Open Access Article
Open Access ArticleExposure to microwave irradiation at constant culture temperature slows the growth of Escherichia coli DE3 cells, leading to modified proteomic profiles†
Sina Atrin Mazinani‡
a,
Nour Noaman‡b,
Melissa R. Pergandec,
Stephanie M. Cologna c,
Jens Coorssen*b and
Hongbin Yan
c,
Jens Coorssen*b and
Hongbin Yan *a
*a
aDepartment of Chemistry and Centre for Biotechnology, Brock University, St. Catharines, Ontario, L2S 3A1, Canada. E-mail: tyan@brocku.ca
bDepartment of Applied Health Sciences, Department of Biological Sciences, Brock University, St. Catharines, Ontario, L2S 3A1, Canada. E-mail: jcoorssen@brocku.ca
cDepartment of Chemistry, University of Illinois at Chicago, 845 W. Taylor St., Chicago, IL 60607, USA
First published on 16th April 2019
Abstract
Despite a few decades of research, interest continues in understanding the potential influences of low energy microwave irradiation on biological systems. In the present study, growth of E. coli DE3 in LB media slowed in the presence of microwave irradiation (max. 10 W) while the temperature of cultures was maintained at 37 °C. Viable cell counts in microwave-irradiated cultures were also significantly lower. When microwave irradiation was ceased, E. coli growth was restored. A top-down proteomic analysis of total proteins isolated from control and microwave-irradiated E. coli cultures revealed differential abundance of 10 resolved protein spots, with multiple proteins identified in each following mass spectrometric analysis. Among these proteins, a number are involved in metabolism, suggesting alterations to metabolic activities following microwave irradiation. Furthermore, four amino acid-tRNA ligases were also identified, pointing to the possibility of stress responses in E. coli under microwave irradiation.
1. Introduction
With increased applications of electromagnetic waves in many aspects of our daily life, determining the effects of microwave irradiation on biological systems is not only essential, but far from fully elucidated. Notions of ‘thermal’ and ‘non-thermal’ effects (the latter is also referred to as ‘microwave-specific effects’) of microwaves have been under repeated scrutiny in the past few decades.1–10 Some recent literature argued that due to the unique manner of energy transfer by microwaves, many observations that are unique to microwave exposure can be attributed to ‘selective heating’ of substances in a system of interest.11,12 As a result, isolation of thermal from microwave-specific effects is challenging,13–15 further complicating the interpretation of experimental results.In this respect, we previously demonstrated that a CEM (Matthews, NC) microwave reactor operating at 2.45 GHz coupled with a Coolmate system allows for systems under investigation to remain typically within ±1 °C throughout experiments (with occasional brief fluctuations of ±2 °C), as measured in situ by a fibre optic temperature probe and an alcohol-based thermometer, when the system is subjected to low-power microwave irradiation while cooled simultaneously. We were particularly interested in studies in which relatively low microwave energy (up to 10 W) is used, as higher microwave output is, presumably, more likely to exacerbate the issue of local heating and generation of hotspots. With these considerations, we demonstrated that while the enzymatic activity of trypsin during digestion of substrates, such as casein, is increased by exposure to microwave irradiation (10 W) at constant bulk temperature,16 a similar impact was not found for α-amylase and phosphatase.17 We further demonstrated that exposure of PC-3 human prostate cancer cells to non-lethal microwave irradiation (10 W) at constant culture temperature (37 °C) likely led to perturbation of cell membrane properties, while no necrosis nor apoptosis was induced.18 Most recently, we showed that uptake of the anticancer compound doxorubicin by PC-3 and human breast cancer MCF-7 cells was significantly enhanced when the cells were exposed to non-lethal microwave irradiation.19
In the present study, we extended our investigation to probe the influence of microwave exposure on bacterial growth while the culture temperature was maintained constant. While many reports show no differences in microorganisms exposed to microwave irradiation as compared with controls,20–26 there is also literature showing that microorganisms respond differently to microwave irradiation of various frequencies, leading to altered growth rate or metabolic activities.21,27–36 Comparisons of results from the literature are compounded by differences in at least the following factors: frequency and power output of the microwave used, duration and pattern of irradiation, choice of biological system, method used in temperature measurement, and the extent of temperature fluctuation.
This study focused on the exposure of E. coli DE3 cells to microwaves at 2.45 GHz, with relatively low power output (up to 10 W), while the culture temperatures were monitored in situ with a fibre optic temperature probe, maintaining the culture temperatures at 37 °C through simultaneous cooling. We report herein that E. coli growth is slowed under these conditions. Furthermore, a top-down proteomic analysis of total proteins suggest that cellular metabolism is modified in bacteria exposed to microwave irradiation.
2. Results and discussions
2.1. Temperature control
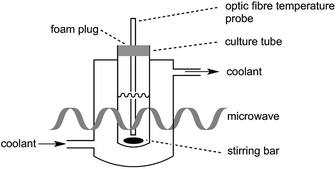
In the present study, great care was taken to ensure that the temperature of the bacterial cultures was controlled through simultaneous cooling and constant stirring; cultures were kept sterile with the use of a foam plug (Fig. 1). Pre-cooled fluid (Galden HT 110) was circulated through the jacket surrounding the culture tube in order to maintain culture temperatures. As we have previously established,17 the temperatures measured by a fibre optic temperature probe and an alcohol-based thermometer are very consistent. For simplicity, a fibre optic temperature probe was used here, as it allowed us to readily keep track of culture temperatures in real-time using the Synergy program. As is shown in Fig. S1,† temperatures of the culture were maintained at 37 °C (typically ±1 °C with occasional brief fluctuations of ±2 °C) throughout the 27.5 h growth period.It is worth noting that as the fibre optic temperature probe provides feedback to the microwave, the microwave power output from the magnetron was not constant. As shown in Fig. S2,† while the maximal power output was set at 10 W, actual power output was below 10 W (>6 W for most of the irradiation period), but did not exceed this pre-set maximal value.
2.2. E. coli growth curve
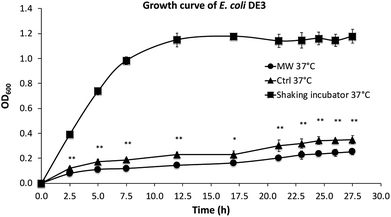
As shown in Fig. 2, the growth rates of E. coli, both in the presence and absence of microwave exposure, were slower than that in a shaking incubator, which is the result of less efficient oxygen transfer in the culture tubes under stirring compared with cultures in a shaking incubator. The growth of E. coli in the presence of maximal 10 W microwave irradiation at 37 °C was found to be significantly slower than control cultures not exposed to microwave irradiation.2.3. Dissolved oxygen (DO) level in tube cultures
Considering the possibility that the slower growth of E. coli in the presence of microwave irradiation might be due to lower DO concentration when cultures are exposed to microwave irradiation, experiments were carried out to compare the DO levels in tube cultures isolated from the environment with foam plugs, in the presence and absence (control) of microwave irradiation.Table S1† shows that the DO levels are slightly lower in the LB media treated with microwave irradiation relative to the control. This observation is consistent with a recent study that demonstrated efficient degassing by microwave exposure, however, at much higher temperatures.37
2.4. Recovery of growth in the E. coli culture exposed to microwave irradiation
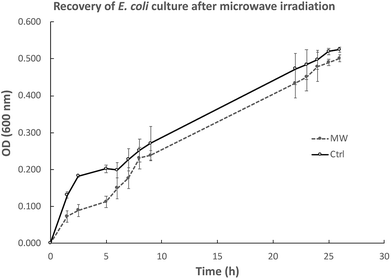
We were also interested in exploring whether the slower growth of E. coli subjected to microwave irradiation is reversible when microwave exposure stops. To this end, after E. coli cultures were exposed to maximal 10 W microwave irradiation at 37 °C for 5 h, the cultures were then incubated at 37 °C in an oil-bath, and growth rate (measured by OD600) was assessed. As depicted in Fig. 3, the difference in cell density between the microwave-treated and control cultures decreased when microwave irradiation was stopped. Viability tests revealed that 21 h after microwave exposure ceased, the difference in viable cell counts in both cultures was not significant (Fig. 4).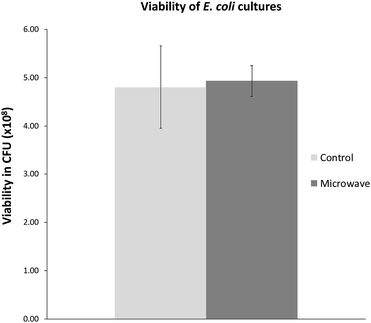 | ||
| Fig. 4 Viable cell counts 21 h after microwave exposure as compared with control. Graph shows mean ± std. dev. | ||
2.5. Two-dimensional gel electrophoresis (2DE) of total proteins isolated from E. coli DE3 cultures
Studies have shown that physical phenomenon such as electromagnetic waves can act as stimuli that may trigger responses in biological systems.38,39 Based on the current observations that E. coli cells grew more slowly in the presence of microwave irradiation, we became interested in studying the protein profiles, which could provide insight into potentially altered biological processes in E. coli under microwave exposure.In order to investigate this scenario, total proteins were isolated from both control and microwave-treated E. coli cultures for 2DE-based proteomic analysis. Proteins from three biological replicates from both control and microwave-treated E. coli cultures were extracted and analyzed by 2DE in technical triplicates (all gel images are available in Fig. S3†). Changes in spot densities between the resolved proteomes of microwave-treated vs. control E. coli were identified by quantitative image analysis. Here, 10 spots were identified as changing significantly, with three decreasing and seven increasing in abundance (Fig. 5).
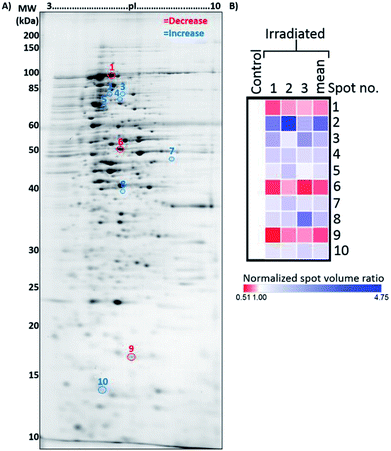 | ||
| Fig. 5 Representative 2DE gel used for the analysis of proteins extracted from control and microwave-treated E. coli cultures. (A) Annotated 2DE gel image showing protein spots which changed in abundance following microwave irradiation. Spots which decreased in abundance are delineated in red; those which increased are delineated in blue. Refer to Table 1 for calibrated pI and molecular weights of each delineated spot. (B) Heat map displaying normalised spot volume ratios in irradiated cultures relative to control cultures, across three biological replicates, averaged from three technical gel replicates each. Mean ratio is also shown. | ||
2.6. Mass spectrometric (MS) analysis
Using the criteria described in the Experimental section, within these 10 resolved spots a total of 42 proteins were identified by comparing the MS data against known E. coli K12 proteins (Table 1). These proteins were grouped into two clusters according to the spot intensity seen in the 2D gels, one containing 16 proteins with overall decreased abundance and another containing 26 proteins with overall increased abundance in the microwave-treated cultures, respectively. Fig. S4† shows the LC-MS chromatograms of the 10 resolved protein spots.| Protein spot | Abundance in irradiated E. coli [fold-change (p-valuea)] | Protein ID | Accession | Coverage (%) | No. of unique peptides | Theoretical MW [kDa]/pI | Calculatedb MW [kDa]/pI | Sequest score |
|---|---|---|---|---|---|---|---|---|
| a Significantly different from control, as displayed in the Delta2D quantitation table (Student’s t-test).b Calculated in Delta2D, calibrated with Bio-Rad 2-D PAGE standards. | ||||||||
| 1 | ↓1.3 (0.004) | Aconitate hydratase B | P36683 | 44 | 38 | 93.4/5.4 | 83.4/4.9 | 846.66 |
| Pyruvate dehydrogenase E1 component | P0AFG8 | 25 | 22 | 99.6/5.7 | 85.02 | |||
| Aldehyde–alcohol dehydrogenase | P0A9Q7 | 17 | 12 | 96.1/6.8 | 71.4 | |||
| 2-Oxoglutarate dehydrogenase E1 component | P0AFG3 | 12 | 9 | 105/6.5 | 23.09 | |||
| NADH-quinone oxidoreductase subunit G | P33602 | 9 | 9 | 100.2/6.3 | 22.31 | |||
| Leucine-tRNA ligase | P07813 | 9 | 7 | 97.2/5.3 | 17.1 | |||
| Elongation factor G | P0A6M8 | 8 | 4 | 77.5/5.4 | 9.61 | |||
| DNA mismatch repair protein MutS | P23909 | 3 | 2 | 95.2/5.55 | 4.3 | |||
| DNA gyrase subunit A | P0AES4 | 2 | 2 | 96.9/5.2 | 4.22 | |||
| 2 | ↑3.0 (<0.001) | GTP-binding protein TypA/BipA | P32132 | 13 | 6 | 67.3/5.3 | 73.4/4.8 | 5.33 |
| Catalase-peroxidase | P13029 | 8 | 5 | 80/5.3 | 5.31 | |||
| Formate acetyltransferase 1 | P09373 | 4 | 3 | 85.3/6.0 | 6.01 | |||
| Polyribonucleotide nucleotidyltransferase | P05055 | 3 | 2 | 77.1/5.2 | 5.21 | |||
| 3 | ↑2.0 (<0.001) | Formate acetyltransferase 1 | P09373 | 16 | 11 | 85.3/6.0 | 72.7/5.3 | 6.01 |
| Phosphoenolpyruvate-dependent phosphotransferase system | P37177 | 3 | 2 | 83.7/5.8 | 5.78 | |||
| 4 | ↑1.6 (<0.001) | Exoribonuclease 2 | P30850 | 14 | 7 | 72.4/5.6 | 70.6/5.2 | 5.62 |
| Chaperone protein ClpB | P63284 | 7 | 5 | 95.5/5.5 | 5.52 | |||
| Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex | P06959 | 5 | 3 | 66.1/5.2 | 5.17 | |||
| Glycine-tRNA ligase beta subunit | P00961 | 2 | 2 | 76.8/5.4 | 5.44 | |||
| Bifunctional polymyxin resistance protein ArnA | P77398 | 3 | 2 | 74.2/6.9 | 6.87 | |||
| 5 | ↑1.4 (0.001) | Chaperone protein HtpG | P0A6Z3 | 49 | 33 | 71.4/5.2 | 67.7/4.7 | 5.21 |
| Lysine-tRNA ligase, heat inducible | P0A8N5 | 26 | 12 | 57.8/5.2 | 5.24 | |||
| Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex | P06959 | 14 | 7 | 66.1/5.2 | 5.17 | |||
| Proline-tRNA ligase | P16659 | 8 | 4 | 63.7/5.2 | 5.24 | |||
| 30S ribosomal protein S1 | P0AG67 | 6 | 4 | 61.1/5.0 | 4.98 | |||
| Polyribonucleotide nucleotidyltransferase | P05055 | 3 | 2 | 77.1/5.2 | 5.21 | |||
| Methionine-tRNA ligase | P00959 | 3 | 2 | 76.2/5.9 | 5.86 | |||
| 6 | ↓1.5 (<0.001) | D-Tagatose-1,6-bisphosphate aldolase subunit GatZ | P0C8J8 | 18 | 7 | 47.1/5.8 | 53.7/5.2 | 5.77 |
| Periplasmic pH-dependent serine endoprotease DegQ | P39099 | 8 | 3 | 47.2/6.0 | 5.95 | |||
| Dihydrofolate synthase/folylpolyglutamate synthase | P08192 | 5 | 2 | 45.4/5.8 | 5.8 | |||
| Xaa-Pro aminopeptidase | P15034 | 6 | 2 | 49.8/5.4 | 5.39 | |||
| Bifunctional protein HldE | P76658 | 5 | 2 | 51.0/5.4 | 5.41 | |||
| 7 | ↑1.5 (0.006) | NADH-quinone oxidoreductase subunit F | P31979 | 12 | 5 | 49.3/6.9 | 49.9/6.6 | 6.86 |
| 8 | ↑1.9 (<0.001) | Aspartate aminotransferase | P00509 | 17 | 7 | 43.5/5.8 | 43.3/5.3 | 5.77 |
| NADPH-dependent curcumin reductase | P76113 | 7 | 2 | 37.6/5.8 | 5.82 | |||
| Aminomethyltransferase | P27248 | 5 | 2 | 40.1/5.6 | 5.57 | |||
| Maltose-binding periplasmic protein | P0AEX9 | 6 | 2 | 43.4/5.7 | 5.71 | |||
| Fructose-bisphosphate aldolase class 2 | P0AB71 | 6 | 2 | 39.1/5.9 | 5.86 | |||
| 9 | ↓1.5 (0.006) | DNA protection during starvation protein | P0ABT2 | 50 | 12 | 18.7/6.1 | 16.9/5.6 | 6.11 |
| 50S ribosomal protein L13 | P0AA10 | 10 | 2 | 16.0/9.9 | 9.91 | |||
| 10 | ↑1.4 (0.011) | 10 kDa chaperonin | P0A6F9 | 65 | 6 | 10.4/5.2 | 13.4/4.9 | 5.24 |
| 30S ribosomal protein S6 | P02358 | 13 | 2 | 15.7/5.0 | 5 | |||
Analysis of the proteins with decreased levels revealed that many proteins are involved in metabolic pathways, including the citric acid cycle, such as aconitate hydratase B, pyruvate dehydrogenase E1 component, aldehyde–alcohol dehydrogenase, 2-oxoglutarate dehydrogenase E1 component, NADH-quinone oxidoreductase subunit G, D-tagatose-1,6-bisphosphate aldolase subunit, periplasmic pH-dependent serine endoprotease, dihydrofolate synthase/folylpolyglutamate synthase and bifunctional protein HldE.
For the 26 proteins with increased levels, four enzymes required for the aminoacyl-tRNA biosynthesis pathway were identified, glycine-tRNA ligase (beta subunit), lysine-tRNA ligase (heat inducible), proline-tRNA ligase and methionine-tRNA ligase.
These results suggest that exposure of E. coli to microwave irradiation under the experimental conditions led to slower growth, likely due to stress imposed by microwave exposure. In response to the stimulation, metabolism of E. coli cells is down-regulated. The finding that four enzymes involved in tRNA synthesis were up-regulated also corroborates this hypothesis, as the literature points to the regulation of cellular tRNA levels as a response to stresses, such as starvation and heat.40–42
Taken together, the proteomic results from this study support the possibility that exposure of E. coli to non-lethal microwave irradiation at constant culture temperatures can be perceived by the microorganism as a stimulus. In adaptation, the bacteria modulate their metabolism and tRNA biosynthesis.
3. Materials and methods
Chemicals were purchased from Sigma-Aldrich unless otherwise stated, and were used without further purification. E. coli DE3 was a gift from Professor Charles Déspres’ lab (Department of Biological Sciences, Brock University).3.1. Microwave irradiation
A CEM Discovery Coolmate microwave system that operates at 2.45 GHz was used for microwave irradiation, and controlled with Synergy software from CEM (Matthews, NC). The standard Reaction Vessel (CEM part number 168302) was used as culture tubes, inserted into the standard Attenuator Assembly (CEM part number 542476), which provides cooling to the culture through circulating coolant. Solvay Solexis H Galden Coolant Zt 130 was pre-cooled in the coolant reservoir of Coolmate by liquid nitrogen. Temperature of cultures was preset as desired in Synergy and maintained through adjustment of the temperature of coolant and flow rates. The instrument and culture set-up are described in Fig. 1. Speed of the stirring bar was determined with an Omega HHT13 tachometer.3.2. Generation of E. coli growth curves from cultures in a shaking incubator
Single bacterial colonies from LB agar plates were grown in LB broth in a shaking incubator at 37 °C and 260 rpm until an OD600 in a range of 0.8–0.9 was reached. Aliquots (200 μl) were taken at appropriate intervals, and absorbance was read at 600 nm using a Biotek 405U microtitre plate reader.3.3. Generation of E. coli growth curves from tube cultures in the presence of microwave and in hotplate oil-bath
100 μl of cultures grown in the shaking incubator was added to fresh LB media (4.9 ml, to make a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 dilution) in the reaction vessel in the CEM microwave reactor and exposed to a maximal 10 W microwave irradiation while the culture temperature was maintained at 37 °C through simultaneous cooling with constant stirring (stirring speed set as “high” at 800 ± 20 rpm). The speed of the stirring bar was measured in rpm by an Omega HHT13-Kit tachometer. The culture tube was kept sterile by a foam plug. Aliquots (200 μl) were taken at appropriate intervals, and absorbance was read at 600 nm using a Biotek 405U microtitre plate reader.
50 dilution) in the reaction vessel in the CEM microwave reactor and exposed to a maximal 10 W microwave irradiation while the culture temperature was maintained at 37 °C through simultaneous cooling with constant stirring (stirring speed set as “high” at 800 ± 20 rpm). The speed of the stirring bar was measured in rpm by an Omega HHT13-Kit tachometer. The culture tube was kept sterile by a foam plug. Aliquots (200 μl) were taken at appropriate intervals, and absorbance was read at 600 nm using a Biotek 405U microtitre plate reader.
For growth recovery experiments, microwave irradiation was terminated after 5 h and the culture tube was transferred to an oil-bath and incubated at 37 °C. Growth was monitored for 26 h after initial inoculation by taking aliquots (200 μl) for OD600 measurement. A total viable cell count test was carried out at the end of each experiment.
As a control, E. coli cultures were incubated in an oil-bath at 37 °C and stirred. The speed of the stirring bar was measured by a tachometer to be 800 ± 20 rpm. Culture growth was monitored for 27.5 h after inoculation. A total viable cell count test as described below was carried out at the end of each experiment.
3.4. Total viable cell counts
To determine the total viable cell count (colony-forming units, CFU, per ml) at the end of each growth curve experiment, bacterial cultures were diluted up to 108-fold by serial dilution. Aliquots (100 μl) of each dilution were streaked on an LB agar plate. Single colonies were counted after 18 h of incubation at 37 °C in a static incubator.3.5. DO measurements
The apparatus used for measuring DO levels was made in-house (Fig. 6). Instead of an external cooling jacket, coolant was circulated through an internal cooling tube inserted into the culture tube that contained uninoculated LB media used in the growth experiments, isolated from the environment by a foam plug. The temperature inside the tube was monitored by a fibre optic temperature probe.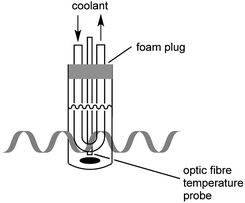 | ||
| Fig. 6 Setup for DO measurement. LB media isolated from the environment with the use of a foam plug were cooled through circulation in an internal cooling tube. | ||
LB media (35 ml) was irradiated with microwave as described above, with stirring. At 1 and 2 h, the foam plug and cooling coil were removed, the vessel was quickly sealed, and DO was measured by inserting a Vernier Micro SD Optical Dissolved Oxygen probe into the vessel. DO levels were recorded after values stabilized (usually after two minutes). DO levels in non-irradiated controls were measured accordingly.
3.6. Bacterial culture and total protein extraction
LB media was inoculated with single colonies of E. coli DE3 and incubated in a shaking incubator at 37 °C and 260 rpm until an OD600 in a range of 1.32–1.36 was reached. A portion of this pre-culture (4.5 ml) was then added to fresh LB media (40.5 ml, to make a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution) in the reaction vessel in CEM microwave reactor and exposed to a maximal 10 W microwave irradiation while the culture temperature was maintained at 37 °C through simultaneous cooling and stirring was set at high. The speed of the stirring bar was measured by a tachometer (Omega HHT13-Kit) to be 800 ± 20 rpm. The control was prepared similarly, except that the reaction vessel was placed in a water bath on a hotplate. The reaction vessels were isolated from the environment by foam plugs. After 5 h, cultures were transferred to 50 ml-Falcon tubes and centrifuged at 4255 RCF for 20 minutes at 4 °C. The supernatants were discarded and the cell pellets were washed three times by resuspension in 1× PBS (30 ml), and centrifugated as above. Supernatant-free cell pellets were flash frozen with liquid nitrogen and was stored at −80 °C. Three biological replicates per condition were prepared for analysis.
10 dilution) in the reaction vessel in CEM microwave reactor and exposed to a maximal 10 W microwave irradiation while the culture temperature was maintained at 37 °C through simultaneous cooling and stirring was set at high. The speed of the stirring bar was measured by a tachometer (Omega HHT13-Kit) to be 800 ± 20 rpm. The control was prepared similarly, except that the reaction vessel was placed in a water bath on a hotplate. The reaction vessels were isolated from the environment by foam plugs. After 5 h, cultures were transferred to 50 ml-Falcon tubes and centrifuged at 4255 RCF for 20 minutes at 4 °C. The supernatants were discarded and the cell pellets were washed three times by resuspension in 1× PBS (30 ml), and centrifugated as above. Supernatant-free cell pellets were flash frozen with liquid nitrogen and was stored at −80 °C. Three biological replicates per condition were prepared for analysis.
Pellets were homogenized via automated frozen disruption and solubilized in protein/kinase/phosphatase inhibitor-supplemented 2DE lysis buffer as described previously.43 Sample aliquots were snap-frozen and stored at −80 °C. Total protein concentrations were determined using a solid-phase dot-blot assay as described previously,44 with BSA as the calibration standard.
3.7. 2DE and in-gel protein detection
All methods for immobilized pH gradient strip (IPG) rehydration, isoelectric focusing (IEF), and subsequent sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) were carried out as described in detail previously.45 Briefly, 100 μg total protein from each sample was reduced and alkylated prior to passive rehydration into a 7 cm pH 3–10 non-linear (ReadyStrip™ IPG Strip (Bio-Rad, Hercules, CA)) for 16 h. IEF in a Protean i12 IEF Cell (Bio-Rad, Hercules, CA) was carried out at 4000 V for 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 VH with several electrode wick changes during linear voltage ramping to facilitate desalting. Focused IPGs were equilibrated with reduction and alkylation prior to further protein resolution in large-format (18 × 17 × 0.1 cm) 12.5% T gels. Two IPGs were adjacently resolved per large gel, yielding ‘mini-tall’ gels (i.e. 7 cm first-dimension and 18 cm long second-dimension). Unstained protein marker (10–200 kDa; New England Biolabs, Ipswich, MA) were resolved in parallel to approximate resolved proteoform molecular weights. Electrophoresis was carried out in a cold room at 4 °C, for 20 min at 300 V then 120 V to completion (∼18 h). Gels were fixed and stained using highly sensitive colloidal Coomassie Brilliant Blue (cCBB) as described previously.46 Three technical replicates per biological replicate were assessed.
500 VH with several electrode wick changes during linear voltage ramping to facilitate desalting. Focused IPGs were equilibrated with reduction and alkylation prior to further protein resolution in large-format (18 × 17 × 0.1 cm) 12.5% T gels. Two IPGs were adjacently resolved per large gel, yielding ‘mini-tall’ gels (i.e. 7 cm first-dimension and 18 cm long second-dimension). Unstained protein marker (10–200 kDa; New England Biolabs, Ipswich, MA) were resolved in parallel to approximate resolved proteoform molecular weights. Electrophoresis was carried out in a cold room at 4 °C, for 20 min at 300 V then 120 V to completion (∼18 h). Gels were fixed and stained using highly sensitive colloidal Coomassie Brilliant Blue (cCBB) as described previously.46 Three technical replicates per biological replicate were assessed.
3.8. Gel imaging and analysis
Destained gels were imaged via transmissive densitometry using the GS-900™ Calibrated Densitometer (Bio-Rad, Hercules) at maximal scanning resolution. 16-Bit greyscale TIFF images were exported for analysis. Quantitative image analysis was performed using Delta2D (Decodon GmbH, Greifswald, Germany) following the manufacturer-recommended workflow (image alignment through to statistical analysis), with automated processes (i.e. warping, consensus spot pattern) manually refined to ensure accuracy.47A Welch’s t-test was carried out for all protein spots across the control and treatment groups using the following parameters: p-value based on all permutations, critical p-value of 0.01, and false discovery rate from permutations to exceed no more than 5 proteins. From the resulting list of significantly altered spots (18), any spot with a relative standard deviation (RSD) of >40% of the normalized volume in either experimental group was excluded. The remaining spots, the majority of which exhibited ≤30% normalised volume RSD across all replicates, were considered genuine after again assessing quality by manual comparison to ensure the highest possible reliability.
For isoelectric point (pI) and molecular weight (kDa) calibration, one mini-tall gel in which 2-D PAGE standards (Bio-Rad, Hercules, CA) were resolved, and one mini-tall gel in which 10 μg E. coli sample spiked with 2-D PAGE standards was resolved (to ensure correct alignment) were prepared as described above. Calibration gel images were imported, warped, and annotated in Delta2D according to manufacturer recommendations for use of the in-built calibration tool.
3.9. Spot picking and in-gel digestion
Protein spots of interest were manually excised from several gels from both experimental groups, pooled, and transferred to microcentrifuge tubes. In-gel tryptic digestion was carried out as described previously47 using 3 ng μl−1 trypsin with digestion performed at 4 °C for 30 min followed by 12 h at ambient temperature. Peptide solutions were recovered into microcentrifuge tubes and dried in a speed vacuum. Samples were shipped at ambient temperature for LC-MS/MS analysis.3.10. Mass spectrometry analysis and protein identification
Mass spectrometry analysis was carried out using a Q-Exactive mass spectrometer. Data-dependant acquisition was preformed on the top 10 peptides per duty cycle with automatic switching between MS and MS/MS. Full-scan MS mode (375–1600 m/z) was operated at a resolution of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 with automatic gain control (AGC) and a target of 1 × 106 ions. Ions selected for MS/MS (fixed first mass 100 m/z) were subjected to the following parameters: resolution 17
000 with automatic gain control (AGC) and a target of 1 × 106 ions. Ions selected for MS/MS (fixed first mass 100 m/z) were subjected to the following parameters: resolution 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, target of 1 × 105 ions, 1.5 m/z isolation window, normalized collision energy 27.0 V and dynamic exclusion 20.0 s. Source ionization parameters were as follows: spray voltage, 1.9 kV; capillary temperature, 280 °C; and s-lens RF level, 50.0. Chromatographic separation of peptides was accomplished using a Zorbax 300SB-C18 column (3.5 μm i.d. × 150 mm, particle size 5 μm, pore size 100 Å, Agilent Technologies, Wilmington, DE). Here, peptides were loaded onto a Zorbax 300SB-C18 trap cartridge at a flow rate of 2 μl per minute for 10 min. After washing with 0.1% formic acid the peptides were eluted using a 5–40% B gradient for 30 min at a flow rate of 250 nl min−1 acid (mobile phase A = 0.1% formic acid; mobile phase B = 0.1% formic acid in acetonitrile) on an Agilent 1260 capillary/nano system.
500, target of 1 × 105 ions, 1.5 m/z isolation window, normalized collision energy 27.0 V and dynamic exclusion 20.0 s. Source ionization parameters were as follows: spray voltage, 1.9 kV; capillary temperature, 280 °C; and s-lens RF level, 50.0. Chromatographic separation of peptides was accomplished using a Zorbax 300SB-C18 column (3.5 μm i.d. × 150 mm, particle size 5 μm, pore size 100 Å, Agilent Technologies, Wilmington, DE). Here, peptides were loaded onto a Zorbax 300SB-C18 trap cartridge at a flow rate of 2 μl per minute for 10 min. After washing with 0.1% formic acid the peptides were eluted using a 5–40% B gradient for 30 min at a flow rate of 250 nl min−1 acid (mobile phase A = 0.1% formic acid; mobile phase B = 0.1% formic acid in acetonitrile) on an Agilent 1260 capillary/nano system.
LC-MS/MS results were searched using Proteome Discoverer (version 2.2, Thermo Scientific against the Uniprot E. coli (strain K12) (4340 entries)) and the Proteome Discoverer contaminants database in which raw files were searched using the Sequest HT algorithm. Peptides produced by digestion with trypsin, with a maximum of two missed cleavages, were matched using precursor and fragment mass tolerances of 10 ppm and 0.04 Da, respectively. Carbamidomethylation of cysteine residues (C) was selected as a static modification. Oxidation (M), deamidation (NQ) and acetyl (protein N-term) were chosen as dynamic modifications. Peptide spectrum matches (PSMs) were verified based on q-values set to 1% false discovery rate (FDR).
4. Conclusions
When E. coli cultures were exposed to up to 10 W microwave irradiation, while temperature was maintained at 37 ± 1 °C with occasional brief fluctuations of ±2 °C, the growth slowed relative to non-irradiated cultures. When microwave irradiation was terminated, however, growth was restored. The identifications of the altered proteins suggest that the microwave exposure likely leads to the alteration of the metabolic profiles of the bacteria, however the long-term effects of such exposure on bacterial physiology remain unclear at this time. In connection with our previous findings, it appears that exposure of biological systems to non-lethal microwave irradiation can lead to alterations of enzymatic activities, membrane properties, and cellular metabolism. Future work will focus on the validation of microwave exposure related biological processes in E. coli and transcriptomic analysis of the proteins involved in these processes.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the Natural Sciences and Engineering Research Council of Canada (HY). JRC notes the support from Brock University. Additional financial support is acknowledged from the Department of Chemistry, College of Liberal Arts and Sciences, University of Illinois at Chicago to SMC. NN was supported by a scholarship from the Molecular Medicine Research Group (Western Sydney University) and was a Visiting Scholar in the Brock University Mentorship Program. MRP was supported by an Abraham Lincoln Fellowship (University of Illinois at Chicago).Notes and references
- N. Kuhnert, Angew. Chem., Int. Ed., 2002, 41, 1863 CrossRef CAS PubMed.
- A. de la Hoz, A. Diaz-Ortiz and A. Moreno, Chem. Soc. Rev., 2005, 34, 164 RSC.
- G. B. Dudley, A. E. Stiegman and M. R. Rosana, Angew. Chem., Int. Ed., 2013, 52, 7918 CrossRef CAS PubMed.
- B. Gutmann, A. M. Schwan, B. Reichart, C. Gspan, F. Hofer and C. O. Kappe, Angew. Chem., Int. Ed., 2011, 50, 7636 CrossRef CAS PubMed.
- C. O. Kappe, Angew. Chem., Int. Ed., 2013, 52, 7924 CrossRef CAS PubMed.
- C. O. Kappe, B. Pieber and D. Dallinger, Angew. Chem., Int. Ed., 2013, 52, 1088 CrossRef CAS PubMed.
- I. Belyaev, Electromagn. Biol. Med., 2005, 24, 375 CrossRef CAS.
- A. Shazman, S. Mizrahi, U. Cogan and E. Shimoni, Food Chem., 2007, 103, 444 CrossRef CAS.
- Non-thermal effects and mechanisms of interaction between electromagnetic fields and living matter, ed. L. Giuliani and M. Soffritti, Ramazzini Institute Eur. J. Oncol. Library 2010, vol. 5 Search PubMed.
- I. Belyaev, 2015, Biophysical mechanisms for non thermal microwave effects, in Electromagnetic Fields in Biology and Medicine, ed. M. S.Markov, CRC Press, New York, pp. 49–67 Search PubMed.
- G. B. Dudley, R. Richert and A. E. Stiegman, Chem. Sci., 2015, 6, 2144 RSC.
- C. P. Kabb, R. N. Carmean and B. S. Sumerlin, Chem. Sci., 2015, 6, 5662 RSC.
- M. A. Herrero, J. M. Kremsner and C. O. Kappe, J. Org. Chem., 2008, 73, 36 CrossRef CAS PubMed.
- S. Hayden, M. Damm and C. O. Kappe, Macromol. Chem. Phys., 2013, 214, 423 CrossRef CAS.
- C. O. Kappe, Chem. Soc. Rev., 2013, 42, 4977 RSC.
- S. A. Mazinani, B. DeLong and H. Yan, Tetrahedron Lett., 2015, 56, 5804 CrossRef CAS.
- S. A. Mazinani and H. Yan, Tetrahedron Lett., 2016, 57, 1589 CrossRef CAS.
- S. A. Mazinani, F. Moradi, J. A. Stuart and H. Yan, ChemistrySelect, 2017, 2, 7983 CrossRef CAS.
- S. A. Mazinani, J. A. Stuart and H. Yan, RSC Adv., 2018, 8, 31465 RSC.
- C. F. Blackman, S. G. Benane, C. M. Weil and J. S. Ali, Ann. N. Y. Acad. Sci., 1975, 247, 352 CrossRef CAS PubMed.
- D. Y. C. Fung and F. E. Cunningham, J. Food Prot., 1980, 43, 641 CrossRef PubMed.
- L. Furia, D. W. Hill and O. P. Gandhi, IEEE Trans. Biomed. Eng., 1986, 33, 993 CAS.
- P. Gos, B. Eicher, J. Kohli and W.-D. Heyer, Bioelectromagnetics, 1997, 18, 142 CrossRef CAS PubMed.
- R. Carta and F. Desogus, AIChE J., 2010, 56, 1270 CAS.
- I. Cohen, R. Cahan, G. Shani, E. Cohen and A. Abramovich, Int. J. Radiat. Biol., 2010, 86, 390 CrossRef CAS PubMed.
- Y. Shamis, R. Croft, A. Taube, R. J. Crawford and E. P. Ivanova, Appl. Microbiol. Biotechnol., 2012, 96, 319 CrossRef CAS PubMed.
- S. J. Webb and D. D. Dodds, Nature, 1968, 218, 374 CrossRef CAS PubMed.
- S. J. Webb and M. E. Stoneham, Phys. Lett. A, 1977, 60, 267 CrossRef.
- W. Grundler and F. Keilmann, Phys. Rev. Lett., 1983, 51, 1214 CrossRef.
- M. S. Dreyfuss and J. R. Chipley, Appl. Environ. Microbiol., 1980, 39, 13 CAS.
- I. Nakouti, G. Hobbs, Y. Teethaisong and D. Phipps, Biotechnol. Prog., 2017, 33, 37 CrossRef CAS PubMed.
- Y. Shamis, A. Taube, N. Mitik-Dineva, R. Croft, R. J. Crawford and E. P. Ivanova, Appl. Environ. Microbiol., 2011, 77, 3017 CrossRef CAS PubMed.
- H. Torgomyan and A. Trchounian, Crit. Rev. Microbiol., 2013, 39, 102 CrossRef CAS PubMed.
- H. Torgomyan, K. Hovnanyan and A. Trchounian, Cell Biochem. Biophys., 2013, 65, 445 CrossRef CAS PubMed.
- V. Berzin, M. Kiriukhin and M. Tyurin, Arch. Microbiol., 2013, 195, 181 CrossRef CAS PubMed.
- H. Torgomyan and A. Trchounian, Cell Biochem. Biophys., 2015, 71, 419 CrossRef CAS PubMed.
- S. Horikoshi, S. Matsuzaki, S. Sakamoto and N. Serpone, Radiat. Phys. Chem., 2014, 97, 48 CrossRef CAS.
- G. Reguera, Trends Microbiol., 2011, 19, 105 CrossRef CAS PubMed.
- O. Kučera and M. Cifra, J. Biol. Phys., 2016, 42, 1 CrossRef PubMed.
- M. Raina and M. Ibba, Front. Genet., 2014, 5, 171 Search PubMed.
- J. Zhong, C. Xiao, W. Gu, G. Du, X. Sun, Q.-Y. He and G. Zhang, PLoS Genet., 2015, 11, e1005302 CrossRef PubMed.
- M. A. Sørensen, A. O. Fehler and S. L. Svenningsen, RNA Biol., 2018, 15, 586 CrossRef PubMed.
- R. Butt and J. R. Coorssen, J. Proteome Res., 2006, 5, 437 CrossRef CAS PubMed.
- N. Noaman and J. R. Coorssen, Anal. Biochem., 2018, 556, 53 CrossRef CAS PubMed.
- R. H. Butt, T. A. Pfeifer, A. Delaney, T. A. Grigliatti, W. G. Tetzlaff and J. R. Coorssen, Mol. Cell. Proteomics, 2007, 6, 1574 CrossRef CAS PubMed.
- N. Noaman, P. S. Abbineni, M. Withers and J. R. Coorssen, Electrophoresis, 2017, 38, 3086 CrossRef CAS PubMed.
- E. P. Wright, M. A. Partridge, M. P. Padula, V. J. Gauci, C. S. Malladi and J. R. Coorssen, Proteomics, 2014, 14, 872 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Copy of original 2D-gel images and mass spectrometry data. See DOI: 10.1039/c9ra00617f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |