Open Access Article
Open Access ArticleWeighted spectral reconstruction method for discrimination of bacterial species with low signal-to-noise ratio Raman measurements
Shanshan Zhu a,
Xiaoyu Cuiad,
Wenbin Xub,
Shuo Chen*ad and
Wei Qianc
a,
Xiaoyu Cuiad,
Wenbin Xub,
Shuo Chen*ad and
Wei Qianc
aSino-Dutch Biomedical and Information Engineering School, Northeastern University, Shenyang, China 110169. E-mail: chenshuo@bmie.neu.edu.cn; Tel: +86 24 83680230
bScience and Technology on Optical Radiation Laboratory, Beijing, China 110854
cCollege of Engineering, University of Texas at El Paso, El Paso, USA 79968
dKey Laboratory of Data Analytics and Optimization for Smart Industry (Northeastern University), Ministry of Education, China
First published on 25th March 2019
Abstract
Raman spectroscopy is a label-free and non-destructive spectroscopic technique that has been explored for bacterial identification. However, noise often interferes with the interesting Raman peaks because the Raman signal is inherently weak, especially for bacterial samples. Although this problem can be solved by increasing the exposure time or the power of the excitation laser, a longer acquisition time is required or the risk of sample damage is increased. In contrast, short exposure time and low laser power often lead to inadequate acquisition of Raman scattering, in which the Raman spectra with low signal-to-noise ratio (SNR) is difficult to be further analyzed. In order to quickly and accurately characterize biological samples by using low SNR Raman measurements, a weighted spectral reconstruction based method was developed and tested on Raman spectra with low SNR from 20 bacterial samples of two species. Principal component analysis followed by support vector machine was applied on the reference Raman spectra and the spectra recovered from the low SNR Raman measurements by the proposed method, the traditional spectral reconstruction method, and four other commonly used de-noising methods for the discrimination of bacterial species. The results showed that a classification accuracy of 90% was achieved based on our method, which was comparable to that of the reference Raman spectra and showed significant advantages over other spectral recovery methods. Therefore, the weighted spectral reconstruction method can preserve the most biochemical information for the bacterial species' identification while removing the noise from the low SNR Raman spectra, in which the advantages of lesser sample damage and shorter acquisition time would promote wider biomedical applications of Raman spectroscopy.
Introduction
The quick identification and discrimination of bacterial species have been a hot point of concern especially for clinicians, which is critical for the treatment of infections.1,2 These concerns are highly relevant in the area of medical applications and aim at fast and precise diagnosis and treatment. Different approaches for bacterial identification include morphological and biochemical tests, such as Gram staining, enzyme activity tests, antibiotic susceptibility profiles as well as 16S ribosomal deoxyribonucleic acid or 16S ribosomal ribonucleic acid analysis.3,4 The conventional examinations can be applied only on pure, isolated bacteria derived from culture plates.5 However, bacterial culture and purification are often time-consuming and complicated, and usually require several days for the clinicians to get the final laboratory report after the sample collection from patients. Although genotypic methods are faster and more accurate than the traditional biochemical tests, DNA extraction from pure cultures and consumption of expensive reagents are still needed.4 Due to the low efficiency of the current bacterial identification methods, empirical therapy usually has to be performed while awaiting laboratory results, which leads to a low treatment effect on the infection and loss of the optimal time for appropriate treatment, even resulting in deterioration of the infection or mortality. Tumbarello et al. indicated that 20.1% patients with bloodstream infections caused by Escherichia coli do not receive proper antimicrobial therapy and have a higher proportion of extended-spectrum β-lactamase bloodstream infections (74.0% versus 15.8%) and higher 21 day mortality rates (40.7% versus 5.6%) compared with patients who receive the appropriate therapy from the beginning.6 Werarak et al. demonstrated that although carbapenem is the most frequently used treatment for hospital-acquired pneumonia and ventilator-associated pneumonia, the patients who are treated by carbapenem in the beginning of infection have more severe complications and high mortality rate.7 Hence, a rapid and accurate identification of the bacterial pathogens is critical for the precise treatment of patients.In recent years, Raman spectroscopy has been explored in the detection and identification of bacteria as a fast, non-destructive, and label-free spectroscopic technique based on measuring the inelastic scattering of photons by vibrating molecules or crystal lattices.8,9 Based on the specific vibrational modes of the molecules, a large amount of qualitative and quantitative biochemical information enables the identification and differentiation of bacterial biochemical components.10,11 Unfortunately, noise often interferes with the interesting Raman peaks because the Raman signal is inherently weak, especially for bacterial samples.12,13 Although this problem can be solved by increasing the exposure time or the power of the excitation laser, a longer acquisition time is required or the risk of sample damage is increased. If a short exposure time and low laser power is applied during the Raman measurement, inadequate acquisition of Raman scattering would lead to low signal-to-noise ratio (SNR) measurements, which makes the further spectral data processing and analysis difficult. Therefore, a method to quickly and accurately discriminate the bacterial species by using low SNR Raman measurements with low laser power and short exposure time can potentially solve the above problems.
In this study, a weighted spectral reconstruction based method was newly developed and tested on Raman spectra with low SNR equal to 0.98 from 20 bacterial samples of two species, i.e., Pseudomonas aeruginosa and Staphylococcus aureus. For identifying the different bacterial species, principal component analysis (PCA) followed by support vector machine (SVM) was applied on the reference Raman spectra (high SNR), low SNR Raman spectra, and low SNR Raman spectra processed by the proposed method, traditional spectral reconstruction method, and four other commonly used de-noising methods, i.e., Savitzky–Golay (SG) algorithm, wavelet transform, finite impulse response (FIR) filtration, and factor analysis. Compared with other methods, the proposed method demonstrated significant improvement in the spectral recovery and higher accuracy in the discrimination of bacterial species, in which the classification accuracy of the Raman spectra recovered by the proposed method was even comparable to that of the reference Raman spectra with high SNR. Therefore, our method demonstrated significant potential in the rapid and accurate bacterial species' identification based on low SNR Raman spectra, wherein the sample damage was lesser and a shorter acquisition time was required.
Experimental
Sample preparation
The bacterial samples of the two species, i.e., Staphylococcus aureus (ATCC 29213) and Pseudomonas aeruginosa (ATCC 9027), were cultured overnight at a fixed temperature of 35 °C in Tryptic Soy Agar Plates. Thereafter, few bacterial colonies were selected and placed in the phosphate buffered saline to reach a final concentration of 1 × 108 CFU mL−1, followed by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for five minutes to concentrate and purify the bacterial samples. The bacterial samples were rinsed and immersed twice in distilled water to wash away the culture medium, and finally resuspended in 100 μL distilled water. To prepare the samples for Raman measurements, 2 μL of the suspension was repeatedly dropped and air dried at the same location on an aluminum foil, in which the bacterial samples were concentrated and a relatively higher Raman signal could be achieved.14
000 rpm for five minutes to concentrate and purify the bacterial samples. The bacterial samples were rinsed and immersed twice in distilled water to wash away the culture medium, and finally resuspended in 100 μL distilled water. To prepare the samples for Raman measurements, 2 μL of the suspension was repeatedly dropped and air dried at the same location on an aluminum foil, in which the bacterial samples were concentrated and a relatively higher Raman signal could be achieved.14
Raman spectra acquisition
Twenty pairs of Raman spectra, i.e., 10 pairs of Raman measurements from Pseudomonas aeruginosa and 10 pairs of Raman measurements from Staphylococcus aureus, were obtained in the wavenumber range from 600 cm−1 to 1600 cm−1 by a confocal Raman microscope (inVia, Renishaw, UK). A 633 nm diode laser was used for excitation and the spectral resolution was 2 cm−1. In each pair of Raman measurements, both the low SNR and high SNR Raman spectra were collected from the exact same position. The exposure time for low SNR Raman measurement was 1 s and accumulated once, whereas that for the corresponding high SNR Raman measurement was 10 s and accumulated 30 times, so that the actual acquisition time for the high SNR Raman measurements was 300 times of that for the low SNR Raman measurements. In this study, the high SNR Raman spectra were treated as the reference to evaluate the proposed method. The noise level was quantified by a published method,15 i.e., by dividing the maximum intensity of the normalized reference Raman spectrum by the root mean square error (RMSE) between the normalized low SNR Raman spectrum and the normalized reference Raman spectrum, in which the normalization for each Raman spectrum was performed by dividing the Raman intensity at each wavenumber by the summation of the Raman intensities at all wavenumbers. Thus, the SNR of the low SNR Raman spectra used in this study was 0.98.Spectral recovery methods
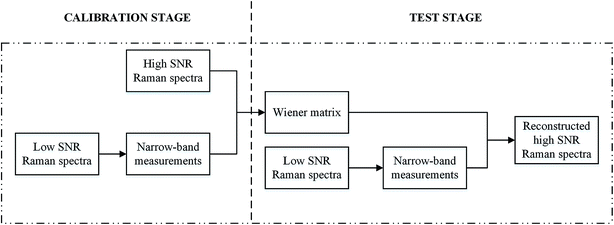
Twenty Raman spectra with low SNR were de-noised by the traditional spectral reconstruction method, weighted spectral reconstruction method, and four other commonly used de-nosing methods, i.e., SG algorithm, wavelet transform, FIR filtration, and factor analysis. The corresponding high SNR Raman spectra were treated as the reference Raman spectra to evaluate the performance of each method.Traditional spectral reconstruction was performed to retrieve the Raman spectra from the low SNR Raman measurements,15–17 in which a calibration data set was required, as shown in Fig. 1. In the calibration data set, both the reference Raman spectra and the narrow-band measurements derived from the low SNR Raman spectra were included, whereas the test data set contained only the low SNR Raman spectra. The narrow-band measurements were numerically calculated by the inner production of low SNR Raman spectra and the transmittance of the non-negative PC based filters,18 in which the first six non-negative principal component (PC) based filters were used. In the calibration stage, the Wiener matrix W was calculated based on the calibration dataset to extract the relation between the narrow-band measurements Ccal and the reference Raman spectra Rref, as shown in eqn (1).
| W = E(RrefCcalT)[E(CcalCcalT)]−1 | (1) |
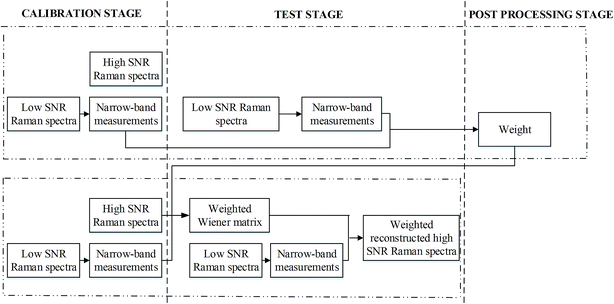
The weighted spectral reconstruction was a newly developed method and different from the traditional spectral reconstruction method, in which the ensemble average was replaced by the weighted average when constructing the weighted Wiener matrix Ŵ, as shown in eqn (2).
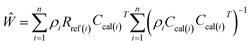 | (2) |
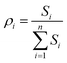 | (3) |
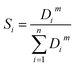 | (4) |
Besides the traditional spectral reconstruction and weighted spectral reconstruction methods, SG algorithm, wavelet transform, FIR filtration, and factor analysis were applied on the same set of low SNR Raman spectra for comparison. For the SG algorithm, each part of the original spectrum with a selected window size was fitted to a polynomial function for smoothing purpose.20 In contrast, the wavelet transform, FIR filtration, and factor analysis commonly remove noise by filtering techniques. For the wavelet transform,21,22 the spectral data were decomposed into the wavelet domain by various wavelet basis and reconstructed after noise removal by certain thresholds. The FIR filtration is a linear filtration technique, in which a window-based FIR filter is designed based on the frame size and cut-off frequency and was subsequently used for noise removal in this study.23 For factor analysis,24 the original spectral information is projected into the linear combination of a certain number of subspectra, and those subspectra related to the noise can be subsequently removed. The parameters of each de-noising method and the range of the parameters are shown in Table 1.
| Method | Parameter | Parameter range |
|---|---|---|
| Traditional spectral reconstruction | Number of non-negative PC scores based filters | 6 |
| Weighted spectral reconstruction | Number of non-negative PC scores based filters | 6 |
| Power | −0.1 to −10 | |
| SG algorithm | Window size | 3 to 729 |
| Polynomial degree | 1 to 9 | |
| Wavelet transform | Wavelet basis | Common wavelet filters built in Matlab |
| Decomposition level | 1 to 10 | |
| Threshold | Soft threshold or hard threshold: threshold value were selected according to the Birge–Massart strategy | |
| FIR filtration | Frame size | 2 to 243 |
| Cut-off frequency | 1 × 10−10 to 1 | |
| Factor analysis | Number of subspectra | 1 to 20 |
After the low SNR Raman spectra were de-noised, the broad and slowly varying fluorescence background was estimated by using the fifth order polynomial fitting and subtracted from the original spectra.25 Normalization was subsequently performed on each Raman spectrum by dividing the Raman intensity at each wavenumber by the summation of the Raman intensities at all the wavenumbers. In order to evaluate the accuracy of the recovered Raman spectra, the above fluorescence background removal algorithm and normalization were applied on the corresponding reference Raman spectra as well and the mean relative RMSE26 was used as the metric.
PCA-SVM based classification methods
After obtaining the recovered Raman spectra from the low SNR Raman spectra by each of the above methods, principal component analysis (PCA) followed by support vector machine (SVM) was applied on the reference Raman spectra, low SNR Raman spectra, and the recovered Raman spectra for bacterial species' discrimination. As a multivariate statistical method, PCA can reduce the large original spectral data into small number of independent variables named as PC scores, which can effectively carry the most important information of the corresponding spectra. Thus, PCA can be used to group the spectra by choosing different combinations of the PC scores to build a new coordinate system.27 In order to achieve a fair comparison of the different spectral recovery methods in this study, the first four PC scores were chosen for bacterial species' discrimination because the highest classification accuracy was achieved based on the reference Raman spectra with high SNR when using the first four PC scores. Support vector machine is a state-of-the-art supervised machine learning method especially suited to classify and identify different bacterial species.28,29 SVM belongs to the group of maximum margin classifiers and can efficiently find the optimal solution for the given parameters. In this paper, a classification model was built by inputting the first four PC scores for SVM to identify the bacterial species. The leave-one-out method was used for cross validation and the performance of the different spectral recovery methods was compared in terms of accuracy, sensitivity, and specificity in identifying the different bacterial species. In addition, the integrated area under the receiver operating characteristic (ROC) curve was used to quantify the performance of the classification model on different groups of Raman spectra.Results and discussion
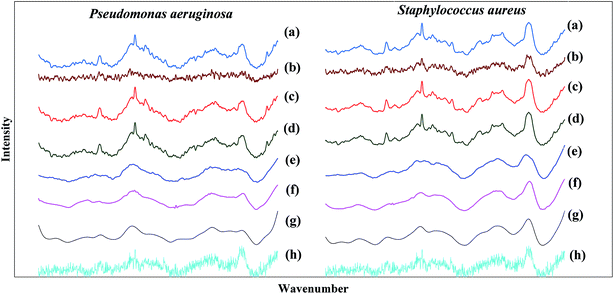
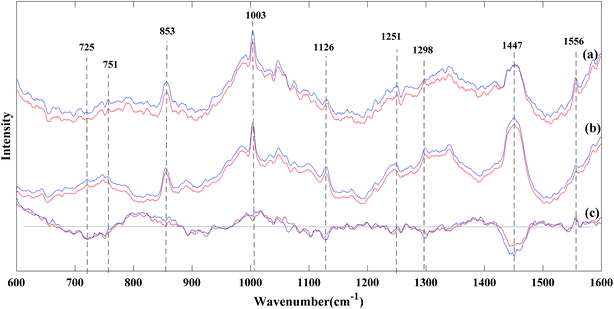
Fig. 3 shows the comparison among the average spectra after the fluorescence background removal and normalization based on the different spectral recovery methods between Pseudomonas aeruginosa and Staphylococcus aureus samples. It was found that both the traditional spectral reconstruction method (see Fig. 3(c)) and weighted spectral reconstruction method (see Fig. 3(d)) showed excellent agreement in the Raman peaks and the spectral shape compared to the reference Raman spectra (see Fig. 3(a)). The excellent performance of the spectral reconstruction based methods can be attributed to two factors. One is that the Raman measurements are integrated along the wavenumber dimension in the procedure of synthesizing the narrow-band measurements from the low SNR Raman spectra, thus, the SNR can be significantly improved by eliminating the shot noise in Raman measurements.30,31 The other factor is that the spectral reconstruction based methods contain prior information about the samples, in which the high SNR Raman spectra are used to associate with the narrow-band measurements in the calibration stage. Furthermore, the weighted spectral reconstruction method shows the best performance in spectral recovery among all the methods, and improves 4.3% in the mean relative RMSE compared to the traditional spectral reconstruction method, as shown in Table 2. The reason is that the weighted spectral reconstruction method takes the advantage of collecting more effective information from the samples with higher similarity when constructing the weighted Wiener matrix. However, the time efficiency of the weighted spectral reconstruction is lower because different weighted Wiener matrices have to be created for each test sample, whereas the traditional spectral reconstruction method only requires a single Wiener matrix when the calibration dataset is fixed. Although the SG algorithm is a commonly used smoothing method for de-noising Raman spectra, the spectral shape was distorted and some of the peak locations were shifted, as shown in Fig. 3(e), because those weak features in the Raman spectra are comparable to the noise level, and thus, can be easily smoothed out during noise removal.20 the wavelet transform (see Fig. 3(f)) and FIR filtration (see Fig. 3(g)) methods showed good performance in noise removal, at the cost of losing some important spectral shape information, e.g., the central wavelengths and bandwidths of the peaks.21 Unfortunately, the factor analysis did not work well when the SNR was extremely low, which could not even smoothen out the noise and lost plenty of useful information during bacterial discrimination, as shown in Fig. 3(h). The reason might be that the factor analysis loses the ability to decompose the noise and signal when the SNR is extremely low because the noise contributes similarly or even more to the Raman spectra compared with the signal.24| Low SNR Raman spectra | Traditional spectral reconstruction | Weighted spectral reconstruction | SG algorithm | Wavelet transform | FIR filtration | Factor analysis | |
|---|---|---|---|---|---|---|---|
| Mean relative RMSE | 1.98 × 10−1 | 8.21 × 10−2 | 7.86 × 10−2 | 1.47 × 10−1 | 1.45 × 10−1 | 1.54 × 10−1 | 1.48 × 10−1 |
Table 3 shows the comparison of the classification accuracy, sensitivity, and specificity of Pseudomonas aeruginosa and Staphylococcus aureus from the Raman spectra after fluorescence background removal and normalization of the reference Raman spectra, low SNR Raman spectra, and Raman spectra recovered by using the traditional spectral reconstruction method, weighted spectral reconstruction method, SG algorithm, wavelet transform, FIR filtration, and factor analysis. For identifying Pseudomonas aeruginosa and Staphylococcus aureus, the Raman spectra recovered by both traditional and weighted spectral reconstruction methods can achieve a classification accuracy of 90%, which was exactly the same as that of the reference Raman spectra and showed significant advantages over the other commonly used de-noising methods as well as the results of the low SNR Raman spectra. Furthermore, the weighted spectral reconstruction method also successfully demonstrated exactly the same sensitivity and specificity compared to the reference Raman spectra, whereas the traditional spectral reconstruction failed. Although the specificity of the traditional spectral reconstruction is the highest, it sacrifices the sensitivity, as shown in Table 3, and we believe some improper prior information is used during the spectral recovery process of the traditional spectral reconstruction. Thus, the higher spectral recovery accuracy of the weighted spectral reconstruction method is indeed critical for better performance in the following spectral data analysis. In practical applications, the choice of the traditional spectral reconstruction and weighted spectral reconstruction should be mainly dependent on its specific applications, in which the compromise between time efficiency and spectral recovery accuracy should be considered. Interestingly, the classification accuracy did not fully comply with the mean relative RMSE, i.e., the agreement between the recovered Raman spectra and the reference Raman spectra. The reason might be that most of the information is preserved whereas some critical information for bacterial identification is lost during the noise removal, especially for the SG algorithm, FIR filtration, and factor analysis methods. The classification results of these three methods are even lower than that of the low SNR Raman spectra, indicating that more information is lost compared to the information gained during the spectral recovery by these three methods. This can be attributed to the fact that the importance of the information cannot be distinguished by these commonly used de-noising methods. For the SG algorithm, the weak features in the Raman spectra comparable to the noise level can be easily smoothed out during noise removal, resulting in some shifted Raman peaks and the distorted spectral shape.20 By the FIR filtration method, some important spectral shape information is lost simultaneously, while the noise is well removed. Although the Raman spectra de-noised by the SG algorithm and FIR filtration methods retain some important information about the peak locations and the spectral shape, the information regarding the discrimination of the two bacterial samples might be removed during noise removal, resulting in relatively low classification accuracy of only 70%. The classification accuracy of the factor analysis method was the lowest among all the methods, whereas the mean relative RMSE was not the worst. The reason is that factor analysis loses the ability to decompose the noise and signal when the SNR is extremely low, thus, plenty of important Raman peaks for bacterial discrimination as well as the noise are smoothed out simultaneously.24
| Reference Raman spectra | Low SNR Raman spectra | Traditional spectral reconstruction | Weighted spectral reconstruction | SG algorithm | Wavelet transform | FIR filtration | Factor analysis | |
|---|---|---|---|---|---|---|---|---|
| Classification accuracy | 90% | 75% | 90% | 90% | 70% | 85% | 70% | 35% |
| Sensitivity | 90% | 80% | 80% | 90% | 70% | 90% | 50% | 40% |
| Specificity | 90% | 70% | 100% | 90% | 70% | 80% | 90% | 30% |
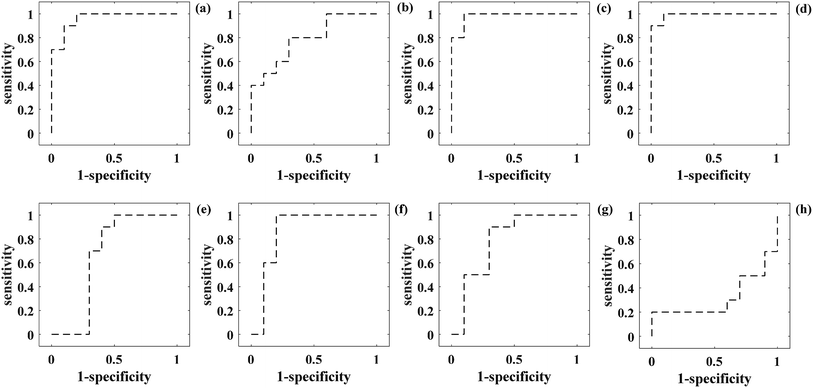
To further evaluate the performance of the PCA-SVM-based classification model for bacterial species, the ROC curves were generated at different threshold levels for different groups of Raman spectra, respectively. The integrated area under the ROC curve (AUC) is a quantitative indicator used to represent the classifier performance, in which the larger AUC value usually means that the classifier has higher prediction accuracy.32 According to the results in Fig. 4, the integrated areas under the ROC curves (AUC) are 0.96, 0.79, 0.98, 0.99, 0.66, 0.86, 0.78, and 0.32 for the reference Raman spectra, low SNR Raman spectra, and Raman spectra recovered from the low SNR Raman measurements using the traditional spectral reconstruction, weighted spectral reconstruction, SG algorithm, wavelet transform, FIR filtration, and factor analysis, respectively. Thus, the Raman spectra recovered from the low SNR Raman measurements using the weighted spectral reconstruction method demonstrates the strongest ability of bacterial species' identification with high sensitivity and specificity, and even outperforms the reference Raman spectra with high SNR. The reason might be that the spectral reconstruction procedure can remove some of the useless information within the reference Raman spectra, which may provide negative impacts on the bacterial species' identification.
Fig. 5 shows the average Raman spectra based on the reference Raman spectra and Raman spectra recovered by the spectral reconstruction method of Pseudomonas aeruginosa and Staphylococcus aureus, respectively. By the visual inspection of the reference Raman spectra (red curve) and the Raman spectra after spectral reconstruction (blue curve) in Fig. 5(a) and (b), it can be noted that the major Raman features were at 853 cm−1 (tyrosine ring breathing vibration of protein), 1003 cm−1 (phenylalanine ring vibration of protein), 1126 cm−1 (C–N, C–C stretching of protein and C–C lipid stretch), 1447 cm−1 (CH2, CH3 lipid, and protein), and 1556 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C vibration of protein).33,34 Moreover, the Raman peaks at 725 cm−1 and 751 cm−1 can be identified and assigned to the adenine and thymine ring breathing vibrations of DNA. From Fig. 5(c), it can be seen that the intensity of these bands were consistently lower in Pseudomonas aeruginosa compared to Staphylococcus aureus, indicating a significant reduction in both the DNA and RNA concentrations in Pseudomonas aeruginosa. In addition, the Raman peaks at 1298 cm−1 and 1447 cm−1, which are assigned to the CH2 and CH3 bending modes were found primarily in proteins and lipids. The intensity of these two peaks notably decreased in Pseudomonas aeruginosa compared to those in Staphylococcus aureus, as shown in Fig. 5(c), which may indicate the decrease of the membranous lipids in Pseudomonas aeruginosa.14 The proteins have a prominent peak at 1556 cm−1, which was assigned to the C
C vibration of protein).33,34 Moreover, the Raman peaks at 725 cm−1 and 751 cm−1 can be identified and assigned to the adenine and thymine ring breathing vibrations of DNA. From Fig. 5(c), it can be seen that the intensity of these bands were consistently lower in Pseudomonas aeruginosa compared to Staphylococcus aureus, indicating a significant reduction in both the DNA and RNA concentrations in Pseudomonas aeruginosa. In addition, the Raman peaks at 1298 cm−1 and 1447 cm−1, which are assigned to the CH2 and CH3 bending modes were found primarily in proteins and lipids. The intensity of these two peaks notably decreased in Pseudomonas aeruginosa compared to those in Staphylococcus aureus, as shown in Fig. 5(c), which may indicate the decrease of the membranous lipids in Pseudomonas aeruginosa.14 The proteins have a prominent peak at 1556 cm−1, which was assigned to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C vibration.35 As in Fig. 5(c), the intensity at this wavenumber increased in Pseudomonas aeruginosa compared to that in Staphylococcus aureus, which indicated the difference in the protein level between these two bacterial samples.35 Although the recovered Raman spectra retained the information about most of the Raman peaks, some Raman information were still lost or overlapped, especially for Raman peaks at around 751 cm−1 (thymine ring breathing vibration of DNA) and 1251 cm−1 (amide III of protein and adenine ring breathing vibration of DNA), as shown in Fig. 5(c). It was found that the difference in the spectra of the two bacterial species at these two peaks after spectral reconstruction were much closer to zero than that of the reference spectra, which demonstrated that some Raman information of DNA and proteins was lost or overlapped by the surrounding peaks. Even though the information in these bands and modes of bacterial components were lost or overlapped, it has very minor impact on the final classification results between Pseudomonas aeruginosa and Staphylococcus aureus (see Table 3). In other words, these bands and modes may not be the key biomarkers responsible for the discrimination in these two bacterial species.
C vibration.35 As in Fig. 5(c), the intensity at this wavenumber increased in Pseudomonas aeruginosa compared to that in Staphylococcus aureus, which indicated the difference in the protein level between these two bacterial samples.35 Although the recovered Raman spectra retained the information about most of the Raman peaks, some Raman information were still lost or overlapped, especially for Raman peaks at around 751 cm−1 (thymine ring breathing vibration of DNA) and 1251 cm−1 (amide III of protein and adenine ring breathing vibration of DNA), as shown in Fig. 5(c). It was found that the difference in the spectra of the two bacterial species at these two peaks after spectral reconstruction were much closer to zero than that of the reference spectra, which demonstrated that some Raman information of DNA and proteins was lost or overlapped by the surrounding peaks. Even though the information in these bands and modes of bacterial components were lost or overlapped, it has very minor impact on the final classification results between Pseudomonas aeruginosa and Staphylococcus aureus (see Table 3). In other words, these bands and modes may not be the key biomarkers responsible for the discrimination in these two bacterial species.
Although only twenty pairs of Raman spectra of Pseudomonas aeruginosa and Staphylococcus aureus were tested in this study, many studies based on a large amount of bacterial samples have verified the feasibility of using Raman spectroscopy as a powerful tool for bacterial species' identification with acceptable Raman signals.34,36,37 In this study, we mainly focused on the information preservation by the proposed weighted spectral reconstruction method during the noise removal from the low SNR Raman spectra. The recovered Raman spectra by the proposed weighted spectral reconstruction method show closest relative RMSE, accuracy, sensitivity, specificity, and AUC value compared to those of the reference Raman spectra, which demonstrates the proposed method's excellent preservation of the most useful spectral information such as Raman peaks and the spectral shape for bacterial species' identification. Thus, we believe that the proposed method can still work well or even better for bacterial species' identification when the amount of bacterial samples increases. In the future study, a large size of sample data set and even other bacterial species will be investigated to validate and confirm these conclusions.
Conclusions
In this study, a weighted spectral reconstruction based method was developed and tested on low SNR Raman spectra from bacterial samples for the discrimination of two bacterial species. PCA combined with SVM classifier was used to identify different bacterial species. Based on the results, it was found that the proposed method could recover the Raman spectra in excellent agreement with the reference Raman spectra and reach a comparable classification accuracy of 90% for bacterial species' identification, which shows significant advantages over other spectral recovery methods. Therefore, the weighted spectral reconstruction based method can excellently recover the Raman signal from the low SNR Raman spectra and preserve the most important information, in which the lesser sample damage and shorter acquisition time would promote the wider application of Raman spectroscopy in biomedical applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to acknowledge the financial support from the National Natural Science Foundation of China (61605025, 61501101), Science and Technology Foundation of National Defense Key Laboratory (61424080209), Program for Innovation Talents in Universities of Liaoning Province (LR2016031), Programs Supported by Ningbo Natural Science Foundation (2018A610365), the Fundamental Research Funds for the Central Universities (N171902001, N171904006), and the 111 Project (B16009).References
- A. M. Caliendo, D. N. Gilbert, C. C. Ginocchio, K. E. Hanson, L. May and T. C. Quinn, et al., Clin. Infect. Dis., 2013, 57, S139–S170 CrossRef PubMed.
- C. A. Cornett, S. A. Vincent, J. Crow and A. Hewlett, Journal of the American Academy of Orthopaedic Surgeons, 2016, 24, 11–18 CrossRef PubMed.
- T. Karre, E. A. Vetter, J. N. Mandrekar and R. Patel, J. Clin. Microbiol., 2010, 48, 1504–1505 CrossRef PubMed.
- A. T. Hassanain, L. Baha and M. Z. Zaini, J. Clin. Microbiol., 2014, 52, 3244–3249 CrossRef PubMed.
- F. Bittar, H. Richet, J. C. Dubus, M. Rwynaud-Gaubert, N. Stremler, J. Sarles, D. Raoult and J. M. Rolain, PLoS One, 2008, 7, S41 Search PubMed.
- M. Tumbarello, T. Spanu, R. Di Bidino, M. Marchetti, M. Ruggeri, E. M. Trecarichi, G. De Pascale, E. M. Proli, R. Cauda, A. Cicchetti and G. Fadda, Antimicrob. Agents Chemother., 2010, 54, 4085–4091 CrossRef CAS PubMed.
- P. Werarak, P. Kiratisin and V. Thamlikitkul, J. Med. Assoc. Thailand, 2010, 93, s126–s138 Search PubMed.
- I. F. Cheng, H. C. Chang, T. Y. Chen, C. Hu and F. L. Yang, Sci. Rep., 2013, 3, 2365 CrossRef PubMed.
- K. Rebrošová, M. Šiler, O. Samek, F. Růžička, S. Bernatová, V. Holá, J. Ježek, P. Zemánek, J. Sokolová and P. Petráš, Sci. Rep., 2017, 7, 14846 CrossRef.
- S. Y. Feng, W. B. Wang, T. I. Tai, G. N. Chen, R. Chen and H. S. Zeng, Biomed. Opt. Express, 2015, 6, 3494–3502 CrossRef.
- K. Lin, W. Zheng, C. M. Lim and Z. W. Huang, Biomed. Opt. Express, 2016, 7, 3705–3715 CrossRef PubMed.
- Z. M. Zhang, S. Chen, Y. Z. Liang, Z. X. Liu, Q. M. Zhang, L. X. Ding, F. Yei and H. Zhou, J. Raman Spectrosc., 2010, 41, 659–669 CrossRef CAS.
- P. Wang and W. R. Yao, Sci. Technol. Food Ind., 2012, 33, 427–430 CAS.
- Y. H. Ong, M. Lim and Q. Liu, Opt. Express, 2012, 20, 22158–22171 CrossRef CAS.
- S. Chen, X. Lin, C. Yuen, S. Padmanabhan, R. W. Beuerman and Q. Liu, Opt. Express, 2014, 22, 12102–12114 CrossRef CAS.
- H. Haneishi, T. Hasegawa, A. Hosoi, Y. Yokoyama, N. Tsumura and Y. Miyake, Appl. Opt., 2000, 39, 6621–6632 CrossRef CAS.
- S. Chen and Q. Liu, J. Biomed. Opt., 2012, 17, 030501 CrossRef.
- R. Piché, J. Opt. Soc. Am. A, 2002, 19, 1946–1950 CrossRef.
- S. Arlot and A. Celisse, Stat. Surv., 2009, 4, 40–79 CrossRef.
- K. Chen, H. Zhang, H. Wei and Y. Li, Appl. Opt., 2014, 53, 5559–5569 CrossRef.
- A. E. Villanueva-Luna, J. Castro-Ramos, S. Y. Montiel, A. Flores-Gil, J. A. D. Atencio and E. E. O. Guillén, Optical Memory and Neural Networks, 2010, 19, 310–317 CrossRef.
- S. Sidhik, Optik, 2015, 126, 5952–5955 CrossRef.
- Č. Martin, M. Pavel and V. Karel, J. Raman Spectrosc., 2007, 38, 1174–1179 CrossRef.
- K. M. Omberg, J. C. Osborn, S. L. Zhang, J. P. Freyer, J. R. Mourant and J. R. Schoonover, Appl. Spectrosc., 2002, 56, 813–819 CrossRef CAS.
- B. D. Beier and A. J. Berger, Analyst, 2009, 134, 1198–1202 RSC.
- S. Chen, G. Wang, X. Y. Cui and Q. Liu, Opt. Express, 2017, 25, 1005–1018 CrossRef CAS.
- S. Wold, K. Esbensen and P. Geladi, Chemom. Intell. Lab. Syst., 1987, 2, 37–52 CrossRef CAS.
- S. Meisel, S. Stöckel, M. Elschner, F. Melzer, P. Rösch and J. Popp, Appl. Environ. Microbiol., 2012, 78, 5575–5583 CrossRef CAS.
- S. Meisel, S. Stöckel, M. Elschner, P. Rösch and J. Popp, Analyst, 2011, 136, 4997–5005 RSC.
- M. J. B. Moester, F. Ariese and J. F. D. Boer, J. Eur. Opt. Soc., 2015, 10, 15022 Search PubMed.
- D. Sompel, E. Garai, C. Zavaleta and S. S. Gambhir, J. Raman Spectrosc., 2013, 44, 841–856 CrossRef.
- S. Y. Feng, R. Chen, J. Lin, J. Pan, G. Chen, Y. Li, M. Cheng, Z. Huang, J. Chen and H. Zeng, Biosens. Bioelectron., 2010, 25, 2414–2419 CrossRef CAS.
- I. Notingher, S. Verrier, H. Romanska, A. E. Bishop, J. Polak and L. L. Hench, Spectroscopy, 2014, 16, 43–51 CrossRef.
- G. Rusciano, P. Capriglione, G. Pesce, P. Abete, V. Carnovale and A. Sasso, Laser Phys. Lett., 2013, 10, 075603 CrossRef.
- A. Rygula, K. Majzner, K. M. Marzec, A. Kaczor, M. Pilarczyk and M. Baranska, J. Raman Spectrosc., 2013, 44, 1061–1076 CrossRef CAS.
- E. E. Rossi, A. L. Pinheiro, O. C. Baltatu, M. T. Pacheco and L. J. Silveria, J. Photochem. Photobiol., B, 2012, 107, 73–78 CrossRef CAS.
- B. Lorenz, C. Wichmann, S. Stöckel, P. Rösch and J. Popp, Trends Microbiol., 2017, 25, 413–424 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2019 |