Open Access Article
Open Access ArticleCell changes and differential proteomic analysis during biodegradation of decabromodiphenyl ether (BDE-209) by Pseudomonas aeruginosa
Yu Liuabc,
Zhe Liuab,
Aijun Gong *ab,
Lina Qiuab,
Weiwei Zhangab,
Jingrui Liab,
Fukai Liab,
Yuzhen Baiab,
Jiandi Liab and
Ge Gaoab
*ab,
Lina Qiuab,
Weiwei Zhangab,
Jingrui Liab,
Fukai Liab,
Yuzhen Baiab,
Jiandi Liab and
Ge Gaoab
aSchool of Chemistry and Biological Engineering, University of Science and Technology Beijing, Beijing 100083, China. E-mail: gongaijun5661@ustb.edu.cn; Fax: +86-10-82375661; Tel: +86-10-82375661
bBeijing Key Laboratory for Science and Application of Functional Molecular and Crystalline Materials, University of Science and Technology Beijing, Beijing 100083, China
cInstitute of Biotechnology, Daqing Branch of Heilongjiang Academy of Science, Daqing 163319, China
First published on 12th August 2019
Abstract
Decabromodiphenyl ether (BDE-209) is a brominated flame retardant widely used in the world which, being an emerging persistent organic pollutant, poses a great potential hazard to both human health and the ecological environment. Microbial biodegradation has been considered as an effective and environment-friendly technique to remediate BDE-209. Pseudomonas aeruginosa, a Gram-negative bacterium capable of degrading BDE-209, was isolated from PBDEs-contaminated soil. To promote microbial biodegradation of BDE-209 and gain further insight into its mechanism, cell changes and differential proteomic analysis of P. aeruginosa during biodegradation were studied. The results showed that high cell surface hydrophobicity of P. aeruginosa make the bacteria absorb BDE-209 more easily. The increase in cell membrane permeability was caused by the P. aeruginosa responding to BDE-209 stress. IR spectra showed that hydroxyl, amide and CH2 groups in the P. aeruginosa cell surface were involved in the interactions between BDE-209 with P. aeruginosa. The apoptotic-like cell changes and cell surface morphology changes were observed by flow cytometry (FCM) and field emission scanning electron microscopy (FESEM), respectively. Differentially expressed protein was analysed by two-dimensional electrophoresis (2-DE) and 40 protein spots were identified to be different after 5 days biodegradation.
1 Introduction
Polybrominated diphenyl ethers (PBDEs), a class of brominated flame retardants (BFRs), are widely used as flame retardants in a variety of consumer products, such as plastics, textiles, electronic goods and construction materials.1 There are three major commercial formulations of PBDEs, which differ in the degree of bromination: penta-BDE, octa-BDE and deca-BDE.2 Penta-BDE and octa-BDE were banned under the Stockholm Convention in 2009. Deca-BDE (mainly BDE-209) was phased out in Europe and completely banned in the US in 2013. However, it is still produced and used in large quantities in some parts of the world.3,4 Due to the widespread use, chemical persistence and bioaccumulation, BDE-209 has been detected as the predominant BDE congener in atmosphere, water, soil, sediment, dust, especially in biota and humans, and its level has been increasing since 1970s.5 Many toxicological studies have demonstrated that exposure to BDE-209 can lead to neurobehavioral toxicity, thyroid hormone disruption, cancer and reproductive toxicity.6 Therefore ongoing human exposure to BDE-209 may cause a potential risk to human health, and there is an urgent need to develop efficient methods for degrading and eliminating BDE-209.Photolytic degradation, zerovalent iron (ZVI) degradation, and biodegradation are currently the major BDE-209 degradation methods.7 Biodegradation have gained much attention due to its being environment-friendly and low-cost. Recent studies on biodegradation of BDE-209 using microbes mostly focus on detecting degradation characteristics and proposing degradation pathways,8,9 but cell changes and differential proteomic analysis during biodegradation of BDE-209 are still unclear and deserve further investigation. Pseudomonas aeruginosa (P. aeruginosa), well capable of degrading BDE-209, could be used in bioremediation of the increasingly serious BDE-209-contamination. In the previous studies, Shi et al. reported the effect of cadmium ion on cell surface hydrophobicity (CSH) and membrane permeability variation during BDE-209 biodegradation by Pseudomonas aeruginosa. Results showed that direct adherence to the cell surface is the first step in biodegradation process,10 but no other changes of cell surface have been reported so far. Moreover, we have reported the biodegradation of BDE-209 by crude enzyme extracted from P. aeruginosa. Our results suggested that enzyme actually play the key role in the biodegradation process,11 but it is unknown which kind of enzyme is responsible for the BDE-209 degradation. Up to now, differentially expressed protein in P. aeruginosa cell during biodegradation of BDE-209 have never been reported.
In this work, to better understand the mechanism of BDE-209 biodegradation in cell level, P. aeruginosa cell changes were examined by focusing on cell activity, cell surface hydrophobicity, cell membrane permeability, cell surface functional group and morphology. Moreover, differentially expressed protein during the biodegradation of BDE-209 by P. aeruginosa was also investigated.
2 Materials and experiments
2.1 Chemicals
Decabromodiphenyl ether (BDE-209, >99% purity) was purchased from Alfa Aesar (Tianjin, China). The single standard of BDE-209 was purchased from Sigma (St. Louis, USA). All other reagents were analytical grade and purchased from Sinopharm Chemical Reagent Beijing Co., Ltd (Beijing, China).2.2 Bacterial strain cultivation
Pseudomonas aeruginosa (P. aeruginosa) was previously isolated from PBDEs-contaminated soil in China, and preserved by our laboratory. P. aeruginosa was enriched in 50 mL of beef extract peptone medium on a rotary shaker at 200 rpm and 30 °C for 30 h, followed by centrifugation at 8000g for 10 min at 4 °C. The harvested live cells were washed, resuspended, and cultivated in the mineral salt medium (MSM) with 20 mg L−1 BDE-209 on a rotary shaker at 200 rpm and 35 °C.The compositions of the beef extract peptone medium and the MSM medium were same as described in our previous research.11
2.3 BDE-209 biodegradation experiments
All BDE-209 biodegradation experiments were conducted in batch with 500 mL Erlenmeyer flasks. In each case, 6.0 mg BDE-209 and 300 mL MSM were added into a flask and mixed ultrasonically for 15 min to get a suspension. Afterwards, 120 mg live P. aeruginosa was transferred to the suspension and cultured on a rotary shaker at 200 rpm and 35 °C for 7 days. It was appropriate to keep cell density around 1 × 108 cells per mL. As for the control, dead P. aeruginosa instead of live cells was used. At the degradation time of 0, 1, 3, 5, 7 days, the quantification of BDE-209 was determined by using a gas chromatograph-mass spectrometer (GC-MS). BDE-209 degradation efficiency was calculated using the following equation:| Degradation efficiency (%) = (1 − C1/C0) × 100, |
2.4 Flow cytometry (FCM) analysis
P. aeruginosa cells cultivated in MSM containing 20 mg L−1 BDE-209 at different biodegradation time were harvested by centrifugation and washed twice with PBS buffer (pH 7.0), then re-suspended in 200 μL binding buffer with 5 μL Annexin V-FITC and 10 μL PI. The mixture was incubated in the dark at room temperature for 20 min to allow for complete staining. Live, apoptotic, and dead cell populations were analyzed by flow cytometry (BD FACSCalibur), and data were analyzed using FlowJo software (v10.0.7). The control was conducted under same condition but in the absence of BDE-209.2.5 Cell surface characteristics analysis
2.6 Differentially expressed protein analysis
P. aeruginosa cultured for 5 days in MSM with or without BDE-209 (20 mg L−1) were harvested by centrifugation at 6000g and 4 °C for 15 min and washed three times with cold PBS. The cell pellet was resuspended in the lysis buffer, and then cells were disrupted by sonication in an ice bath. The protein extracts underwent ultracentrifugation for 45 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and 4 °C. Afterwards, the obtained supernatants were purified using 2-D Clean-Up Kit (Amersham Biosciences, USA) and the protein concentration was determined by 2-D Quant Kit (Amersham Biosciences, USA) with bovine serum albumin as a standard. Two-dimensional electrophoresis (2-DE) was performed based on the method described by Liu et al.14 For the first dimension, isoelectric focusing (IEF), 500 μg of protein was loaded onto an immobilized pH gradient (IPG) strip (24 cm, pH 4–7). After IEF separation, the IPG strips were equilibrated in two equilibration buffer for 15 min as described by Song et al.15 Then the proteins were further separated on 12.5% SDS-polyacrylamide gels for the second dimension. After the completion of 2-DE, gels were stained with Coomassie brilliant blue, scanned using ImageScanner III (GE Healthcare) and analyzed with ImageMaster 2D Platinum software (Version 7.0, GE Healthcare). Altered abundances of spots were standardized and then compared based on their volume percentages in the total spot volume over the whole gel image. Significantly changed spots were selected by the rate increased/decreased ≥ 2-fold or complete appearance and disappearance. Differential expressed proteins were excised from the gels and digested with trypsin and analyzed by a 4800 Plus MALDI TOF/TOF mass spectrometer (Applied Biosystems, Foster City, USA). Both the MS and MS/MS data were integrated and processed using the GPS Explorer V3.6 software (Applied Biosystems, USA) and searched against the NCBI database using Mascot V2.1 software (Matrix Science, UK). Protein scores greater than 65 were significant and accepted.
000g and 4 °C. Afterwards, the obtained supernatants were purified using 2-D Clean-Up Kit (Amersham Biosciences, USA) and the protein concentration was determined by 2-D Quant Kit (Amersham Biosciences, USA) with bovine serum albumin as a standard. Two-dimensional electrophoresis (2-DE) was performed based on the method described by Liu et al.14 For the first dimension, isoelectric focusing (IEF), 500 μg of protein was loaded onto an immobilized pH gradient (IPG) strip (24 cm, pH 4–7). After IEF separation, the IPG strips were equilibrated in two equilibration buffer for 15 min as described by Song et al.15 Then the proteins were further separated on 12.5% SDS-polyacrylamide gels for the second dimension. After the completion of 2-DE, gels were stained with Coomassie brilliant blue, scanned using ImageScanner III (GE Healthcare) and analyzed with ImageMaster 2D Platinum software (Version 7.0, GE Healthcare). Altered abundances of spots were standardized and then compared based on their volume percentages in the total spot volume over the whole gel image. Significantly changed spots were selected by the rate increased/decreased ≥ 2-fold or complete appearance and disappearance. Differential expressed proteins were excised from the gels and digested with trypsin and analyzed by a 4800 Plus MALDI TOF/TOF mass spectrometer (Applied Biosystems, Foster City, USA). Both the MS and MS/MS data were integrated and processed using the GPS Explorer V3.6 software (Applied Biosystems, USA) and searched against the NCBI database using Mascot V2.1 software (Matrix Science, UK). Protein scores greater than 65 were significant and accepted.
2.7 Extraction and analytical methods
For the extraction procedure, we refer to previous work.11 The analytical method for BDE-209 was the same as in the literature16 and analysis by 7890-5975c GC-MS (Agilent, USA) equipped with a DB-5 MS column (60 m × 0.25 mm × 0.25 μm). The quantification was performed with a five-point standard curve of BDE-209.2.8 Quality assurance and quality control
All experiments were performed in triplicate and the mean values were given. The standard deviations ranged from 1.0% to 5.0%.3 Results
3.1 BDE-209 biodegradation by P. aeruginosa
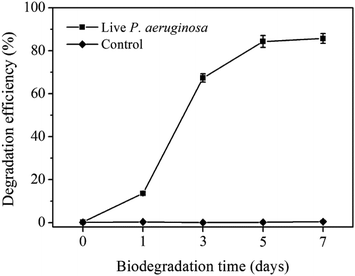
Fig. 1 shows the degradation efficiency of BDE-209 by P. aeruginosa during biodegradation time. As shown, the degradation efficiency of BDE-209 rose slowly in day 1 and then ascended sharply from day 3, finally levelled off after 5 days. This trend was in accordance with Shi's work,10 but changes in cellular properties during the biodegradation of BDE-209 by P. aeruginosa were not discussed deeply. Hence, in the following, we focused on cell changes of P. aeruginosa during degradation time.3.2 Apoptotic-like cell changes of P. aeruginosa during BDE-209 biodegradation
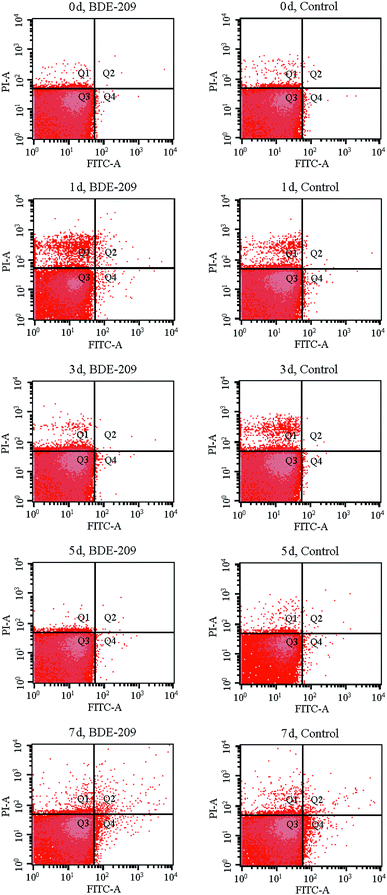
To quantitatively gain insight into apoptotic-like cell changes of P. aeruginosa during BDE-209 biodegradation, a display of PI versus Annexin V-FITC fluorescence was measured by FCM analysis (Fig. 2). As shown in Fig. 2, four groups of cells were clearly seen in each acquired data plot: dead cells (Q1, positive for PI and negative for Annexin V-FITC), late apoptotic cells (Q2, positive for PI and Annexin V-FITC), live cells (Q3, negative for PI and Annexin V-FITC), early apoptotic cells (Q4, negative for PI and positive for Annexin V-FITC). From the results it can be observed, P. aeruginosa cells treated with 20 mg L−1 BDE-209 in day 1 and day 7 exhibited an increase of apoptosis and death compared to day 0. The total apoptosis rate first increased to 1.17% in the day 1 as compared to that in day 0 (0.22%), then declined in day 3 and day 5, finally increased to 2.63% in day 7. Moreover, cells treated with BDE-209 showed the highest population of dead cells (5.84%) in day 1. For P. aeruginosa cells in control, cell death increased in day 1 (3.23%) and day 3 (4.11%), then cell apoptosis began to increase from day 5 (0.46%), and reached 1.97% in day 7.3.3 Cell surface changes during BDE-209 biodegradation
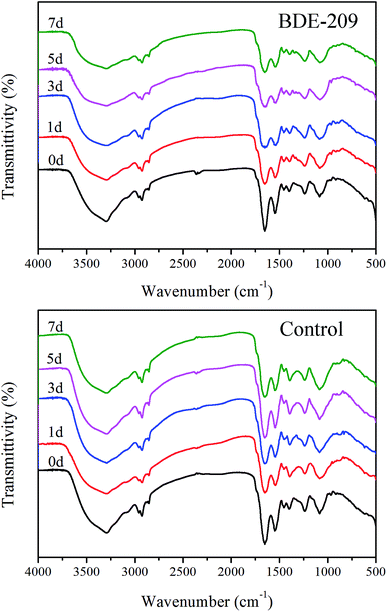
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of phosphate groups, 1398 cm−1 ascribed to C–O stretching vibration of –COOH, 1455 cm−1 ascribed to C–H bending vibration of –CH2, 1544 cm−1 ascribed to N–H and C–N in amide groups, 1651 and 1741 cm−1 ascribed to C
O stretching vibration of phosphate groups, 1398 cm−1 ascribed to C–O stretching vibration of –COOH, 1455 cm−1 ascribed to C–H bending vibration of –CH2, 1544 cm−1 ascribed to N–H and C–N in amide groups, 1651 and 1741 cm−1 ascribed to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration, 2925 and 2958 cm−1 ascribed to C–H stretching vibration, 3296 cm−1 ascribed to O–H stretching vibration.17 For P. aeruginosa cells treated with BDE-209, the peaks at 3296, 1651 and 1544 cm−1 become broader, and the intensity of peaks 2925 cm−1 decreased gradually. In control, less obvious changes with time were observed.
O stretching vibration, 2925 and 2958 cm−1 ascribed to C–H stretching vibration, 3296 cm−1 ascribed to O–H stretching vibration.17 For P. aeruginosa cells treated with BDE-209, the peaks at 3296, 1651 and 1544 cm−1 become broader, and the intensity of peaks 2925 cm−1 decreased gradually. In control, less obvious changes with time were observed.
3.4 Differentially expressed protein during BDE-209 biodegradation
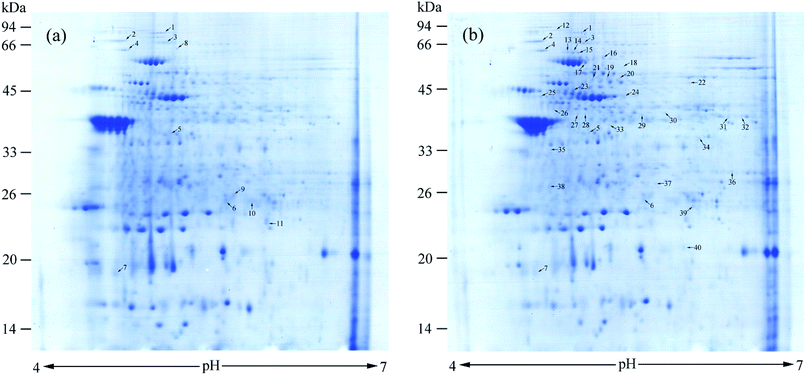
To investigate protein changes in P. aeruginosa cells during BDE-209 biodegradation, a comparative proteomic approach was utilized to identify differentially expressed proteins from P. aeruginosa cultured in MSM without BDE-209 and with 20 mg L−1 BDE-209. Representative maps of the proteins separated on a 2-DE gel were shown in Fig. 6. Approximately 40 protein spots were identified to be different after 5 days biodegradation. Out of them, seven protein spots in the same location of Fig. 6a and b exhibited significant changes at the level of protein expression, they were named as 1–7. Spots 8–11, and spots 12–40 were those spots that only appeared in Fig. 6a and b, respectively. The 40 spots were analysed by MALDI-TOF-TOF with a significant mascot score > 65 (p-value < 0.05), among which 12 identified spots are shown in Table 1. Table 1 summarizes details of these identified proteins including their protein names, accession numbers and other MS data.| Spots no | Protein name | Accession number | MW | PI | Mascot score | Peptides matched | Coverage (%) |
|---|---|---|---|---|---|---|---|
| 1 | Peroxiredoxin | gi|1279400824 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 643 643 |
5.89 | 760 | 93 | 65 |
| 2 | Molecular chaperone GroEL | gi|1279402470 | 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 107 107 |
5.04 | 536 | 47 | 9 |
| 3 | 30S ribosomal protein S1 | gi|1279398492 | 61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 946 946 |
4.83 | 2432 | 203 | 45 |
| 12 | Outer membrane protein assembly factor BamA | gi|1279401791 | 88![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 150 |
5.06 | 151 | 12 | 8 |
| 16 | NAD-dependent succinate-semialdehyde dehydrogenase | gi|1279399219 | 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 875 875 |
5.60 | 7433 | 400 | 62 |
| 18 | Isocitrate dehydrogenase (NADP(+)) | gi|518511667 | 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 689 689 |
5.10 | 1629 | 11 | 28 |
| 23 | 2-Phosphoglycerate dehydratase | gi|13124125 | 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 338 338 |
5.04 | 3059 | 202 | 68 |
| 24 | Agmatine deiminase | gi|1279399248 | 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 564 564 |
4.84 | 761 | 39 | 20 |
| 32 | L-Asparaginase | gi|1279400388 | 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 620 620 |
6.67 | 3026 | 249 | 52 |
| 35 | Electron transfer flavoprotein subunit beta | gi|518513080 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 535 535 |
4.85 | 1912 | 164 | 64 |
| 38 | ATP synthase subunit alpha | gi|1279400601 | 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 530 530 |
5.33 | 845 | 67 | 29 |
| 40 | dTDP-4-dehydrorhamnose 3,5-epimerase | gi|81622016 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 924 924 |
5.74 | 542 | 76 | 61 |
4 Discussion
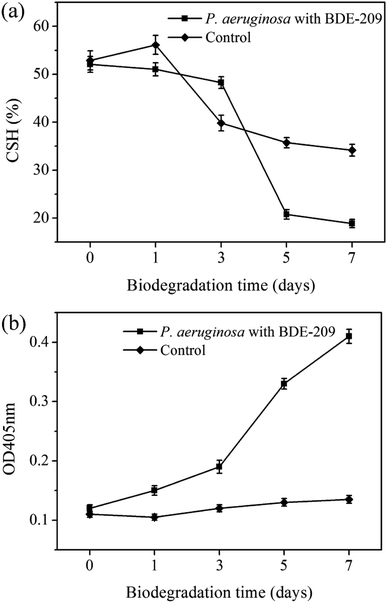
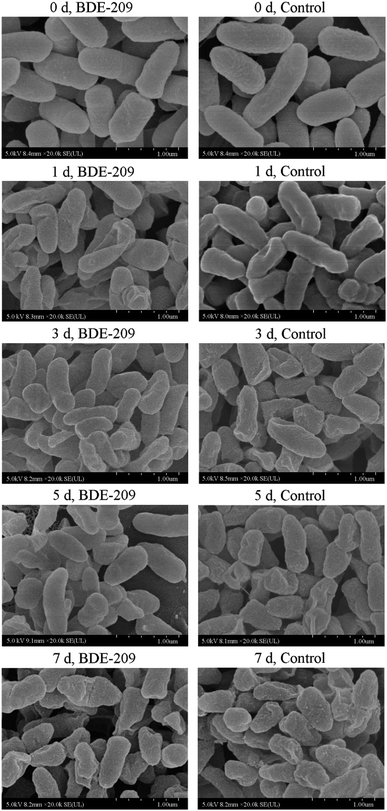
In the biodegradation process of harmful and toxic organic contaminants, microbes resist organic contaminants at the beginning, and then the interaction between microbes and contaminants leads to a series of cellular characteristic changes.18 In this work, we have found that P. aeruginosa had a series of cellular characteristic changes during the biodegradation of BDE-209. When BDE-209 biodegradation had just began (day 0), P. aeruginosa treated with and without BDE-209 both grew well and had normal cell morphology. In day 1, BDE-209 degradation efficiency was low. Compared with control, P. aeruginosa treated with BDE-209 caused a larger population of apoptotic and dead cells, accompanied by the appearance of plicated and irregular shaped cells. Apoptosis is a form of programmed cell death (PCD) that is essential in eliminating old, unnecessary or damaged cells to maintain appropriate cell numbers and protect bacteria against poor environment.19,20 So this is probably because the initial exposure of P. aeruginosa to BDE-209 lead to resistance, some cells become deformed, and then apoptosis appears to protect bacteria against BDE-209 stress.21 In day 3 and 5, BDE-209 degradation efficiency increased apparently, apoptotic and dead cell decreased, cell morphology return to normal. This is probably because P. aeruginosa has adapted to BDE-209 gradually, followed by degrading BDE-209 as carbon source for metabolism.22 As for control, these changes were not observed. In day 7, no matter P. aeruginosa was treated with or without BDE-209, cell apoptosis and death appeared, and cells became rough and atrophic. This is probably because the quantity of nutrients in the culture medium declined and growing environment became unfavorable.23One of most common mechanisms explains that hydrophobic BDE-209 first adheres to the hydrophobic regions of the cell surface, then enters the cell through cell membrane, finally is degraded by intracellular enzymes.24,25 For this reason, CSH and cell membrane permeability are the most important factors in determining whether BDE-209 can be successfully adsorbed onto the surface of cells and enter into the cells. In this experiment, we have found that CSH maintained at a high level till day 3, making P. aeruginosa adsorb hydrophobic BDE-209 more easily. Then CSH decreased markedly from day 5. Meanwhile, the cell membrane permeability increased gradually from day 1 then sharply from day 5. The increase in cell membrane permeability was caused by the P. aeruginosa responding to BDE-209 stress. The membrane structure was changed due to the sharply increased cell membrane permeability and the hydrophobic regions was disrupted, which may be part of the reason for the decline of CSH of P. aeruginosa.20
P. aeruginosa is a Gram-negative bacterium. Its cell wall is mainly composed of polysaccharides, phospholipids and proteins, characteristic of carboxyl, phosphate, and amino groups, which can be assigned to in IR spectra (Fig. 4).26 During degradation, the changed peaks suggested that hydroxyl, amide and CH2 groups were involved in the interactions between BDE-209 and P. aeruginosa. In control, these peaks did not change obviously with time. All the above results indicated that polysaccharides and proteins in the cell walls of P. aeruginosa may play critical roles in the binding of BDE-209.27
2-DE is an effective tool for gaining insight into physiological changes at cellular level.28 Thus analysis and identification of differentially expressed proteins during BDE-209 biodegradation could give us an insight into cell changes in this process. In this experiment, 12 differentially expressed proteins were identified and listed in Table 1. Peroxidase can degrade BDE-209, Li et al. found that manganese peroxidase plays an important role in BDE-209 depletion.29 30S ribosomal protein S1, which can bind to a specific site of mRNA, starts protein synthesis.30 Electron transfer flavoprotein can transfer the electrons to the main respiratory chain.31 Outer membrane protein can form a stable folding intermediate, which keeps the Gram-negative bacterium survive under stress conditions.32 Molecular chaperone works to prevent outer membrane protein being adsorbed to outer membrane by capturing and stabilizing its folding intermediates.33 Therefore, these enzymes help P. aeruginosa adapt to restricted BDE-209 conditions, reduce toxicity, and maintain the normal metabolism of cells.34 NAD-dependent succinate-semialdehyde dehydrogenase, isocitrate dehydrogenase and 2-phosphoglycerate dehydratase were probably attributed to the resistance systems of P. aeruginosa to the oxidative stress induced by BDE-209.35 ATP synthase generates energy in the form of ATP, which was confirmed to play an important role in BDE biodegradation.36 The roles of agmatine deiminase, L-asparaginase and dTDP-4-dehydrorhamnose 3,5-epimerase in BDE-209 degradation is not known yet and further study is still needed.
5 Conclusions
Collectively, both cell surface and intracellular enzyme changes in the biodegradation of BDE-209 by P. aeruginosa. The apoptotic-like cell changes and cell surface morphology changes were observed during the biodegradation of BDE-209 by P. aeruginosa. The results showed that high CSH of P. aeruginosa facilitated the attachment of BDE-209 to cell surface. The increase in cell membrane permeability caused by P. aeruginosa responding to BDE-209 stress favored BDE-209 penetrating into cells and being degraded. IR spectra showed that polysaccharides and proteins in the cell walls of P. aeruginosa may participate in binding BDE-209. Differentially expressed protein was analysed through 2-DE, among which peroxidase and ATP synthase play a critical role in BDE-209 degradation. As far as we know, this is the first report on investigating cell changes and differentially expressed proteins during biodegradation of BDE-209 by P. aeruginosa. These explorations offer new insights into the mechanism of BDE-209 biodegradation by P. aeruginosa.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by National Key R&D Program of China (No. 2017YFF0106006) and Chinese National Natural Science Foundation (No. 51701016).References
- C. A. de Wit, Chemosphere, 2002, 46, 583–624 CrossRef CAS PubMed.
- R. Quiroz, L. Arellano, J. O. Grimalt and P. Fernandez, J. Chromatogr., A, 2008, 1192, 147–151 CrossRef CAS PubMed.
- A. Francois, R. Techer, M. Houde, P. Spear and J. Verreault, Environ. Toxicol. Chem., 2016, 35, 2215–2222 CrossRef CAS PubMed.
- H. W. Zhu, Y. Wang, X. W. Wang, T. G. Luan and N. F. Tam, Sci. Total Environ., 2014, 468–469, 130–139 CrossRef CAS PubMed.
- H. Stiborova, J. Vrkoslavova, P. Lovecka, J. Pulkrabova, P. Hradkova, J. Hajslova and K. Demnerova, Chemosphere, 2015, 118, 315–321 CrossRef CAS PubMed.
- M. Li, Z. Liu, L. Gu, R. Yin, H. Li, X. Zhang, T. Cao and C. Jiang, Front. Genet., 2014, 5, 1–6 CAS.
- B. Z. Wu, Y. J. Sun, Y. H. Chen, H. K. Yak, J. J. Yu, W. S. Liao, K. H. Chiu and S. M. Peng, Chemosphere, 2016, 157, 115–123 CrossRef CAS PubMed.
- M. Lu, Z. Z. Zhang, X. J. Wu, Y. X. Xu, X. L. Su, M. Zhang and J. X. Wang, Bioresour. Technol., 2013, 149, 8–15 CrossRef CAS PubMed.
- G. Y. Xu and J. B. Wang, Chemosphere, 2014, 110, 70–77 CrossRef CAS PubMed.
- G. Y. Shi, H. Yin, J. S. Ye, H. Peng, J. Li and C. L. Luo, J. Hazard. Mater., 2013, 263, 711–717 CrossRef CAS PubMed.
- Y. Liu, A. J. Gong, L. N. Qiu, J. R. Li and F. K. Li, Int. J. Environ. Res. Public Health, 2015, 12, 11829–11847 CrossRef CAS PubMed.
- D. Y. Ren, C. Li, Y. Q. Qin, R. L. Yin, X. Li, M. Y. Tian, S. W. Du, H. H. Guo, C. X. Liu, N. Zhu, D. D. Sun, Y. Li and N. Y. Jin, Anaerobe, 2012, 18, 508–515 CrossRef CAS PubMed.
- L. L. Wang, L. T. Tang, R. Wang, X. Y. Wang, J. S. Ye and Y. Long, Environ. Sci. Pollut. Res., 2016, 23, 5166–5178 CrossRef CAS PubMed.
- S. S. Liu, C. L. Guo, Z. Dang and X. J. Liang, Ecotoxicol. Environ. Saf., 2017, 137, 256–264 CrossRef CAS PubMed.
- Q. Q. Song, P. F. Zheng, L. G. Qiu, X. Jiang, H. W. Zhao, H. L. Zhou, Q. Han and X. P. Diao, Toxicol. Lett., 2016, 240, 185–195 CrossRef CAS PubMed.
- Y. Liu, A. J. Gong, L. N. Qiu, J. R. Li and F. K. Li, MicrobiologyOpen, 2017, 6, 1–10 CrossRef CAS PubMed.
- N. Bai, S. Wang, R. Abuduaini, M. N. Zhang, X. F. Zhu and Y. H. Zhao, Sci. Total Environ., 2017, 590–591, 343–351 CrossRef CAS PubMed.
- S. S. Liu, C. L. Guo, X. J. Liang, F. J. Wu and Z. Dang, Ecotoxicol. Environ. Saf., 2016, 129, 210–218 CrossRef CAS PubMed.
- N. Allocati, M. Masulli, C. Di Ilio and V. De Laurenzi, Cell Death Dis., 2015, 6, e1609 CrossRef CAS PubMed.
- S. Tang, H. Yin, S. Chen, H. Peng, J. Chang, Z. Liu and Z. Dang, J. Hazard. Mater., 2016, 308, 335–342 CrossRef CAS PubMed.
- M. C. Ferrante, G. M. Raso, E. Esposito, G. Bianco, A. Iacono, M. T. Clausi, P. Amero, A. Santoro, R. Simeoli, G. Autore and R. Meli, Toxicol. Lett., 2011, 202, 61–68 CrossRef CAS PubMed.
- L. L. Liu, Y. C. Zhang, R. H. Liu, Z. P. Wang, F. Xu, Y. L. Chen and K. F. Lin, Environ. Sci. Pollut. Res., 2016, 23, 3925–3933 CrossRef CAS PubMed.
- H. Lee and D. G. Lee, J. Microbiol. Biotechnol., 2019, 29, 1014–1021 CrossRef PubMed.
- L. Shapiro, Cell, 1993, 73, 841–855 CrossRef CAS PubMed.
- Z. Wu, M. Xie, Y. Li, G. Gao, M. Bartlam and Y. Wang, Amb. Express, 2018, 8, 27 CrossRef PubMed.
- S. Luo, L. L. Li, A. W. Chen, Q. R. Zeng, H. Xia and J. D. Gu, Chemosphere, 2017, 178, 187–196 CrossRef CAS PubMed.
- N. Bai, S. Wang, R. Abuduaini, M. Zhang, X. F. Zhu and Y. H. Zhao, Sci. Total Environ., 2017, 590, 343–351 CrossRef PubMed.
- Z. W. Cheng, Z. Y. Chen, X. Yan, Y. W. Bian, X. Deng and Y. M. Yan, J. Proteomics, 2018, 170, 1–13 CrossRef CAS PubMed.
- X. Li, A. Y. Chen, Y. Wu, L. Wu, L. Xiang, H. M. Zhao, Q. Y. Cai, Y. W. Li, C. H. Mo, M. H. Wong and H. Li, Sci. Total Environ., 2018, 634, 417–426 CrossRef CAS PubMed.
- M. P. Kalapos, H. Paulus and N. Sarkar, Biochimie, 1997, 79, 493–502 CrossRef CAS PubMed.
- C. L. Hall and J. D. Lambeth, J. Biol. Chem., 1980, 255, 3591–3595 CAS.
- K. Chourabi, F. Torrella, S. Kloula, J. A. Rodriguez, I. Trabelsi, S. Campoy, A. Landoulsi and A. Chatti, Arabian J. Geosci., 2017, 10, 1–7 CrossRef.
- R. N. Reusch, FEBS J., 2012, 279, 894–909 CrossRef CAS PubMed.
- J. Xu, H. Q. Wang and D. K. Kong, IOP Conf. Ser. Earth Environ. Sci., 2018, 111, 012032 CrossRef.
- A. K. Felux, D. Spiteller, J. Klebensberger and D. Schleheck, Proc. Natl. Acad. Sci., 2015, 112, E4298–E4305 CrossRef CAS PubMed.
- M. Feng, H. Yin, H. Peng, X. T. Liu, P. P. Yang, G. N. Lu and Z. Dang, Ecotoxicol. Environ. Saf., 2017, 142, 388–398 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |