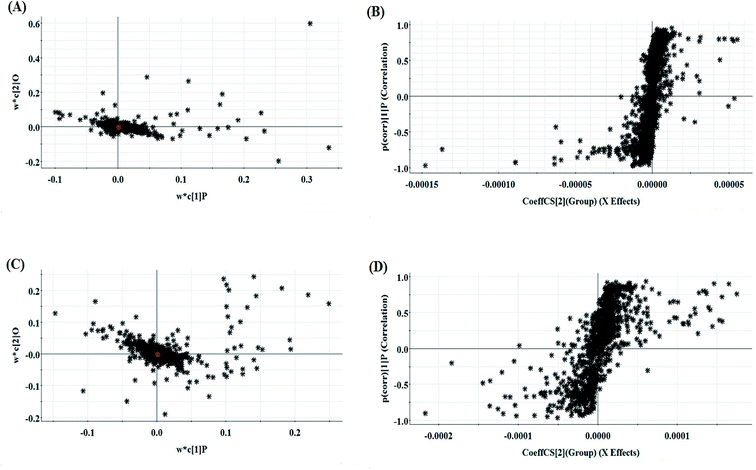
The relative standard deviation (RSDs) values of precision were no more than 4.12%, indicating that the method was precise. The RSDs of repeatability obtained were less than 2.80%. For the stability, the RSDs were no more than 3.45%. These results verified that all analytes were stable for 24 h at room temperature, which demonstrates that the UPLC-MS method could be applied to discover and identify the potential biomarkers that lead to the biological difference between HBEC and NSCLCC. The cell samples were analyzed in both positive and negative ionization modes by UPLC/MS. The trajectory analysis of the PCA score plots for the control and NSCLC groups reflects a clear separation in the positive ionization mode (Fig. 2A) and the negative ionization mode (Fig. 2C). It was indicated that cell metabolite alteration and biochemical changes happened. To maximize the differences between the different groups and determine the variables that contribute to discrimination, OPLS-DA, a more sophisticated supervised pattern recognition method, was further employed for the metabolic data (Fig. 2B and D), which exhibited good 3D separation in the control and NSCLC cells. When one PLS component and one orthogonal component were analyzed, the cumulative R2Y and Q2 were measured, respectively. In addition, the lack of over-fitting was displayed based on the results of permutation tests (Fig. S1A and B†). In the corresponding loading-plots (Fig. 3A and C), the ions furthest away from the origin may be considered as the differentiating metabolites. S-plots in positive mode and negative mode (Fig. 3B and D) are used to evaluate the causative factors resulting from diverse clustering on the score plots.Differential metabolite discovery and identification
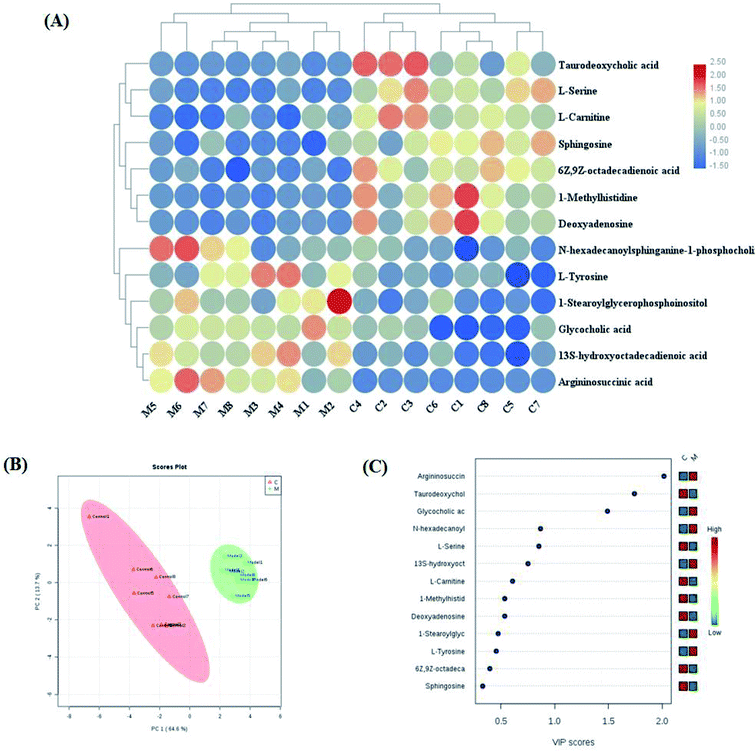
Variables from the OPLS-DA model with a VIP > 1 and an independent sample t-test with p < 0.05 were considered as differentially regulated metabolites that distinguish healthy cells and NSCLC cells (Fig. S2A and B†). 13 metabolites were identified that reveal the pathological mechanism of NSCLC (Table S1†). In hierarchical cluster analysis, it was shown that six of these were up-regulated, including 1-stearoylglycerophosphoinositol, 13S-hydroxyoctadeca-dienoic acid, L-tyrosine, N-hexadecanoylsphinganine-1-phosphocholine, glycocholic acid and argininosuccinic acid, and seven were down-regulated, including sphingosine, L-serine, 6Z,9Z-octadecadienoic acid, 1-methylhistidine, L-carnitine, deoxyadenosine and taurodeoxycholic acid, compared to healthy controls cells according to Pearson's linear correlation analysis in the heatmap (Fig. 4A). As shown in Fig. 4B, the corresponding targeted analysis PCA score plot presents clear separation of the two groups. The relative concentration of these metabolites is shown in the cluster diagram from OPLS-DA, which revealed that the two groups could be separated completely by 3 metabolites with VIP score > 1.0, namely argininosuccinic acid, taurodeoxycholic acid and glycocholic acid (Fig. 4C). The relative signal intensities of the cell metabolites identified by UPLC-MS are demonstrated in Fig. S3.† The results show that several metabolites, such as aspartate, glutamate, methionine, alanine and malonyl carnitine, were deemed to indicate apoptotic processes during cell culture obtained in fast high-throughput screening. Metabolomic features might be applied in investigating the mechanisms of cancer.21Targeted metabolite pathway analysis
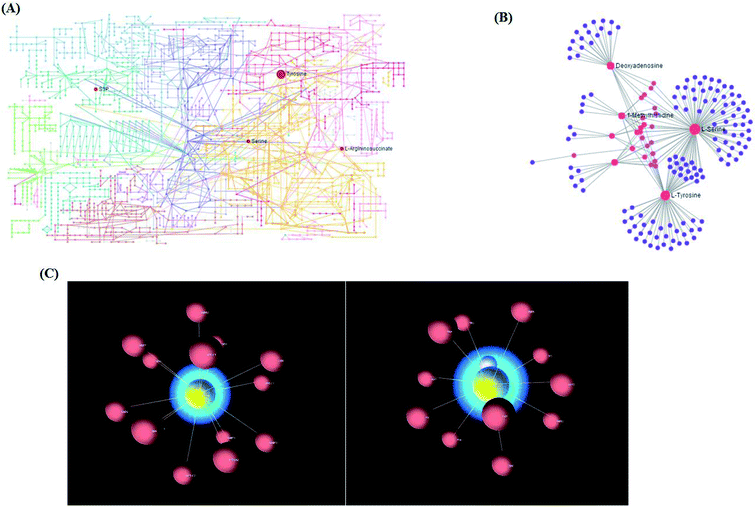
Using the “pathway analysis” module within the MetaboAnalyst software, the above-mentioned 13 metabolites were further analyzed to gain insight into the metabolic mechanism of NSCLC, and the results show that 11 kinds of metabolic alteration, including glycine, serine and threonine metabolism, aminoacyl-tRNA biosynthesis, tyrosine metabolism and sphingolipid metabolism with impact indices of 0.1360, 0.056, 0.04 and 0.0257, were involved in NSCLC as shown in Fig. S4.† Then, integrated network analysis of NSCLCC was performed to outline the biochemical relationships. From the KEGG global metabolic network, which can map metabolites in NSCLC cells and enzymes/Kos (Fig. 5A), it is suitable to integrate the results from joint metabolomics and metagenomics studies mainly referring to amino acid metabolism. The metabolite–metabolite interaction network based on reactions from similar chemical structures and similar molecular activities that highlight potential functional relationships between a wide set of annotated metabolites is shown in Fig. 5B for 4 metabolites, including L-tyrosine, L-serine, deoxyadenosine and 1-methylhistidine. The relationships between genes and the main cell metabolites of NSCLC in three-dimensional (3D) space are demonstrated in Fig. 5C by OmicsNet, a novel network visual analytics system. The enhanced metabolism of glycolysis, glutaminolysis, and amino acids, leading to elevated ATP and protein content, could make a difference to the death of Bcr-Abl cells cultivated in the presence of imatinib. Due to the higher energy demand and turnover of anabolic metabolism during tumor cell proliferation, some rapidly growing metabolites, such as those in glycolysis and the tricarboxylic acid (TCA) cycle, as well as most of the amino acids, were explored in the process of tumor invasion and metastasis of murine osteosarcoma cells. In this study, it is well known that the deficiency of argininosuccinic acid catenase is related to high mortality and morbidity rates characterized by anorexia, irritability, rapid breathing, lethargy, vomiting and other symptoms, which may be an important clue to resist NSCLC. Amino acid metabolism was the top altered pathway in lung cancer.22 Serine can become essential under certain conditions, and is thus important in maintaining health and preventing disease. It is very likely that the predominant source of L-serine will be very different in different tissues and during different stages of human development. In the biosynthetic pathway, the glycolytic intermediate 3-phosphoglycerate is converted into phosphohydroxypyruvate, in a reaction catalyzed by 3-phosphoglycerate dehydrogenase. Phosphohydroxypyruvate is metabolized to phosphoserine by phosphohydroxypyruvate aminotransferase and, finally, phosphoserine is converted into L-serine by phosphoserine phosphatase.Proteomics confirmation
In order to prove the altered metabolic pathways in NSCLC cells uncovered by the metabolomic analysis, targeted proteomics was performed to analyze the changes of key enzymes in the pathways. For targeted proteomics, in the argininosuccinic acid pathway, the protein expression of argininosuccinic acid catenase was prominently decreased in NSCLC cells compared with the control cells (Fig. S5A†). 3-Phosphoglycerate dehydrogenase and phosphoserine phosphatase are the rate-limiting enzymes for serine. We found that 3-phosphoglycerate dehydrogenase and phosphoserine phosphatase were decreased in NSCLC cells with respect to the control (Fig. S5B and C†). This result was no different to the decrease in L-serine in our metabolomic analysis. Tyrosinase controls the production of melanin. Compared with the control cells, tyrosinase was significantly decreased, indicating the upregulation of oxidation–reduction reactions in NSCLC cells, which was in line with the accumulation of hazardous substances found in the metabolomic analysis (Fig. S5D†).Cell metabolomics prospects
Metabolomics, a relatively young branch, integrates many related disciplines, including analytical chemistry, molecular biology, biochemistry, and computational bioinformatics for big data in science. There is a major challenge in the concurrent extraction, separation, and detection of cells because the endogenous metabolites vary widely in their physical and chemical properties in comprehensive characterization of the cell metabolome.23,24 The chemical properties of a great number of metabolites are still unknown, leading to some difficulty in the problem of standardizing quenching and extraction methods. Some standard compounds are not yet readily available. Most of the theoretical research is still focused on the sample preparation techniques and the development of adherent mammalian cells. Another major concern for cell line metabolomics is optimizing the cell culture growth conditions and sample preparation protocols in an attempt to keep the cells in a stable metabolic state.25 There has been much probability of the application of cell metabolomics in the context of cancer. Cancer cells are characterized by several specific metabolic traits, which have been explored for targeted therapies.26–28 Agents that promote apoptosis, as a powerful tool for cancer therapies, have been recognized by more and more people in modern society. Metabolomic features might be applied in testing the efficacy of agents that result in apoptosis in cell culture. For example, metabolic biomarkers of apoptosis can differentiate it from necrosis in HEK and HepG2 cells.29The application of cell metabolomics to lung cancer has only recently been performed. Metabolic markers of lung cancer, which have been screened from a limited number of cases and lack large-scale clinical validation, are still at the scientific research stage.30–37 Most studies of metabolic markers are concentrated on qualitative comparisons, and quantitative analysis of tissues, cells, blood and urine is lacking.38–41 To explore the mechanism of lung cancer, joint platforms of multiple technologies, such as omics and imageology, are still not available, and a platform in the context of big data integration and collection methods is still lacking for precision medicine. With the development of modern analytical techniques, metabolomics will attract more attention in the early diagnosis of NSCLC, molecular mechanisms and precision medicine.
Conclusion
This study was focused on differentiating metabolic changes in cell samples from healthy controls and NSCLC to reveal the pathological mechanism by non-targeted and targeted metabolomics approaches. These data indicated that metabolomics analysis could be utilized to detect metabolic alterations closely related to lung cancer progression, which possess potential clinical usefulness. Compared to healthy controls cells, many cell metabolites were obviously altered in NSCLC cells. The advances in the molecular understanding of the roles of metabolic pathways associated with particular metabolites, such as amino acids and fatty acids, in the NSCLC process will bring about more possibilities for the development of novel therapeutic tools against NSCLC.Conflicts of interest
There are no conflicts to declare.References
- S. Shah and L. Boucai, Effect of Age on Response to Therapy and Mortality in Patients With Thyroid Cancer at High Risk of Recurrence, J. Clin. Endocrinol. Metab., 2018, 103(2), 689–697 CrossRef PubMed.
- L. Abar, A. R. Vieira and D. Aune, et al., Height and body fatness and colorectal cancer risk: an update of the WCRF-AICR systematic review of published prospective studies, Eur. J. Nutr., 2018, 57(5), 1701–1720 CrossRef PubMed.
- M. Kim, K. S. Choi and M. Suh, et al., Risky Lifestyle Behaviors among Gastric Cancer Survivors Compared with Matched Non-cancer Controls: Results from Baseline Result of Community Based Cohort Study, Cancer Res. Treat., 2018, 50(3), 738–747 CrossRef PubMed.
- I. Radványi, A. Csikós and S. Balogh, et al., The significance of early diagnosis of cancer-related hypercalcaemia, Orv. Hetil., 2013, 154(35), 1367–1373 CrossRef PubMed.
- H. Wakelee, K. Kelly and M. J. Edelman, 50 Years of progress in the systemic therapy of non-small cell lung cancer, American Society of Clinical Oncology Educational Book, 2014, 177–189 CrossRef PubMed.
- C. Gridelli, A. Rossi and D. P. Carbone, et al., Non-small-cell lung cancer, Nat. Rev. Dis. Primers, 2015, 1, 15009 CrossRef PubMed.
- T. E. Stinchcombe, Y. Zhang and E. E. Vokes, et al., Pooled Analysis of Individual Patient Data on Concurrent Chemoradiotherapy for Stage III Non-Small-Cell Lung Cancer in Elderly Patients Compared With Younger Patients Who Participated in US National Cancer Institute Cooperative Group Studies, J. Clin. Oncol., 2017 Sep 1, 35(25), 2885–2892 Search PubMed.
- J. Kutkowska, I. Porębska and A. Rapak, et al., Non-small cell lung cancer - mutations, targeted and combination therapy, Postepy Hig. Med. Dosw., 2017, 71(0), 431–445 Search PubMed.
- N. Dong, X. Liu and T. Zhao, et al., Apoptosis-inducing effects and growth inhibitory of a novel chalcone, in human hepatic cancer cells and lung cancer cells, Biomed. Pharmacother., 2018, 105, 195–203 CrossRef CAS PubMed.
- J. Troisi, L. Sarno and A. Landolfi, Metabolomic Signature of Endometrial Cancer, J. Proteome Res., 2018, 17(2), 804–812 CrossRef CAS PubMed.
- R. J. DeBerardinis, N. Sayed and D. Ditsworth, et al., Brick by brick: metabolism and tumor cell growth, Curr. Opin. Genet. Dev., 2008, 18(1), 54–61 CrossRef CAS PubMed.
- M. G. Vander Heiden, L. C. Cantley and C. B. Thompson, Understanding the Warburg effect: the metabolic requirements of cell pro-liferation, Science, 2009, 324(5930), 1029–1033 CrossRef CAS PubMed.
- C. Ma, Y. Li and H. Wu, et al., Metabolomics analysis of the potential anticancer mechanism of annonaceous acetogenins on a multidrug resistant mammary adenocarcinoma cell, Anal. Biochem., 2018, 553, 1–6 CrossRef CAS PubMed.
- A.-H. Zhang, J.-B. Yu, H. Sun, L. Kong, X.-Q. Wang, Q.-Y. Zhang and X.-J. Wang, Identifying quality-markers from Shengmai San protects against transgenic mouse model of Alzheimer's disease using chinmedomics approach, Phytomedicine, 2018, 45, 84–92 CrossRef CAS PubMed.
- H. Sun, A. Zhang and Q. Song, et al., Functional metabolomics discover pentose and glucuronate interconversion pathways as promising targets for Yang Huang syndrome treatment with Yinchenhao Tang, RSC Adv., 2018, 8(64), 36831–36839 RSC.
- A. Zhang, H. Sun and G. Yan, et al., Chinmedomics: A Powerful Approach Integrating Metabolomics with Serum Pharmacochemistry to Evaluate the Efficacy of Traditional Chinese Medicine, Engineering, 2019, 5(1), 60–68 CrossRef.
- A. Zhang, H. Sun and G. Yan, et al., Metabolomics Analysis of Marker Metabolites for Patients with Pancreatic Cancer, Med. Chem., 2014, 4, 506–510 Search PubMed.
- X. Wang, H. Lv and A. Zhang, et al., Metabolite profiling and pathway analysis of acute hepatitis rats by UPLC-ESI MS combined with pattern recognition methods, Liver Int., 2014, 34(5), 759–770 CrossRef CAS PubMed.
- X. Zhang, X. Zhu and C. Wang, et al., Non-targeted and targeted metabolomics approaches to diagnosing lung cancer and predicting patient prognosis, Oncotarget, 2016, 7(39), 63437–63448 Search PubMed.
- S. Rangaraj and R. Venkatachalam, In vitro and in vivo characteristics of biogenic high surface silica nanoparticles in A549 lung cancer cell lines and Danio rerio model systems for inorganic biomaterials development, Artif. Cells, Nanomed., Biotechnol., 2018, 46(7), 1415–1424 CrossRef CAS PubMed.
- N. Kozar, K. Kruusmaa and M. Bitenc, et al., Data on metabolomic profiling of ovarian cancer patients' serum for potential diagnostic biomarkers, Data in Brief, 2018, 181825–181831 Search PubMed.
- S. T. Pan, Z. W. Zhou and Z. X. He, et al., Proteomic response to 5,6-dimethylxanthenone 4-acetic acid (DMXAA, vadimezan) in human non-small cell lung cancer A549 cells determined by the stable-isotope labeling by amino acids in cell culture (SILAC) approach, Drug Des., Dev. Ther., 2015, 9937–9968 Search PubMed.
- Z. Wang, Y. Zhang and Q. Liu, et al., Investigation of the mechanisms of Genkwa Flos hepatotoxicity by a cell metabolomics strategy combined with serum pharmacology in HL-7702 liver cells, Xenobiotica, 2018, 1–11 Search PubMed.
- Z. León, J. C. García-Cañaveras and M. T. Donato, et al., Mammalian cell metabolomics: experimental design and sample preparation, Electrophoresis, 2013, 34(19), 2762–2775 Search PubMed.
- N. Zhou, K. Hu and Z. Guo, et al., Thermo-Sensitive PLGA-PEG-PLGA Tri-Block Copolymer Hydrogel as Three-Dimensional Cell Culture Matrix for Ovarian Cancer Cells, J. Nanosci. Nanotechnol., 2018, 18(8), 5252–5255 CrossRef CAS PubMed.
- A. Zhang, H. Sun and H. Xu, et al., Cell metabolomics, OMICS, 2013, 17(10), 495–501 CrossRef CAS PubMed.
- H. Sun, A. H. Zhang and S. B. Liu, et al., Cell metabolomics identify regulatory pathways and targets of magnoline against prostate cancer, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2018, 1102–1103, 143–151 CrossRef CAS PubMed.
- M. Ding, F. Li, B. Wang, G. Chi and H. Liu, A comprehensive analysis of WGCNA and serum metabolomics manifests the lung cancer-associated disordered glucose metabolism, J. Cell. Biochem., 2019 DOI:10.1002/jcb.28377.
- R. R. Hafidh, S. Z. Hussein, M. Q. MalAllah, A. S. Abdulamir and F. Abu Bakar, A high-throughput quantitative expression analysis of cancer-related genes in human HepG2 cells in response to limonene, a potential anticancer agent, Curr. Cancer Drug Targets, 2018, 18(8), 807–815 CrossRef CAS PubMed.
- P. Ulivi, L. Mercatali and G. L. Casoni, et al., Multiple marker detection in peripheral blood for NSCLC diagnosis, PLoS One, 2013, 8(2), e57401 CrossRef CAS PubMed.
- M. I. Toki, N. Mani and J. W. Smithy, et al., Immune Marker Profiling and Programmed Death Ligand 1 Expression Across NSCLC Mutations, J. Thorac. Oncol., 2018, 13(12), 1884–1896 CrossRef PubMed.
- B. Callejón-Leblic, T. García-Barrera and A. Pereira-Vega, et al., Metabolomic study of serum, urine and bronchoalveolar lavage fluid based on gas chromatography mass spectrometry to delve into the pathology of lung cancer, J. Pharm. Biomed. Anal., 2019, 163, 122–129 CrossRef PubMed.
- A. Zhang, H. Sun and X. Wang, Urinary metabolic profiling of rat models revealed protective function of scoparone against alcohol induced hepatotoxicity, Sci. Rep., 2014, 4, 6768 CrossRef PubMed.
- X. Liu, A. Zhang and H. Fang, et al., Serum metabolomics strategy for understanding the therapeutic effects of Yin-Chen-Hao-Tang against Yanghuang syndrome, RSC Adv., 2018, 8(14), 7403–7413 RSC.
- A. Zhang, H. Sun and G. Yan, et al., Metabolomics study of type 2 diabetes using ultra-performance LC-ESI/quadrupole-TOF high-definition MS coupled with pattern recognition methods, J. Physiol. Biochem., 2014, 70(1), 117–128 CrossRef CAS PubMed.
- Q. Song, A. Zhang and G. Yan, et al., Technological advances in current metabolomics and its application in tradition Chinese medicine, RSC Adv., 2017, 7(84), 53516–53524 RSC.
- A. Zhang, H. Sun and W. Sun, et al., Metabolomics and Proteomics Annotate Therapeutic Mechanisms of Geniposide, Chinmedomics, Academic Press, 2015, pp. 157–173 Search PubMed.
- Q. Zhao, A. Zhang and W. Zong, et al., Chemometrics strategy coupled with high resolution mass spectrometry for analyzing and interpreting comprehensive metabolomic characterization of hyperlipemia, RSC Adv., 2016, 6(113), 112534–112543 RSC.
- X. Wang, Y. Han, A. Zhang, et al., Metabolic profiling provides a system for the understanding of Alzheimer's disease in rats post-treatment with Kaixin San, Chinmedomics, Academic Press, 2015, pp. 347–362 Search PubMed.
- A. Zhang, H. Wang and H. Sun, et al., Metabolomics strategy reveals therapeutical assessment of limonin on nonbacterial prostatitis, Food Funct., 2015, 6(11), 3540–3549 RSC.
- J. L. Ren, A. H. Zhang and L. Kong, et al., Advances in mass spectrometry-based metabolomics for investigation of metabolites, RSC Adv., 2018, 8(40), 22335–22350 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra00987f |
| This journal is © The Royal Society of Chemistry 2019 |