Open Access Article
Open Access ArticleAntibody-free colorimetric detection of chlorothalonil in cucumber via the inhibition of an enzyme-triggered reaction†
Qingju Liu ab,
Ping Hanab,
Hui Wangab,
Wenwen Gongab and
Xiaoyuan Feng*ab
ab,
Ping Hanab,
Hui Wangab,
Wenwen Gongab and
Xiaoyuan Feng*ab
aBeijing Research Center for Agriculture Standards and Testing, Beijing 100097, China. E-mail: fengxiaoyuan2014@126.com; Fax: +86 10 51503792; Tel: +86 10 51503792
bRisk Assessment Lab for Agro-products (Beijing), Ministry of Agriculture, Beijing 100097, China
First published on 29th March 2019
Abstract
Currently known rapid determination of fungicides usually relies on antibody-based immunoassay. This paper reports a simple antibody-free colorimetric assay for chlorothalonil via the inhibition of an enzyme-triggered reaction. The enzymatic activity of glyceraldehyde-3-phosphate dehydrogenase was significantly inhibited by chlorothalonil, and the color change of NBT-PMS system induced from NADH formation was suppressed, which could be used indirectly to assay chlorothalonil. The limit of detection (LOD) was 0.05 μM with a linear range from 0.5 to 10 μM, and the detection of 1 μM chlorothalonil in solution was achieved with a naked-eye readout. In addition, the colorimetric measurement results of the cucumber samples showed a good recovery rate, although the sensitivity was less effective than the instrumental method. Nevertheless, the results demonstrates that the chlorometric method provides potential opportunities for reliable, cost-effective quantitative detection for chlorothalonil residues in vegetables.
Introduction
As a broad-spectrum fungicide widely used on fruits and vegetables,1–3 chlorothalonil (2,4,5,6-tetrachloroisophthalonitrile, CTL) residue in raw fruits and vegetables and their processed products has drawn wide public attention. Due to its highly toxic to aquatic species and marine organisms, significant cumulative toxicity3,4 and more toxic and persistent of its main metabolites (4-OH-CTL) in the environment, CTL and its metabolites are considered as a probable human carcinogen by the United States Environmental Protection Agency (USEPA).12The most popular approaches for accurate and precise determination of CTL are based on highly sensitive detectors equipped with chromatographic units such as gas chromatography (GC) coupled with mass spectrometry (MS)13–15 or electron capture detectors,16,17 and HPLC with DAD detection18 or coupled with MS.6 However, susceptibility to factors such as pH8 and matrix components19 makes CTL a nasty pesticide of poor recovery for sample preparation prior to chromatographic analysis. In addition, the need of specialized persons and high cost limit their application for on-site screening. Antibody-based immunoassay systems such as enzyme-linked immunosorbent assay20–23 (ELISA), immunosensor,7 immunochip24 and immune-strip25–28 offer potential opportunities for rapid detection of CTL on-site and substantial investigations have been reported and there are already some test kits availability on the market. Despite its high specificity and sensitivity, the preparation process of antibodies is high cost and time-consuming. Moreover, susceptibility to degradation and denaturation of antibodies remains a great challenge during its application. It should be noted that the use of nanoparticles29 and self-propelled micromotors30 with inherent enzymatic (artificial enzymes) activity display attractive performance in sensing of phenylenediamines isomers. In spite of its facile synthesis and stable performance, the lack of specific recognition limits their target scope. Unfortunately, there are frequent reports about detection of CTL residue in many types of vegetables and fruits.31–33 Thus, consumers need a simple, and inexpensive sensor system to monitor the risk of CTL in fruits and vegetables.
It is well known that enzyme-based signal amplification is commonly used in biochemical assays because of their high catalytic efficacy. Long known as a key enzyme in glycolysis, glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.22.12; GAPDH) has recently been assigned numerous other cellular functions.34 Due to its unique characteristics such as good stability, rapid response, high catalytic efficiency, low cost and biocompatibility, GAPDH is extensively employed in clinical diagnosis and drug design.35–37 GAPDH catalyze the oxidative phosphorylation of glyceraldehyde-3-phosphate with concomitant reduction of NAD+ into NADH.38
Herein, we report an antibody-free method for visual detection of CTL utilizing a commercially available enzyme GAPDH coupled with phenazine methosulphate (PMS)-β-nicotinamide adenine dinucleotide (NADH)-nitroblue tetrazolium (NBT) chromogenic system. In addition, we have successfully applied this method for detection of CTL in cucumber with good recovery, demonstrating that quantification of CTL concentration by measuring the color change based on the inhibition of enzyme-triggered reaction has a further potential application for detection of CTL on-site.
Experimental
Reagents
All chemical used were commercially available. CTL was purchased from Aladdin. Glyceraldehyde 3-phosphate (GAP) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were obtained from Sigma (U.S.). NAD+, nitrotetrazolium blue chloride (NBT), sodium pyrophosphate, sodium dihydrogen phosphate, and dibasic sodium phosphate were got from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China). Phenazine methosulfate (PMS) was bought from Sangon Biotech (Shanghai, China). Pentachloronitrobenzene (PCB), pentachloroaniline (PCA), chlorothaldimethyl (CDM), dichlobenil (DCB), phthalide (PTL), quintozene (QTZ) were got from Shanghai Aladdin Bio-chem Technology Co., LTD. Hexachlorobenzene (HCB) and pentachlorophenol (PCP) were purchased from J&K Scientific Ltd.Measurements and apparatus
UV-vis absorption spectra were acquired on a TU-1901 spectrophotometer (China). The chromatographic analysis of CTL was performed by a 1260 HPLC system (Agilent) equipped with a VWD detector. The photographs were taken by a IXUS-190 camera (Canon, Japan).Colorimetric detection of CTL through enzymatic-triggered reaction
GAPDH (5 U) was preincubated at 37 °C in 50 mM phosphate buffer (pH 5.0) containing noted concentrations of CTL under stirring for 45 min. Following the preincubation, the reaction catalyzed by GAPDH with NAD+ as cofactor coupled with NBT-PMS was assay at room temperature in NaPPi buffer (pH 8.5) containing 0.5 mM GAP, 0.35 mM NAD+, 120 μM NBT, 75 μM PMS in a total volume of 200 μL. At that time, the color of the solution was recorded by photograph and determined by following the change in absorbance at 560 nm via UV-vis spectrophotometry.Pretreatment of cucumber samples
Cucumbers purchased from a market (Haidian, Beijing), were used for recovery examinations. After addition of 10 mL phosphoric acid solution (phosphoric acid/water = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v), the sample (100 g) were homogenized by a blender immediately. Then, each sample (100 g) was spiked with some known concentrations of CTL standards and allowed to stand at room temperature for 2 h.
1 v/v), the sample (100 g) were homogenized by a blender immediately. Then, each sample (100 g) was spiked with some known concentrations of CTL standards and allowed to stand at room temperature for 2 h.
For colorimetric determination, methanol (25 mL) was added to the samples (5 g) in a 50 mL tube and the tube was vigorously shaken for 30 min on an orbital shaker to extract CTL. After centrifuged at 3000 rpm for 5 min, the supernatant (1 mL) was downloaded into SPE cartridge. After washing the cartridge with 1 mL of methanol–water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, v/v), the eluate was collected for colorimetric sensor. If needed, the extract solution was further diluted with methanol–water (9
9, v/v), the eluate was collected for colorimetric sensor. If needed, the extract solution was further diluted with methanol–water (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v).
1, v/v).
Results and discussion
Sensing mechanism of the colorimetric method
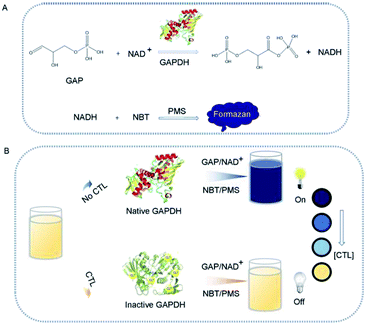
Fig. 1 illustrates the sensing mechanism for colorimetric sensor of CTL based on enzymatic reaction catalyzed by GAPDH coupled with PMS-NBT chromogenic system. This method relies on the fact that CTL is an inhibitor of specific nicotinamide adenine dinucleotide (NAD) thiol-dependent glycolytic enzyme-GAPDH,39,40 leading to a decrease in production of NADH, which fails to induce a color change from yellow to blue of PMS-NBT chromogenic system. According to previous reports, the native GAPDH can catalyze the oxidation of GAP38 into gluconolactone-6-phosphate (1,3-DPG), accompanied with reduction of NAD+ into NADH.38,41 The simultaneously generated NADH participates in a cycling reaction that exhausts NBT yielding a purple coloured formazan (λmax: 560 nm) with PMS as an electron carrier.42–45 The absorbance intensity is proportional to the concentration of NADH. Whereas, previous investigations39 have confirmed that inhibition of GAPDH by CTL was via forming a co-valently bond with amino acid residue Cys 149 in the active pocket (Fig. S1†) that required for NAD binding. The inhibition of activity of GAPDH by CTL leads to quantitative color change depending on the amount of CTL, which can be easily distinguished visually or quantified by the spectrometric measurement. With the absence of CTL, the solution of enzyme-triggered reaction of GAP-NBT-PMS system rapidly proceeds and turns blue, and the system is “on”. However, when CTL is pre-mixed with GAPDH, the enzyme-triggered reaction is suppressed and the solution remain yellow, and the system is “off”. Thus, we devise a colorimetric method based on the catalytic capability of GAPDH coupled with PMS-NBT chromogenic system for detection of CTL. This colorimetric sensor is free from utilization of antibody or modification process, and the results can be obtained by either the naked eye or the spectrometer.Optimization of experimental conditions
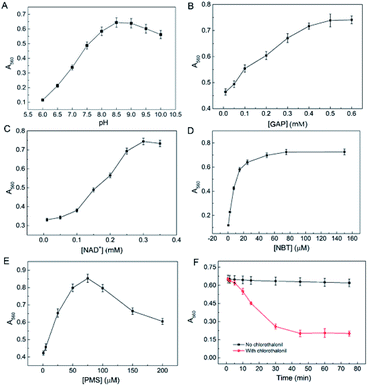
To achieve a highly sensitive response, it's necessary to optimize the experimental conditions. pH is a key factor which can greatly affect the enzyme activity and the chromogenic reaction, thus pH of the reaction system was optimized firstly. As shown in Fig. 2A and S3A,† A560 of the generated formazan products sharply increased with the increasing value of pH and reached to a peak value at about pH 8.5. However, when pH was above 8.5, the absorbance at 560 nm dropped gradually with the increase of pH values, indicating the activity of enzyme was decreased. This was probably because the active-site residues within the catalytic domain center needed appropriate pH value to maintain its protonation state for product formation, and higher pH values led to deprotonation of active-site residues, resulting in the loss of enzyme activity. Therefore, pH 8.5 was selected as the optimal pH value.GAP is catalyzed to form 1,3-diphosphoglycerate by GAPDH, accompanied by formation of NADH with NAD+ as an electron-transferring coenzyme. Therefore, concentrations of NAD+ and GAP would influence the catalytic activity of GAPDH. The results showed that the maximum response was obtained when the concentration of GAP was 0.5 mM and the response slightly decreased with the increase of concentration when concentration of GAP was higher than 0.5 mM (Fig. 2B and S3B†). Similarly, the most remarkable color change was observed when the concentration of NAD+ was 0.35 mM, and the color change was moderately decreased when the concentration was above 0.35 mM (Fig. 2C and S3C†). The above results were in accordance with the theory of enzyme kinetics that there is an optimal concentration of the substrates and the reaction is inhibited when the substrate concentration is too high. Thus, 0.5 mM GAP and 0.35 mM NAD+ were selected as the optimal conditions for further investigations.
As reactants in the second step, the concentrations of NBT and PMS were closely connected with the generation of blue products, thus, their concentrations were further optimized. As depicted in Fig. 2D, E, S3D and S3E,† the absorbance at 560 reached to a plateau when the NBT concentration was 100 μM. In the concentration range of 0–75 μM, the color change was enhanced with the increase of concentration of PMS, and the change was declined when PMS was above than 75 μM. As a result, concentrations of NBT and PMS were selected 100 and 75 μM, respectively.
Finally, the incubation time of CTL with GAPDH was investigated in the same way as the above. Compared with control, A560 increased with the incubation time, and it reached to a minimum value at 45 min, but it remained unchanged with a further increase of the time (Fig. 2F). In consideration of the requirement of rapid determination, we selected 45 min for further investigations.
Sensitivity of the colorimetric method
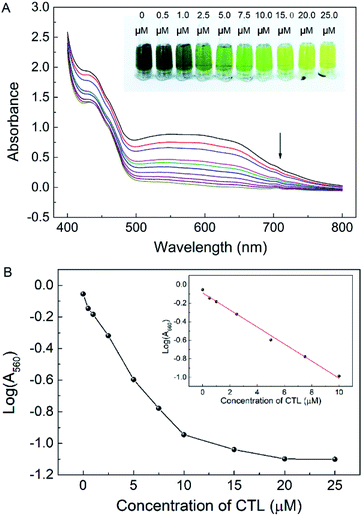
To examine the sensitivity of the enzymatic inhibition strategy-based colorimetric assay for the determining CTL, a series of different concentration of CTL standard solution were added to the GAPDH/GAP-NBT/PMS chromogenic system under the optimized condition. As shown in Fig. 3A, the absorbance band at 560 nm (A560) decreased progressively with the concentration of CTL ranging from 0 to 25 μM. Meanwhile, an obvious color change from blue to yellow was also be found (the inset portion of Fig. 3A) and a sensitivity of about 1 μM CTL was achieved with naked eyes. In addition, the logarithm of A560 (log(A560)) decreased correspondingly over the CTL concentration. With concentration ranging from 0.5 to 10 μM, the log(A560) displayed a linear correlation with R2 = 0.993 (Fig. 3B). The limit to detection (LOD) was estimated to be 0.05 μM (3σ). The high sensitivity and relative broad linear range is attributed to the high catalytic property of GAPDH.As summarized in Table 1, we draw a comparison between the proposed colorimetric detection and other previously reported instrumental chromatographic analysis or antibody-based immunoassay. Compared to instrumental chromatographic analysis, limit of detection for CTL in this work was much higher than that of GC-MS or LC-MS. Also, the sensitivity of the proposed colorimetric assay was not remarkably more effective than ELISA. Since the MRLs of CTL in vegetables was set from 5 mg kg−1 in China (5 mg kg−1 in USA and 1 mg kg−1 in Europe), the sensitivity of the proposed analysis was effective enough to detect CTL residues in vegetables. Moreover, the approach avoided the use of expensive and complex instruments apparatus required in instrumental analysis and utilization of expensive antibody of the immunoassay, which undoubtedly improves their application in real samples.
| Methods | Linear range (μM) | Limit of detection (μM) | Use of large instrument | Use of antibody | Ref. |
|---|---|---|---|---|---|
| GC-MS | 0.19–37.6 | 0.19 | Yes | No | 5 |
| HPLC-MS | 0.001–1.12 | 0.004 | Yes | No | 6 |
| Immunosensor | 0.03–0.16 | 0.09 | No | Yes | 7 |
| ELISA | 0.0007–0.005 | 0.0002 | No | Yes | 8 |
| IC-ELISA | 0.0003–0.004 | 0.001 | No | Yes | 9 |
| IC-ELISA | 0.004–0.022 | 0.001 | No | Yes | 10 |
| Colloidal gold-based strip test | 0.005–0.01 | 0.35 | No | Yes | 11 |
| Colorimetric assay | 0.5–10 | 0.05 | No | No | This study |
Selectivity of the colorimetric method
To determine the selectivity of the colorimetric assay for CTL, CTL (5 μM) and several analogues of closely related molecular structure at the level of 50 μM were carried out to undergo the chromogenic reaction. As shown in Fig. 4, it was observed that only in the presence of CTL, the solutions turned yellow and showed a decrease in A560. However, the other pesticides with the level 10 times higher than CTL stay blue, similarly to the blank sample, and color and A560 of the solution had no notable change, revealing that the inhibition of the reaction was not occurred. The other pesticides yielded only negligible response comparing to that of CTL. CTL inhibits the activity of GAPDH by covalent interaction with cysteine-149 located at the active center responsible for the binding of GAP. Due to difference in the molecular size and chemical nature of the reactive toxiphores, pesticides or chemical compounds with similar chemical structure do not inhibit the enzyme. It should be noted that certain trichloromethyl sulfonyl fungicides suppress activity of GAPDH. However, CTL differed from those fungicides in that it did not react with non-thiol groups of either GAPDH and had a different reaction rate with the GAPDH. This indicated that other pesticides didn't inhibit the catalytic activity of GAPDH and didn't interrupt the chromogenic system of GAPDH coupled with NBT/PMS. The colorimetric method has excellent specificity to CTL and the colorimetric system is suitable for selective detection of CTL.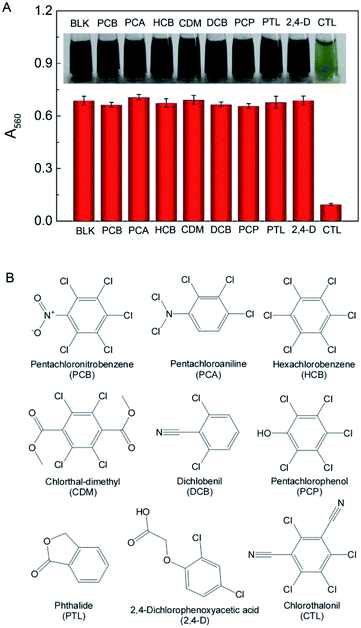 | ||
| Fig. 4 (A) A560 of mixture spiked with CTL and other compounds (5 μm). Inset portion showed the corresponding picture. (B) Chemical structures of the selected compounds. | ||
Application of cucumber samples
As a proof of its applicability, cucumber was chosen as a representative specie to perform the spiked test. Different concentrations of CTL standard solutions were added into the cucumber sample (final concentration 50.0, 100.0, 150.0 μg kg−1, respectively). As listed in Table 2, the recovery values were 82.0%, 90.2% and 84.9%, respectively, which was consistent with that obtained by HPLC (Table S1†). Therefore, this facile, antibody-free assay can used for sample with simple matrixes directly with satisfactory results. However, better sample clean-up procedures are necessary to eliminate interference for sample with more complicated matrixes such as vegetables and fruits. The reason is that substances such as pigment or sulphur-containing compounds in the real samples can interfere the chromogenic reaction, which has distinct impact on the quantification of analyte. We will make efforts to apply the colorimetric procedures to determine CTL in other agro-products in future work.| Sample | Added (μg kg−1) | Visual assay | ||
|---|---|---|---|---|
| Found (μg kg−1) | Recovery (%) | RSD (%) | ||
| Cucumber 1 | 50.0 | 41.0 ± 1.6 | 82.0 | 3.9 |
| Cucumber 2 | 100.0 | 90.2 ± 4.3 | 90.2 | 4.8 |
| Cucumber 3 | 150.0 | 127.4 ± 6.9 | 84.9 | 5.4 |
Conclusions
In summary, we developed a facile, sensitive colorimetric biosensor for CTL detection with straightforward readout and implementation. This strategy was relied on the inhibition of GAPDH activity by CTL, resulting in color change of NBT/PMS system, which could be visualized by naked eyes or quantification via the UV-vis spectrophotometer. The linear range of the method was from 0.5 to 10 μM, and 1 μM CTL could be distinguished with naked eyes. Compared with the existing techniques for CTL detection, the established method was large instrument-free and antibody-free. In addition, we successfully applied the method to the determination of CTL in cucumber. Therefore, it has great potential application as a selective and sensitive platform for detection of CTL in other agro-products.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work were sponsored by National Key Research and Development Program of China (2016YFD0400902), the Construction of Science and Technology Innovation Team of Beijing Academy of Agriculture and Forestry (JNKST201620), the Special Projects of Construction of Science and Technology Innovation Ability of Beijing Academy of Agriculture and Forestry Sciences (KLCX20190405) and Beijing Postdoctoral Research Foundation (2018-ZZ-059).Notes and references
- P. Lv, J. Zhang, T. Z. Shi, L. L. Dai, X. Q. Li, X. W. Wu, X. D. Li, J. Tang, Y. Wang, Q. X. Li and R. M. Hua, Appl. Catal., B, 2017, 217, 137–143 CrossRef CAS.
- L. M. Du Gas, P. S. Ross, J. Walker, V. L. Marlatt and C. J. Kennedy, Environ. Toxicol. Chem., 2017, 36, 1354–1364 CrossRef CAS PubMed.
- Q. He, X. H. Xu, F. Zhang, Y. K. Tai, Y. F. Luo, J. He, Q. Hong, J. D. Jiang and X. Yan, J. Sci. Food Agric., 2017, 97, 2582–2587 CrossRef CAS PubMed.
- M. Y. Zhang, Y. Teng, Z. H. Xu, J. Wang, P. Christie and Y. M. Luo, J. Soils Sediments, 2016, 16, 1754–1763 CrossRef CAS.
- F. Hou, L. W. Zhao and F. M. Liu, Food Anal. Method, 2016, 9, 656–663 CrossRef.
- A. Yamamoto, L. Miyamato, M. Kitagawa, H. Moriwaki, H. Miyakoda, H. Kawasaki and R. Arakawa, Anal. Sci., 2009, 25, 693–697 CrossRef CAS PubMed.
- Y. Hirakawa, T. Yamasaki, E. Watanabe, F. Okazaki, Y. Murakami-Yamaguchi, M. Oda, S. Iwasa, H. Narita and S. Miyake, J. Agric. Food Chem., 2015, 63, 6325–6330 CrossRef CAS PubMed.
- E. Watanabe, S. Miyake, S. Ito, K. Baba, H. Eun, M. Ishizaka and S. Endo, J. Chromatogr. A, 2006, 1129, 273–282 CrossRef CAS PubMed.
- L. Q. Liu, H. J. Yan, X. Zhang, H. Kuang and C. L. Xu, Food Agric. Immunol., 2015, 26, 410–419 CrossRef CAS.
- F. Okazaki, Y. Hirakawa, Y. Yamaguchi-Murakami, A. Harada, E. Watanabe, S. Iwasa, H. Narita and S. Miyake, Food Hyg. Saf. Sci., 2014, 55, 65–72 CrossRef CAS.
- X. D. Yang, G. P. Zhang, F. Y. Wang, Y. B. Wang, X. F. Hu, Q. M. Li, G. C. Jia, Z. Liu, Y. Wang, R. G. Deng and X. Y. Zeng, Food Agric. Immunol., 2015, 26, 729–737 CrossRef CAS.
- T. Bedassa, A. Gure and N. Megersa, Food Anal. Method, 2015, 8, 2020–2027 CrossRef.
- A. Peruga, M. Barreda, J. Beltran and F. Hernandez, Food Addit. Contam., Part A: Chem., Anal., Control, Exposure Risk Assess., 2013, 30, 298–307 CrossRef CAS PubMed.
- N. B. Valles, M. Retamal, M. A. Martinez-Uroz, M. Mezcua, A. R. Fernandez-Alba and A. de Kok, Analyst, 2012, 137, 2513–2520 RSC.
- M. M. Rahman, J. H. Park, A. M. Abd El-Aty, J. H. Choi, H. R. Bae, A. Yang, K. H. Park and J. H. Shim, Biomed. Chromatogr., 2013, 27, 416–421 CrossRef CAS PubMed.
- M. H. S. Kurz, F. F. Goncalves, M. B. Adaime, I. E. D. da Costa, E. G. Primel and R. Zanella, J. Braz. Chem. Soc., 2008, 19, 1129–1135 CrossRef CAS.
- X. Zhou, X. H. Zang, D. Y. Wang, P. L. Cui and Z. Wang, Chin. J. Anal. Chem., 2009, 37, 41–45 CAS.
- M. Catala-Icardo, C. Gomez-Benito, E. F. Simo-Alfonso and J. M. Herrero-Martinez, Anal. Bioanal. Chem., 2017, 409, 243–250 CrossRef CAS PubMed.
- M. Anastassiades, K. Mastovska and S. J. Lehotay, J. Chromatogr. A, 2003, 1015, 163–184 CrossRef CAS PubMed.
- J. M. Yeung and W. H. Newsome, Bull. Environ. Contam. Toxicol., 1995, 54, 444–450 CrossRef CAS PubMed.
- C. Jahn and W. Schwack, J. Agric. Food Chem., 2001, 49, 1233–1238 CrossRef CAS PubMed.
- E. Watanabe, S. Miyake, S. Ito, K. Baba, H. Eun, M. Ishizaka and S. Endo, J. Chromatogr. A, 2006, 1129, 273–282 CrossRef CAS PubMed.
- F. Okazaki, Y. Hirakawa, Y. Yamaguchi-Murakami, A. Harada, E. Watanabe, S. Iwasa, H. Narita and S. Miyake, Shokuhin Eiseigaku Zasshi, 2014, 55, 65–72 CrossRef CAS PubMed.
- M. Lan, Y. Guo, Y. Zhao, Y. Liu, W. Gui and G. Zhu, Anal. Chim. Acta, 2016, 938, 146–155 CrossRef CAS PubMed.
- L. J. Yao, L. Q. Liu, S. S. Song, H. Kuang and C. L. Xu, Food Agric. Immunol., 2017, 28, 639–651 CrossRef CAS.
- L. Q. Liu, S. Suryoprabowo, Q. K. Zheng, S. S. Song and H. Kuang, Food Agric. Immunol., 2017, 28, 427–438 CrossRef CAS.
- Z. J. Xie, D. Z. Kong, L. Q. Liu, S. S. Song and H. Kuang, Food Agric. Immunol., 2017, 28, 439–451 CrossRef CAS.
- Z. X. Wang, Z. J. Xie, G. Cui, L. Q. Liu, S. S. Song, H. Kuang and C. L. Xu, Food Agric. Immunol., 2017, 28, 476–488 CrossRef.
- B. F. Shi, Y. B. Su, L. L. Zhang, M. J. Huang, X. F. Li and S. L. Zhao, Nanoscale, 2016, 8, 10814–10822 RSC.
- R. Maria-Hormigos, B. Jurado-Sanchez and A. Escarpa, Anal. Chem., 2018, 90, 9830–9837 CrossRef CAS PubMed.
- A. M. Mozzachio, J. A. Rusiecki, J. A. Hoppin, R. Mahajan, R. Patel, L. Beane-Freeman and M. C. Alavanja, Environ. Res., 2008, 108, 400–403 CrossRef CAS PubMed.
- T. V. Yudina, N. E. Fedorova, M. V. Larkina, O. E. Egorchenkova and C. K. Rogacheva, Gig. Sanit., 2016, 95, 1108–1112 CAS.
- P. Du, X. Wu, H. He, Y. Zhang, J. Xu, F. Dong, Y. Zheng and X. Liu, Sci. Rep., 2017, 7, 8694 CrossRef PubMed.
- A. B. Seabra, M. Ouellet, M. Antonic, M. N. Chretien and A. M. English, Nitric Oxide, 2013, 35, 116–122 CrossRef CAS PubMed.
- D. W. Darnall and L. V. Murray, Biochem. Biophys. Res. Commun., 1972, 46, 1222–1227 CrossRef CAS PubMed.
- J. W. Yun, S. K. Kim and H. Kim, Cancer Invest., 2011, 29, 93–101 CrossRef CAS PubMed.
- P. Huitorel and D. Pantaloni, Eur. J. Biochem., 1985, 150, 265–269 CrossRef CAS PubMed.
- K. J. Kennedy, J. C. Bressi and M. H. Gelb, Bioorg. Med. Chem. Lett., 2001, 11, 95–98 CrossRef CAS PubMed.
- J. W. Long and M. R. Siegel, Chem.-Biol. Interact., 1975, 10, 383–394 CrossRef CAS PubMed.
- R. W. Tillman, M. R. Siegel and J. W. Long, Pestic. Biochem. Physiol., 1973, 3, 160–167 CrossRef CAS.
- E. V. Schmalhausen, V. I. Muronetz and N. K. Nagradova, FEBS Lett., 1995, 375, 18–20 CrossRef CAS PubMed.
- H. Y. Zhang, Z. Zhang, X. H. Ji and Z. K. He, Chin. J. Anal. Chem., 2014, 42, 1276–1280 CAS.
- B. Bekdeser, M. Ozyurek, K. Guclu and R. Apak, Anal. Chem., 2011, 83, 5652–5660 CrossRef CAS PubMed.
- M. N. Patel, H. N. Joshi and C. R. Patel, J. Organomet. Chem., 2012, 701, 8–16 CrossRef CAS.
- K. Paradowska, B. Polak, A. Chomicki and G. Ginalska, J. Enzyme Inhib. Med. Chem., 2016, 31, 1712–1717 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra00291j |
| This journal is © The Royal Society of Chemistry 2019 |