Open Access Article
Open Access ArticleRe-Du-Ning inhalation solution exerts suppressive effect on the secretion of inflammatory mediators via inhibiting IKKα/β/IκBα/NF-κB, MAPKs/AP-1, and TBK1/IRF3 signaling pathways in lipopolysaccharide stimulated RAW 264.7 macrophages
Yi Zhanga,
Brian Chi-Yan Chengbc,
Ran Xied,
Bing Xua,
Xiao Yan Gaoa and
Gan Luo *a
*a
aSchool of Chinese Materia Medica, Beijing University of Chinese Medicine, Beijing 100102, China. E-mail: luna049@126.com
bCollege of Professional and Continuing Education, Hong Kong Polytechnic University, Hong Kong 999077, China
cQuality Healthcare Medical Services, Hong Kong 999077, China
dInstitute of Chinese Materia Medica, China Academy of Chinese Medical Science, Beijing 100700, China
First published on 18th March 2019
Abstract
Background: Re-Du-Ning inhalation solution (RIS) is a novel preparation derived from the Re-Du-Ning injection, which has been clinically used to treat respiratory diseases such as pneumonia for more than twenty years in China. However, scant reports have been issued on its anti-inflammatory mechanisms. Aim: we investigated the suppressive effect of RIS on inflammatory mediators and explored the underlying mechanism of action. Methods: RIS freeze dried powder was characterized by HPLC analysis. Lipopolysaccharide (LPS)-stimulated RAW 264.7 macrophage was selected as the cell model. The cell viability was determined by using the MTT assay. Moreover, the production of nitric oxide (NO) was measured by the Griess reaction. The protein secretions from inflammatory mediators were determined by the enzyme-linked immunosorbent assay (ELISA). The protein levels and enzyme activities were examined by Western blotting. The nuclear translocation of nuclear factor-kappa B (NF-κB), AP-1, and IRF3 was further explored by immunofluorescence assay. Results: the viability of the RAW 264.7 cells was not significantly changed after 24 h incubation with RIS concentration up to 400 μg mL−1. The RIS remarkably reduced the production of NO and prostaglandin E2 (PGE2), and downregulated the expression of iNOS and COX-2. The concentrations of cytokines (IL-1β, IL-6, and TNF-α) and chemokines (MCP-1, CCL-5, and MIP-1α) in the culture medium were significantly decreased by the RIS treatment. Furthermore, the phosphorylation of IκB-α, IKKα/β, TBK1, ERK, p38, JNK, NF-κB, AP-1, and IRF3 was downregulated by the RIS treatment. The nuclear translocation of NF-κB, AP-1, and IRF3 was also inhibited after the RIS treatment. Conclusion: the suppressive effect of RIS is associated with the regulated NF-κB, AP-1, and IRF3 and their upstream proteins. This study provides a pharmacological basis for the application of RIS in the treatment of inflammatory disorders.
Introduction
Inflammatory responses play an important role in immune homeostasis and are essential for host survival.1 However, hyperactive and chronic inflammation represents a central driving force in many disorders such as asthma,2 pneumonia,3 rheumatoid arthritis,4 and chronic obstructive pulmonary disease.5 These diseases impose a huge burden on the society in terms of incurred disability and premature mortality.6–8 Multiple innate immune cells contribute to the pathophysiological processes of inflammatory diseases, with macrophages playing a significant role.9 Lipopolysaccharide (LPS) is a pathogenic endotoxin present in the outer membrane of Gram-negative bacteria, which can activate the toll-like receptor (TLR)-4 and the downstream signaling pathways including IKKα/β/IκBα/NF-κB, MAPKs/AP-1, and TBK1/IRF3.10 This in turn activates an inflammatory cascade with the production of inflammation-related molecules such as nitric oxide (NO), interleukin-6 (IL-6), and tumour necrosis factor (TNF-α), providing a positive feedback to the TLR4 signaling pathways, which results in persistent inflammation and favours the induction of autoimmunity.11–13 Therefore, targeting the TLR4 signaling is one of the focuses in developing effective anti-inflammatory agents for acute and chronic inflammatory diseases.14Traditional Chinese Medicine (TCM) treatment has been recently applied to offer a different approach to prevent or cure inflammatory diseases.15 The Re-Du-Ning inhalation solution (RIS) is a novel preparation derived from the Re-Du-Ning injection (RDNI). The RDNI has been clinically used as an aerosol to treat respiratory diseases such as pneumonia for more than twenty years in the People's Republic of China.16 This preparation is composed of the flowers of Lonicera japonica, the fruits of Gardenia jasminoides, and the dried aerial parts of Artemisia apiacea in the ratio of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15
15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12.17 It has been reported that these herbs are of low toxicity and have been used in China for hundreds of years to treat early febrile diseases, exogenous fever, and boils.18–21 Previous studies have showed that the main compounds in the flowers of Lonicera japonica (e.g. chlorogenic acid),22,23 the fruits of Gardenia jasminoides (e.g. geniposide)10,24 and Artemisia apiacea (e.g. artemisinin)25 have strong anti-inflammatory activities via inhibition of the TLR4 signaling pathway. However, as a new aerosol preparation, there are no reports in the literature about the pharmacological properties of RIS. In order to provide pharmacological justification for the use of RIS as an anti-inflammatory agent, we employed a classic inflammatory cell model called the LPS-stimulated RAW 264.7 macrophages, to investigate the suppressive effect of RIS on inflammatory mediators and to explore the underling mechanisms.
12.17 It has been reported that these herbs are of low toxicity and have been used in China for hundreds of years to treat early febrile diseases, exogenous fever, and boils.18–21 Previous studies have showed that the main compounds in the flowers of Lonicera japonica (e.g. chlorogenic acid),22,23 the fruits of Gardenia jasminoides (e.g. geniposide)10,24 and Artemisia apiacea (e.g. artemisinin)25 have strong anti-inflammatory activities via inhibition of the TLR4 signaling pathway. However, as a new aerosol preparation, there are no reports in the literature about the pharmacological properties of RIS. In order to provide pharmacological justification for the use of RIS as an anti-inflammatory agent, we employed a classic inflammatory cell model called the LPS-stimulated RAW 264.7 macrophages, to investigate the suppressive effect of RIS on inflammatory mediators and to explore the underling mechanisms.
Materials
Modified Griess reagent, 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT), lipopolysaccharide (LPS, Escherichia coli O55:B5), and bovine serum albumin were bought from Sigma Chemical Co. (St. Louis MO, USA). Penicillin-streptomycin solution was obtained from Caisson labs (Smithfield, UT, USA). Dulbecco's Modified Eagle Medium (DMEM) was acquired from Corning Cellgro (Manassas, VA, USA). Fetal bovine serum (FBS) was purchased from Biological Industries Co. (Beth-Haemek, Israel). Tumor necrosis factor α (TNF-α), interleukin-6 (IL-6), interleukin 1β (IL-1β), macrophage inflammatory protein (MIP)-1α, monocyte chemoattractant protein (MCP)-1, and chemokine (C–C motif) ligand 5 (CCL-5) ELISA kits were purchased from Thermo Fisher Scientific (San Diego, CA). Prostaglandin E2 (PGE2) ELISA kit was provided by Enzo Life Sciences (Exeter, UK). IκB kinase α (IKKα) and sp1 monoclonal antibodies were supplied by Santa Cruz Biotechnology (Santa Cruz, CA, USA). Interferon regulatory factor 3 (IRF3) was purchased from Abcam (Cambridge, UK). Phospho-NF-κB p65 (Ser536), NF-κB p65, phospho-c-Jun (Ser73), c-Jun, cyclooxygenase-2 (COX-2), inducible nitric oxide synthase (iNOS), phospho-IRF3 (Ser396), phospho-IκBα (Ser32), IκBα, phospho-IKKα/β (Ser176/180), phospho-ERK (Thr202/Tyr204), extracellular signal-regulated kinase (ERK), phospho-JNK (Thr183/Tyr185), c-Jun N-terminal kinase (JNK), phospho-p38 (Thr180/Tyr182), p38 mitogen-activated protein kinase (p38), TANK-binding kinase 1 (TBK1), phospho-TBK1 (Ser172), anti-rabbit IgG HRP linked anti-body, and Alexa Fluor 488-conjugated secondary antibody were bought from Cell Signaling Technology (Boston, MA, USA).Methods
Preparation of RIS powder
RIS (batch number: 170803) was provided by Jiangsu Kanion Pharmaceutical Corporation (Jiangsu, China). RIS (200 mL) was concentrated by rotary evaporation under reduced pressure to remove the solvent. The concentrated extracts were rapidly frozen at −80 °C, and then dried in a freeze-dryer (FD-1D-80, Biocool, Beijing, China). The yield of the powdered RIS was 3.87%. To characterize the RIS powder, 0.3542 g of the RIS powder was dissolved with 50% ethanol and diluted to 25 mL. Then, the diluent was filtered through a 0.45 μm Millipore membrane filter for HPLC analysis. To prepare the sample solution for bioassays, RIS was freshly dissolved in DMSO, and then diluted with cell culture medium to various concentrations.Content determination of RIS powder
For the quality control of the prepared RIS, HPLC analysis was performed using an LC-20a HPLC system (Shimadzu Corporation, Tokyo, Japan) consisting of a vacuum degasser, a quaternary pump, an auto sampler, a thermostatic column compartment, and a diode array detector. The separation was conducted on the Agilent Zorbax SB C18 column (250 mm × 4.6 mm with 5 μm particle size) with a gradient mobile phase of solvent A (methanol) and solvent B (0.1% phosphoric acid in water). The HPLC elution profile was as follows: 0–10 min, 15–25% A; 10–30 min, 25–35% A; 30–60 min, 35–50% A. The flow rate was maintained at 0.8 mL min−1 and the column temperature was set at 30 °C. The chromatograms were monitored with the DAD detector at wavelengths of 237 nm and 324 nm to detect geniposide and chlorogenic acid, respectively. Each sample of 10 μL volume was injected for analysis.HPLC fingerprint analysis of RIS
In order to obtain comprehensive chemical information of the RIS powder, fingerprint analysis was carried out using a liquid chromatography with the Agilent 1260 LC system (Agilent Technologies, California, USA) equipped with a quaternary pump, autosampler injector, and a DAD detector adjusted to 225 nm. The chromatographic separation was performed on a Phenomenex Luna C18 column (4.6 × 150 mm, 5 μm; Phenomenex, Los Angeles, USA) at room temperature. The mobile phase constituted of A (methanol) and B (0.1% phosphoric acid). The gradient elution program was as follows: 0–50 min, 20–60% A; 50–60 min, 60% A. The flow rate was set at 0.8 mL min−1. Each sample of 5 μL volume was injected for analysis.Cell culture
The RAW 264.7 murine macrophage cell line was obtained from American Type Culture Collection (Manassa, VA, USA). Cells were incubated in the DMEM medium at 37 °C with 5% CO2. The penicillin/streptomycin antibiotics (1%) and heat-inactivated FBS (10%) were added into the DMEM medium. For each experiment, the cells were seeded in plates and allowed to attach for 24 h. Next, the fresh medium supplemented with RIS or the vehicle was substituted in the culture medium.Cell viability assay
The effect of RIS on the RAW 264.7 cells' viability was measured by the MTT assay. Briefly, 6 × 103 cells (100 μL) per well were seeded in 96 well plates and allowed to adhere overnight. Various concentrations of RIS (12.5–400 μg mL−1) were added into the cells, incubated for 1 h, and then treated in the presence or absence of LPS (1 μg mL−1) for another 24 h. Subsequently, 10 μL of the MTT solution (5 mg mL−1) was added in each well and incubated for 3 h at 37 °C. The supernatant was then removed and the remaining formazan crystals were dissolved with 100 μL of DMSO in each well. The absorbance at 570 nm was determined using a microplate spectrophotometer (BMG SPECTROstar Nano, Germany). The value of cell viability in the control group was set as 100%. Six replicate wells were used.Determination of nitric oxide (NO) production
NO production was assayed by measuring nitrite (a stable degradation product of NO) in the supernatants of cultured RAW 264.7 cells using the Griess modified reagent. Briefly, 2 × 105 cells per well were seeded in 24 well plates. The volume of cell inoculation was 1 mL. After 24 h incubation, the cells were pre-treated with RIS for 1 h, followed by 1 μg mL−1 of LPS, and then incubated in a humidified incubator at 37 °C for 24 h. 100 μL of the supernatant was collected, mixed with an equal volume of Griess reagent, and incubated for 10 min at room temperature. The absorbance measured at 540 nm was used as an indication of the nitrite concentration. Sodium nitrite (NaNO2) was used to produce a standard curve of nitrite concentrations. Four replicate wells were used.Enzyme-linked immunosorbent assay (ELISA)
To measure the production of MIP-1α, MCP-1, TNF-α, IL-6, IL-1β, MCP-1, CCL-5, and PGE2, the cells were incubated with different concentrations of RIS in either the presence or absence of LPS for 24 h. 100 μL of the culture medium supernatant was collected and the concentrations of pro-inflammatory cytokines and chemokines in the cell culture medium were calculated, based on the concentrations of the standard solution, using ELISA kits according to the manufacturer's instructions. Four replicate wells were used.Cytoplasmic and nuclear proteins' extraction
Cytoplasmic and nuclear proteins were extracted using the nuclear extraction kit (Solarbio, Beijing, China) according to the manufacturer's instructions. Briefly, the RAW 264.7 cells were seeded in 100 mm diameter culture dishes (1 × 106 cells, 10 mL), incubated for 24 h, and then treated with 100 and 200 μg mL−1 of RIS for 1 h. LPS (1 μg mL−1) was then added into the culture medium and incubated at 37 °C for 60 min. The cells were harvested and washed with PBS. The ice-cold cytoplasmic protein extraction solution was added to separate the cytoplasmic and nuclear proteins after centrifugation. The same was then added to the nuclear fraction for protein extraction. Three replicates were used.Western blot analysis
Western blot was performed to explore the underlying mechanism of anti-inflammatory action of RIS. Briefly, the RAW 264.7 cells were seeded in 60 mm culture dishes (4 × 105 cells, 3 mL) and treated with 100 and 200 μg mL−1 of RIS for 1 h. LPS (1 μg mL−1) was then added into the culture medium and incubated at 37 °C for 30 min or 60 min. The cells were washed with cold PBS, and then lysed with cold Radio Immunoprecipitation Assay (RIPA) protein extraction buffer containing 1% protease and phosphatase inhibitors (Beyotime Biotechnology, Beijing, China) for 30 min on ice. An aliquot of 20 μg of the supernatant protein from each of the samples was heated with 4 × sodium dodecyl sulfate (SDS) sample buffer at 95 °C for 8 min, and separated electrophoretically on a 10% SDS-polyacrylamide gel. Subsequently, the proteins were transferred onto 0.45 μm pore size PMSF membranes for 3 h and blocked for 1 h. The PMSF membranes were then exposed to indicate the primary antibodies in the blocking buffer at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 or 1
500 or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilutions overnight at 4 °C. The membranes were then incubated with the anti-rabbit IgG secondary antibody conjugated with horseradish at 1
1000 dilutions overnight at 4 °C. The membranes were then incubated with the anti-rabbit IgG secondary antibody conjugated with horseradish at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 dilutions for 1 h. The visualization was performed using Tanon 5200 Multi chemiluminescent imaging system (Tanon Science & Technology Co., Ltd., Shanghai, China) with enhanced-chemiluminescence substrate, and the blots were analyzed using the Image J software (National Institutes of Health [NIH], Bethesda, MD, USA). The protein levels were corrected with the values determined from the β-actin blots. Three replicates were used.
2000 dilutions for 1 h. The visualization was performed using Tanon 5200 Multi chemiluminescent imaging system (Tanon Science & Technology Co., Ltd., Shanghai, China) with enhanced-chemiluminescence substrate, and the blots were analyzed using the Image J software (National Institutes of Health [NIH], Bethesda, MD, USA). The protein levels were corrected with the values determined from the β-actin blots. Three replicates were used.
Immunofluorescence staining
Immunofluorescence staining was conducted to detect the NF-κB/p65, AP-1/c-Jun, and IRF3 localization after the RIS treatment. Briefly, with an inoculation volume of 1 mL, 1 × 104 cells were cultured on coverslips (Cat. 177399; Thermo Fisher Scientific) overnight, treated with RIS (200 μg mL−1), and then fixed with formaldehyde in PBS (w/v, 4%) for 15 min. Then, the cells were washed with PBS and permeabilized with Triton X-100 (0.25%) in 30 min at 37 °C and blocked for 1 h with BSA (2%). The cells were then incubated with NF-κB/p65 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300), c-Jun (1
300), c-Jun (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300), and IRF3 (1
300), and IRF3 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) overnight at 4 °C, respectively. After washing with cold PBS, the cells were incubated with Alexa Fluor 488-conjugated secondary antibody (1
100) overnight at 4 °C, respectively. After washing with cold PBS, the cells were incubated with Alexa Fluor 488-conjugated secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) for 1 h at room temperature. The coverslips were mounted using Fluoroshield with DAPI (YESEN, Shanghai, China) and visualized using the Nikon A1R Eclipse Ti confocal microscope (Nikon Corp., Tokyo, Japan).
500) for 1 h at room temperature. The coverslips were mounted using Fluoroshield with DAPI (YESEN, Shanghai, China) and visualized using the Nikon A1R Eclipse Ti confocal microscope (Nikon Corp., Tokyo, Japan).
Statistical analysis
All values were given as the mean ± standard deviation with at least three replicates for each assay. The statistical differences were determined using one-way ANOVA followed by Turkey's multiple comparisons test. When the value of p was below 0.05, the average values of the different groups were considered significantly different.Results
Characterization of RIS powder
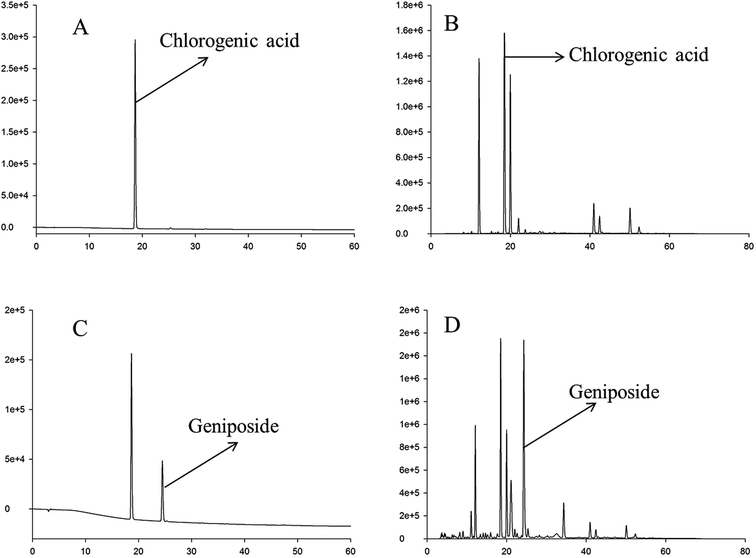
In this study, the HPLC chromatogram showed that geniposide and chlorogenic acid were present in the RIS powder (see Fig. 1). The mean contents of chlorogenic acid and geniposide in the RIS powder were 70.03 and 120.50 mg g−1, respectively.HPLC fingerprint analysis of RIS
In the HPLC fingerprint analysis of RIS, geniposide was selected as the reference substance. As is shown in Fig. 2, peak S was geniposide. Except geniposid, other 13 common peaks were confirmed. Table 1 shows the relative retention times and peak area ratios of 13 common peaks and geniposide. Relative retention time means the ratio of the retention time of a common peak to geniposide. Similarly, the peak area ratio is the ratio of the peak area of a common peak to geniposide.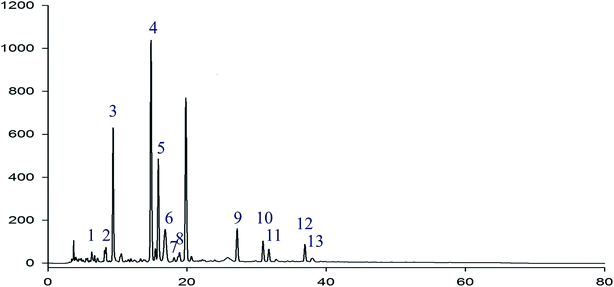 | ||
| Fig. 2 HPLC fingerprint chromatogram of RIS Peak S is geniposide. Except geniposide, other 13 common peaks were detected at 225 nm. | ||
| Peak no. | Relative retention time | Peak area ratio |
|---|---|---|
| 1 | 0.286 | 0.03 |
| 2 | 0.400 | 0.05 |
| 3 | 0.462 | 0.62 |
| 4 | 0.759 | 1.28 |
| 5 | 0.810 | 0.48 |
| 6 | 0.835 | 0.11 |
| 7 | 0.912 | 0.01 |
| 8 | 0.956 | 0.03 |
| S | 1.000 | 1.00 |
| 9 | 1.374 | 0.10 |
| 10 | 1.613 | 0.08 |
| 11 | 1.639 | 0.05 |
| 12 | 1.934 | 0.08 |
| 13 | 1.957 | 0.01 |
Effect of RIS on cell viability
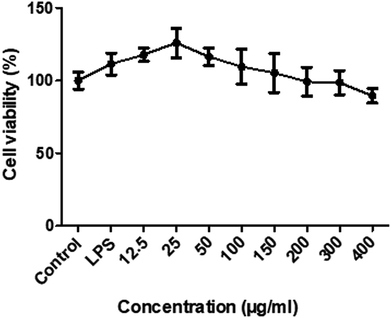
To determine the sub-cytotoxic concentrations of RIS, the MTT assay was conducted. As shown in Fig. 3, the viability of the RAW 264.7 cells was not significantly changed (p > 0.05) up to 400 μg mL−1 concentration of RIS after 24 h incubation with LPS of 1 μg mL−1. Therefore, we chose the concentrations of RIS as 25, 50, 100, and 200 μg mL−1 in the following experiments.Effect of RIS on the production of NO and PGE2 and the expression of iNOS and COX-2
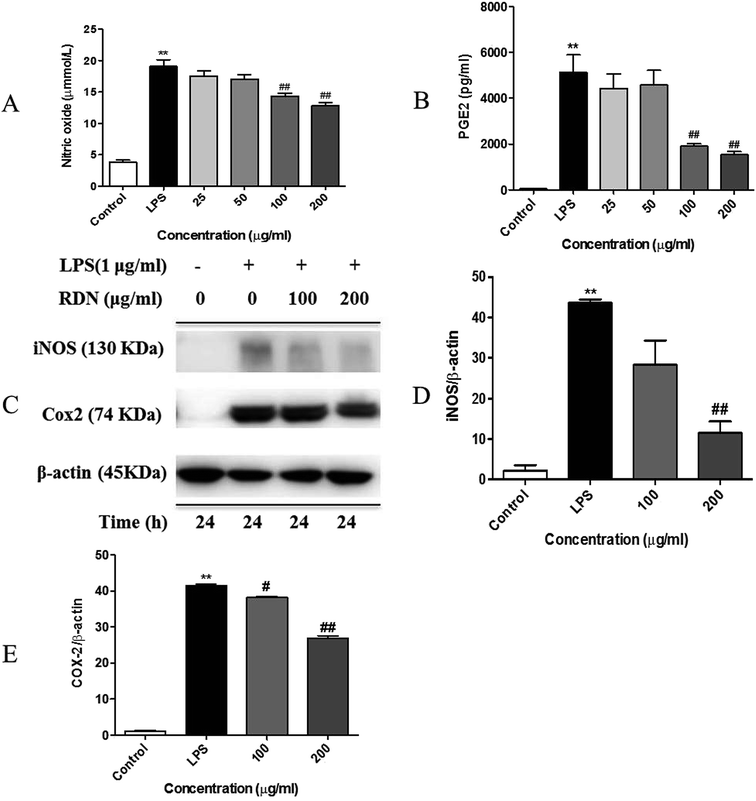
Next, we determined the inflammatory mediators including NO and PGE2 inhibitory activity by RIS in LPS-stimulated RAW 264.7 macrophages. As shown in Fig. 4A and B, the production of NO and PGE2 was strongly inhibited by RIS in a concentration-dependent manner up to 33% and 70%, respectively, in the RAW 264.7 cells (p < 0.01). Furthermore, we detected the expression of key enzymes responsible for the NO and PGE2 production including iNOS and COX-2. The LPS-stimulated RAW 264.7 cells strongly upregulated the protein levels of iNOS and COX-2, whereas this effect was notably (p < 0.01) blocked by RIS (100 and 200 μg mL−1) in a concentration-dependent manner (Fig. 4C and E).Effect of RIS on the production of pro-inflammatory cytokines and chemokines
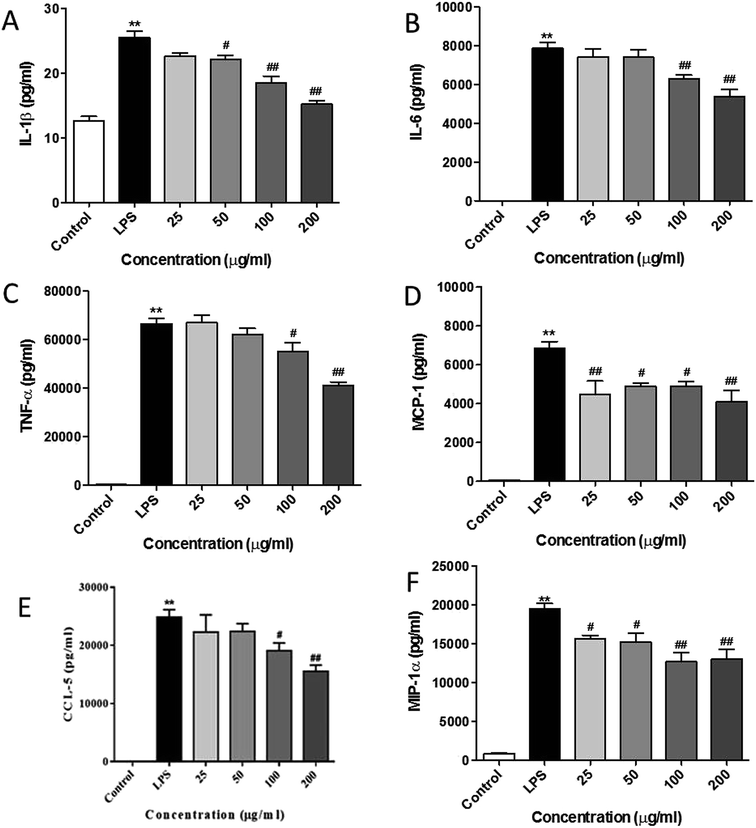
It has been reported that LPS activates the TLR4 signaling pathway and subsequently produces pro-inflammatory cytokines and chemokines in the macrophage cells.15,26,27 In line with the previous studies, we observed that LPS remarkably promoted the release of pro-inflammatory cytokines (see Fig. 5A–C) such as IL-1β, IL-6, and TNF-α as compared with the control cells. RIS efficiently suppressed the elevated cytokine levels in LPS-treated macrophages in a concentration-dependent manner. Moreover, as shown in Fig. 5D–F, the production of chemokines including MIP-1α, CCL-5, and MCP-1 was significantly elevated after the LPS treatment (p < 0.01), indicating that RIS decreased the LPS-induced production of these chemokines (p < 0.05 or p < 0.01) in an effective and concentration-dependent manner.Effect of RIS on the nuclear translocation of NF-κB, AP-1, and IRF3 in RAW 264.7 cells
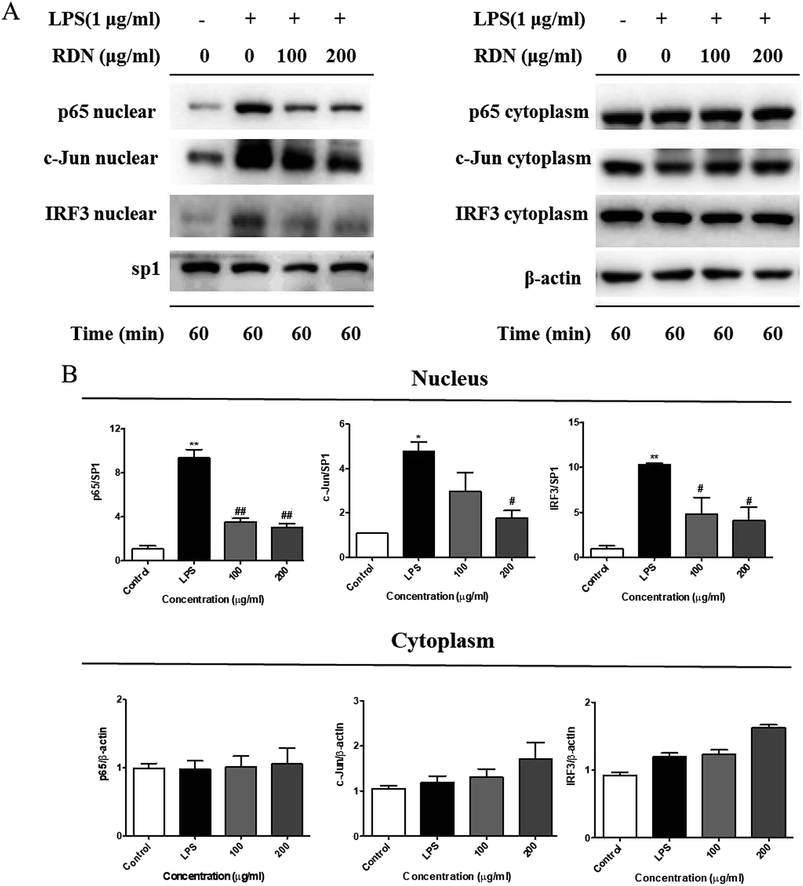
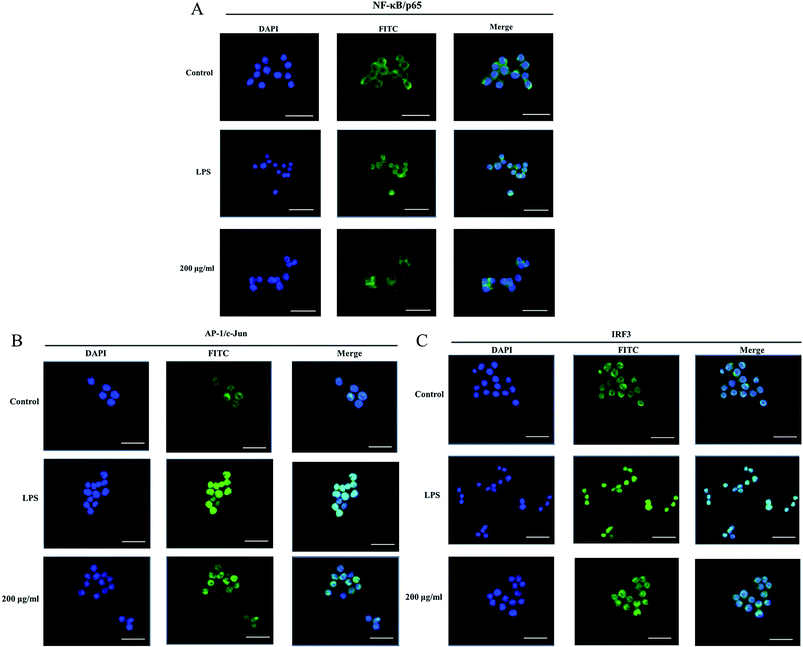
Following LPS stimulation, NF-κB, AP-1, and IRF3 are activated by post-translational modifications, and translocate into the nucleus to induce the transcription of the pro-inflammatory cytokines and chemokines.28,29 Therefore, we firstly investigated the nuclear translocation of these three transcription factors by Western blot analysis. As shown in Fig. 6, the nuclear protein levels of NF-κB/p65, AP-1/c-Jun, and IRF3 were significantly up-regulated after the LPS stimulation, and the RIS treatment markedly reduced the nuclear accumulation of NF-κB/p65, AP-1/c-Jun, and IRF3. The cytoplasmic proteins of these transcriptional factors were not significantly changed after the RIS treatment.Next, we performed an immunofluorescence assay to examine the nuclear localization of NF-κB/p65, AP-1/c-Jun, and IRF3. We observed that NF-κB/p65, AP-1/c-Jun, and IRF3 were translocated from the cytoplasm to the nucleus after the LPS treatment, whereas RIS prevented the LPS-induced nuclear translocation in the RAW 264.7 cells (Fig. 7).
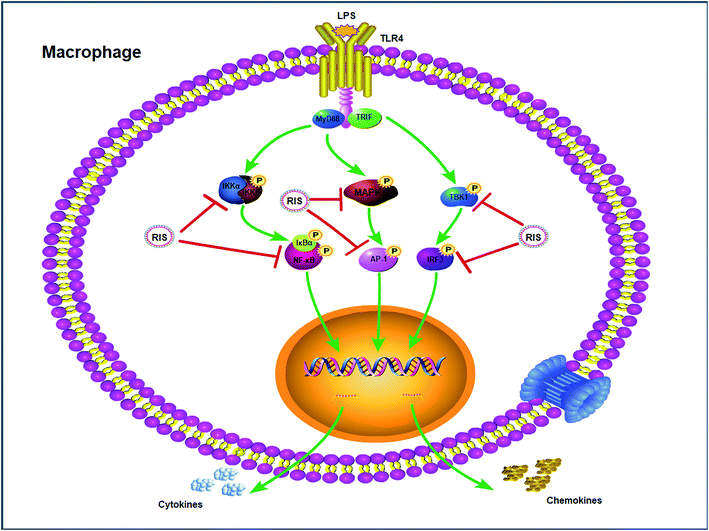
Effect of RIS on the IKKα/β/IκBα/NF-κB, MAPKs/AP-1, and TBK1/IRF3 signaling pathways
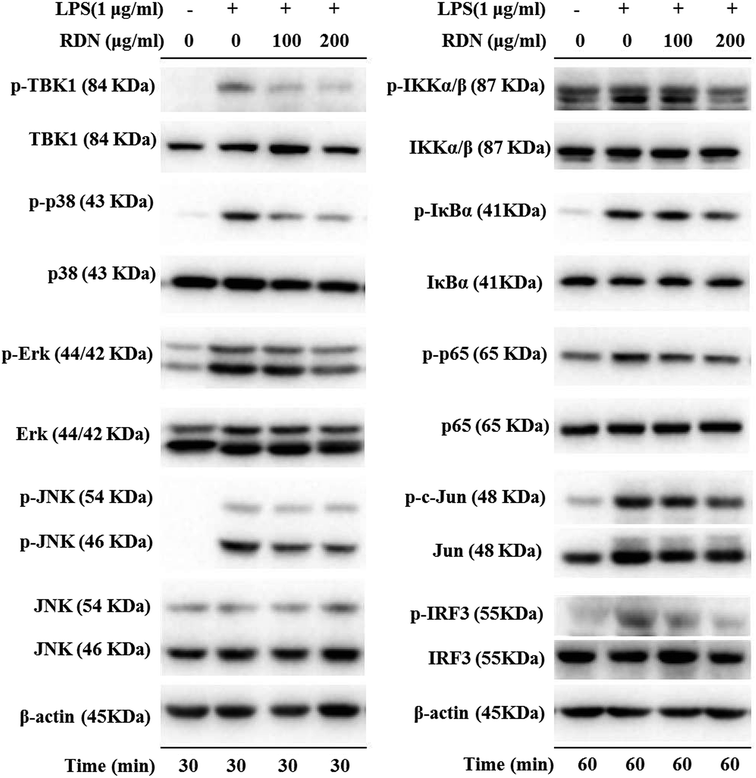
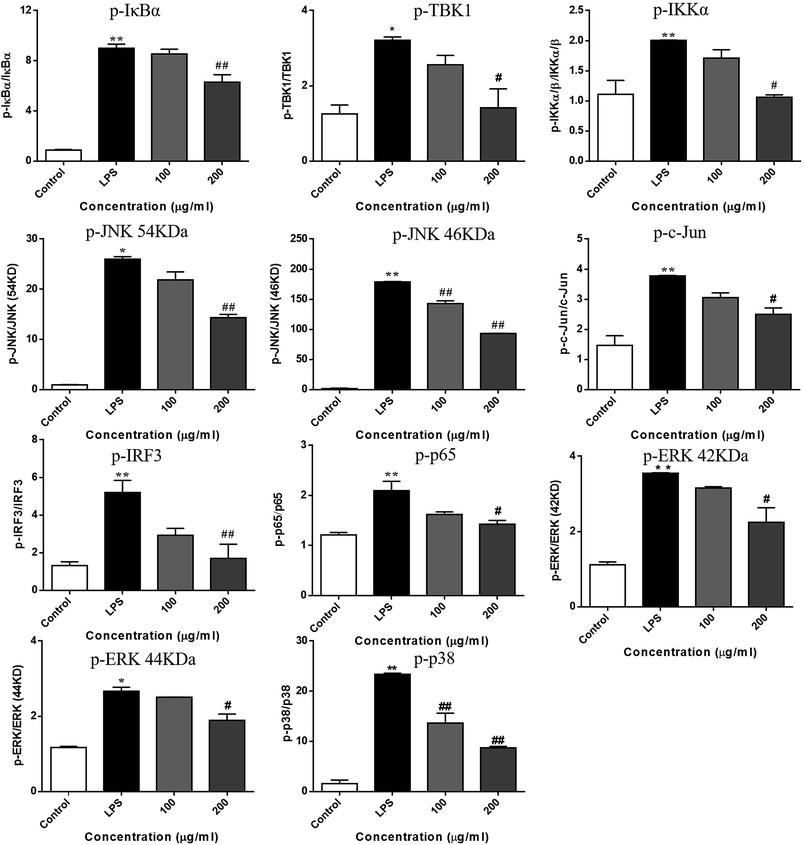
LPS binds to TLR4 and leads to the activation of NF-κB, AP-1, and IRF3 through the recruitment and activation of the upstream proteins.24,27 Therefore, we examined the expression of molecular components of TLR4 related pathways in the RAW 264.7 cells. As shown in Fig. 7 and 8, the phosphorylation levels of IKKα/β and IκBα were significantly up-regulated by the LPS treatment. RIS markedly decreased the LPS-induced elevation of the phosphorylated IKKα/β and IκBα in a concentration-dependent manner. Moreover, the phosphorylation of NF-κB subunit p65 was significantly increased after the LPS treatment, whereas RIS strongly exerted the down-regulative effect on the phosphorylated p65, suggesting that RIS inhibited the IKKα/β/IκBα/NF-κB signaling pathway. Furthermore, the phosphorylation levels of MAPKs including p38, ERK, and JNK were significantly increased after the LPS stimulation in the RAW 264.7 cells. However, RIS dramatically inhibited the LPS-induced upregulation of these three key protein kinases in a concentration-dependent manner. RIS also reduced the phosphorylation level of the down-stream protein of MAPKs such as the AP-1 subunit c-Jun, indicating that it has a potential inhibitory effect on the MAPKs/AP-1 signaling. In addition, we also observed that RIS notably blocked the LPS-induced, upregulated protein expression of phosphorylated TBK1. The phosphorylation level of IRF3 and the downstream protein of TBK1 were also strongly suppressed by the RIS treatment after LPS activation, suggesting that the TBK1/IRF3 signaling pathway was also modulated by RIS (Fig. 8 and 9).Discussion
The anti-inflammatory effects and mechanisms of RDNI have been previously reported.30 In this study, RDNI showed significant suppressive effect on the production of inflammatory mediators in the concentration range of 810–6500 μg mL−1 in the LPS-stimulated RAW 264.7 cells.30 However, RIS exhibited an effective inhibitory effect on the secretion of inflammatory mediators in RAW 264.7 cells in the concentration range of 100–200 μg mL−1. Thus, it could be concluded that RIS had a better anti-inflammatory performance than RDNI.Moreover, we did some exploratory work more deeply and comprehensively based on the former research.30 The nuclear translocation of NF-κB, AP-1, and IRF3 were all studied. The effects of RIS on the three signalling pathways of TLR4 (IKKα/β/IκBα/NF-κB, MAPKs/AP-1, and TBK1/IRF3) in the LPS induced RAW 264.7 cells were investigated in our study as well.
In the present study, chlorogenic acid and geniposide were identified in freeze dried RIS powder using HPLC analysis. It has been reported that chlorogenic acid inhibits the production of cytokines such as IL-1β, TNF-a, and IL-6 in the LPS-stimulated RAW 264.7 cells.31 It also suppressed the carrageenin-induced paw oedema in rats, which may be related to the inhibitory action on the release of inflammatory mediators.32 Moreover, geniposide also exerted potential anti-inflammatory activity via inhibition of the NF-κB signaling pathway.33 Hence, the suppressive effects of RIS on inflammatory mediators may be, at least in part, due to the presence of these two compounds. Further studies will be conducted to identify other active components responsible for the anti-inflammatory effect of RIS.
Current drug treatment options against inflammation such as NSAID are not satisfactory because of their serious adverse effects and toxicity.34 Therefore, researchers are seeking new therapeutic agents with high efficacy and safety. In our study, the cell viability was not altered up to 400 μg mL−1 of RIS concentration, suggesting that RIS has relatively low toxicity. Moreover, the upregulation of the inflammatory mediators including NO and PGE2 in the RAW 264.7 cells was suppressed by RIS. Previous reports have shown that the excessive production of NO and PGE2 by macrophages promotes inflammation, immune mediated disorders as well as the symptoms of redness, swelling, and pain.35,36 Therefore, the inhibition of NO and PGE2 by RIS strongly supports its effectiveness and common use in the treatment of inflammatory diseases. Furthermore, the key enzymes responsible for the synthesis of NO and PGE2 are iNOS and COX-2, respectively.31,37 In our present study, RIS potentially reduced the LPS induced upregulation of iNOS and COX-2 in the RAW 264.7 cells, showing that the inhibitory effect on the NO and PGE2 production was closely related to the suppression of iNOS and COX-2 expression, respectively.
Several studies have proposed novel strategies for the treatment of inflammatory diseases by regulating the activities of macrophages as well as the levels of cytokines (e.g. TNF-α, IL-6, and IL-1β)38 and chemokines (MCP-1, MIP1-α, and CCL-5).39 The TLR4 signaling pathway, which regulates the release of pro-inflammatory cytokines and chemokines, is believed to be one of the novel therapeutic targets in the treatment of inflammatory diseases.40–42 Our results showed that RIS effectively decreased the overproduction of TNF-α, IL-6, IL-1β, MCP-1, MIP1-α, and CCL-5 induced by the LPS-treatment in RAW 264.7 cells, suggesting the inhibitory effect of RIS on inflammatory mediators. This may contribute to its regulative action on the TLR4 signaling transduction.
Upon ligand binding, TLR4 recruits intracellular adapters including MyD88 and activates the IKK complex (including IKKα, IKKβ and IKKγ), which phosphorylates the IκB proteins and lead to its ubiquitination and degradation.43 Subsequently, the NF-κB/p65 complexes are released from IκB and translocate from the cytoplasm to the nucleus. This activates the transcription of a variety of genes participating in the immune and inflammatory responses.44 Our results indicated that RIS significantly reduced the nuclear translocation and phosphorylation of NF-κB and its upstream kinases including IKKα/β and IκBα in the LPS-treated RAW 264.7 cells, showing that RIS suppressed the IKKα/β/IκBα/NF-κB signaling pathway. MAPKs are mainly composed of three characterized subfamilies, namely, ERK1/2, JNK, and p38 MAPK, which stimulate many cellular functions in physiological conditions and are involved in the pathogenesis of many diseases.45 In response to LPS stimulation, MAPKs are activated through dual phosphorylation of tyrosine and threonine residues, and lead to the activation of AP-1.46 Our current study suggested that the phosphorylation of MAPKs (ERK1/2, JNK, and p38 MAPK) was suppressed after the RIS treatment. The nuclear translocation and phosphorylation of AP-1 (c-Jun) was also inhibited by the RIS treatment, revealing that RIS inhibited the MAPKs/AP-1 signaling transduction. In addition to the MyD88-dependent pathways, the TBK1/IRF3 signaling cascade is the main pathway responsible for the induction of type-I interferon production.47 TBK1 can phosphorylate IRF3 and promotes it nuclear translocation, leading to the expression of IFN-β and CCL-5.27 Our data represent that the nuclear and phosphorylation levels of IRF3 as well as the phosphorylated TBK1 were decreased by the RIS treatment, indicating that RIS impeded the LPS-induced TBK1/IRF3 axis.
Conclusion
The result demonstrates that the suppressive capabilities of RIS on inflammatory mediators may be attributed to the inhibition of IKKα/β/IκBα/NF-κB, MAPKs/AP-1, and TBK1/IRF3 signaling pathways in the LPS-stimulated RAW 264.7 cells (Fig. 10). To further investigate the contribution of the TLR4 related pathways in the anti-inflammatory effects of RIS, we will establish animal models to validate it.Chemical compounds studied in this article
Lipopolysaccharide (PubChem CID: 11970143)Dimethyl sulfoxide (PubChem CID: 679)
Penicillin (PubChem CID: 5904)
Streptomycin (PubChem CID: 19649)
Abbreviations
| CCL-5 | Chemkine (C–C motif) ligand 5 |
| AP-1 | Activator protein-1 |
| DMEM | Dulbecco's modified eagle medium |
| ERK | Extracellular signal-regulated kinase |
| FBS | Fetal bovine serum |
| IKK | IκB kinase |
| IL | Interleukin |
| IFN | Interferon |
| IRF3 | IFN regulatory factor 3 |
| JNK | c-Jun N-terminal kinase |
| LPS | Lipopolysaccharide |
| MAPK | Mitogen-activated protein kinase |
| MCP | Monocyte chemoattractant protein |
| MIP | Macrophage inflammatory protein |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenylthiazolium bromide |
| MyD88 | Myeloid differentiation primary response 88 |
| NF-κB | Nuclear factor-κB |
| p38 | p38 mitogen-activated protein kinase |
| TBK1 | TANK-binding kinase 1 |
| TRIF | TIR-domain-containing adaptor protein-inducing IFN-β |
| TNF | Tumour necrosis factor |
| TLR | Toll-like receptor |
| COX-2 | Cyclooxygenase-2 |
| iNOS | Inducible nitric oxide synthase |
Conflicts of interest
The authors declare no conflicts of interest regarding the publication of this paper.Acknowledgements
This study was supported by National Natural Science Foundation of China (81803793) and the Fundamental Research Funds for the Central Universities (2018-JYBZZ-XJSJJ008, 2017-JYB-JS-057).References
- W. Liao, X.-J. He, Z.-W. Yi, W. Xiang and Y. Ding, Biomed. Pharmacother., 2018, 107, 1151–1159 CrossRef CAS PubMed.
- I. Sulaiman, L. C. Woei, S. H. Liong and J. Stanslas, Pulm. Pharmacol. Ther., 2016, 40, 52–68 CrossRef CAS PubMed.
- D. Droemann, S. P. Aries, F. Hansen, M. Moellers, J. Braun, H. A. Katus and K. Dalhoff, Chest, 2000, 117, 1679–1684 CrossRef CAS PubMed.
- J.-G. Chu, X.-J. Wang, H. J. Bi, L.-F. Li, M.-G. Ren and J.-W. Wang, Int. Immunopharmacol., 2018, 59, 174–180 CrossRef CAS.
- Z. Vitenberga, M. Pilmane and A. Babjoniševa, Pathol., Res. Pract., 2019, 215, 97–105 CrossRef CAS PubMed.
- H.-B. Guo and M.-X. Chen, Ecotoxicol. Environ. Saf., 2018, 161, 184–189 CrossRef CAS PubMed.
- India State-Level Disease Burden Initiative CRD Collaborators, Lancet Global Health, 2018, 6, 1363–1374 CrossRef.
- E. Midouhas, E. Flouri, E. Papachristou and T. Kokosi, Intelligence, 2018, 68, 30–36 CrossRef.
- C. Li, Q. Liu and L.-Q. Xie, Biochem. Biophys. Res. Commun., 2018, 507, 22–29 CrossRef CAS PubMed.
- X.-C. Zheng, D. Yang, X. Liu, N. Wang, B. Li, H.-W. Cao, Y.-L. Lu, G. Wei, H. Zhou and J. Zheng, Int. Immunopharmacol., 2010, 10, 1209–1219 CrossRef CAS PubMed.
- F. Liu, Y. Wen, J.-Y. Kang, C.-Y. Wei, M.-H. Wang, Z.-Q. Zheng and J.-T. Peng, Int. J. Mol. Med., 2018, 42, 1436–1444 CAS.
- A. A. Shalmani, M. H. Ghahremani, F. Jeivad, A. Shadboorestan, G. Hassanzadeh, A. Beh-Pajooh, M. Ganbari-Erdi, S. Kasirzadeh, M. Mojtahedzadeh and O. Sabzevari, Life Sci., 2018, 215, 152–158 CrossRef CAS PubMed.
- M. Zhang, J. Wu, J. Han, H. Shu and K. Liu, Chem. Cent. J., 2018, 12, 109 CrossRef PubMed.
- W.-X. Ding, J.-Y. Gu, L. Cao, N. Li, G. Ding, Z.-Z. Wang, L.-R. Chen, X.-J. Xu and W. Xiao, J. Ethnopharmacol., 2014, 155, 589–598 CrossRef CAS PubMed.
- B. C.-Y. Cheng, X.-Q. Ma, H. Y. Kwan, K. W. Tse, H.-H. Cao, T. Su, X. Shu, Z.-Z. Wu and Z.-L. Yu, J. Ethnopharmacol., 2014, 153, 922–927 CrossRef PubMed.
- Y.-Z. He, Chinese Medicine Modern Distance Education of China, 2015, 9, 123–125 Search PubMed.
- China Food and Drug Administration, National drug standards of Re-Du-Ning injection: YBZ08202005-2015Z, 2015, pp. 1–8 Search PubMed.
- T.-F. Tzeng, Y.-C. Tzeng, Y.-J. Cheng, S.-S. Liou and I.-M. Liu, Nutrients, 2015, 7, 8670–8684 CrossRef CAS PubMed.
- Y.-J. Lin, C.-C. Lai, C.-H. Lai, S.-C. Sue, C.-W. Lin, C.-H. Hung, T.-H. Lin, W.-Y. Hsu, S.-M. Huang, Y.-L. Hung, N. Tien, X. Liu, C.-L. Chen and F.-J. Tsai, Eur. J. Med. Chem., 2013, 62, 206–213 CrossRef CAS PubMed.
- E. Hsu, Trans. R. Soc. Trop. Med. Hyg., 2006, 100, 505–508 CrossRef CAS PubMed.
- F. V. D. Kooy and S. E. Sullivan, J. Ethnopharmacol., 2013, 150, 1–13 CrossRef PubMed.
- S. H. Park, E. Roh, H. S. Kim, S. I. Baek, N. S. Choi, N. Kim, B. Y. Hwang, S. B. Han and Y. Kim, Biochem. Biophys. Res. Commun., 2013, 442, 183–188 CrossRef CAS PubMed.
- S.-J. Kim, S.-J. Yoon, Y.-M. Kim, S.-W. Hong, S. H. Yeon, K.-I. Choe and S.-M. Lee, J. Ethnopharmacol., 2014, 155, 256–266 CrossRef PubMed.
- Q.-H. Shi, J.-J. Cao, L. Fang, H.-Y. Zhao, Z.-X. Liu, J.-H. Ran, X.-C. Zheng, X.-L. Li, Y. Zhou, D. Ge, H.-M. Zhang, L. Wang, Y. Ran and J.-F. Fu, Int. Immunopharmacol., 2014, 20, 298–306 CrossRef CAS PubMed.
- K. H. Park, Y. D. Yoon, S.-B. Han, S. J. Oh, J. Yun, C. W. lee, K. Lee, S.-K. Park, H. M. Kim and J. S. Kang, Int. Immunopharmacol., 2012, 14, 580–584 CrossRef CAS PubMed.
- Y. Zhang, H. Guo, B. C.-Y. Cheng, T. Su, X.-Q. Fu, T. Li, P.-L. Zhu, K.-W. Tse, S.-Y. Pan and Z.-L. Yu, Drug Des., Dev. Ther., 2018, 12, 2731–2748 CrossRef PubMed.
- G. Luo, B. Cheng, H. Zhao, X.-Q. Fu, R. Xie, S.-F. Zhang, S.-Y. Pan and Y. Zhang, Molecules, 2018, 23, 3319 CrossRef PubMed.
- H.-T. Shi, L. Dong, X.-Y. Dang, Y.-P. Liu, J. Jiang, Y. Wang, X.-L. Lu and X.-Y. Guo, Inflammation Res., 2013, 62, 581–587 CrossRef CAS PubMed.
- A.-Y. Kim, H.-J. Shim, S. Y. Kim, S. Heo and H.-S. Youn, Int. Immunopharmacol., 2018, 64, 1–9 CrossRef CAS PubMed.
- J. Liu, K. Sun, C. Zheng, X. Chen, W. Zhang, Z. Wang, P. A. Shar, W. Xiao and Y. Wang, PLoS One, 2015, 10, e0123109 CrossRef PubMed.
- S. J. Hwang, Y.-W. Kim, Y. Park, H. J. Lee and K.-W. Kim, Inflammation Res., 2014, 63, 81–90 CrossRef CAS PubMed.
- M. D. d. Santos, M. C. Almeida, N. P. Lopes and G. E. P. d. Souza, Biol. Pharm. Bull., 2006, 29, 2236–2240 CrossRef.
- R. Deng, F. Li, H. Wu, W.-Y. Wang, L. Dai, Z.-R. Zhang and J. Fu, Front. Pharmacol., 2018, 9, 105 CrossRef PubMed.
- J. R. Perkins, M. Sanak and G. Canto, Trends Pharmacol. Sci., 2015, 36, 172–180 CrossRef CAS PubMed.
- X.-H. Jin, K. Ohgami, K. Shiratori, Y. Suzuki, Y. Koyama, K. Yoshida, I. Ilieva, T. Tanaka, K. Onoe and S. Ohno, Exp. Eye Res., 2006, 82, 860–867 CrossRef CAS PubMed.
- R. M. Hinson, J. A. Williams and E. Shacter, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 4885–4890 CrossRef CAS.
- J. Y. Park, M. H. Pillinger and S. B. Abramson, Clin. Immunol., 2006, 119, 229–240 CrossRef CAS PubMed.
- T.-T. Zhang, T. Hu, J.-G. Jiang, J.-W. Zhao and W. Zhu, RSC Adv., 2018, 8, 7134–7141 RSC.
- P. M. Murphy, Chemokines and Chemokine Receptors, Clinical Immunology, ed. R. R.Rich, T. A.Fleisher, W. T.Shearer, H. W.Schroeder, A. J.Frew and C. M.Weyand, Content Repository Only!, London, 5th edn, 2019, vol. 10, pp. 157–170 Search PubMed.
- T. Baluchnejadmojarad, Z. Kiasalari, S. A. Majd, Z. Ghasemi and M. Roghani, Eur. J. Pharmacol., 2017, 794, 69–76 CrossRef CAS PubMed.
- L.-H. Xu, Y.-B. Liu, Y.-H. Sun, H. Li, W.-D. Mi and Y.-G. Jiang, Biomed. Pharmacother., 2018, 107, 526–533 CrossRef CAS PubMed.
- H. Capiralla, V. Vingtdeux, H.-T. Zhao, R. Sankowski, Y. A. Abed, P. Davies and P. Marambaud, J. Neurochem., 2012, 120, 461–472 CrossRef CAS PubMed.
- S. Akia and K. Takeda, Nat. Rev. Immunol., 2004, 4, 499 CrossRef PubMed.
- G. M. Barton and R. Medzhitov, Science, 2003, 300, 1524 CrossRef CAS PubMed.
- Q.-H. Shao, X.-L. Zhang, Y. Chen, C.-G. Zhu, J.-G. Shi, Y.-H. Yuan and N.-H. Chen, Mol. Immunol., 2018, 99, 115–123 CrossRef CAS PubMed.
- T. Kawasaki and T. Kawai, Front. Immunol., 2014, 5, 461 Search PubMed.
- S. Wei, D.-B. Yang, J.-F. Yang, X.-S. Zhang, J.-L. Zhang, J.-C. Fu, G.-B. Zhou, H.-J. Liu, Z.-X. Lian and H.-B. Han, Eur. J. Cell Biol., 2019, 98, 36–50 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |