Open Access Article
Open Access ArticleTime-based LC-MS/MS analysis provides insights into early responses to mechanical wounding, a major trigger to agarwood formation in Aquilaria malaccensis Lam†
Muhammad Syahmi Hishamuddina,
Shiou Yih Leea,
Nurulfiza Mat Isab,
Dhilia Udie Lamasudinbc,
Syafiq Asnawi Zainal Abidind and
Rozi Mohamed *a
*a
aForest Biotechnology Laboratory, Department of Forest Management, Faculty of Forestry, Universiti Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia. E-mail: rozimohd@upm.edu.my
bDepartment of Cell and Molecular Biology, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia
cHalal Products Research Institute, Universiti Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia
dLiquid Chromatography Mass Spectrometry (LCMS) Platform, Jeffrey Cheah School of Medicine and Health Sciences, Monash University Malaysia, Jalan Lagoon Selatan, 47500, Bandar Sunway, Selangor, Malaysia
First published on 11th June 2019
Abstract
Mechanical wounding is the major trigger for the formation of agarwood in the tropical tree taxon Aquilaria. To understand the molecular mechanism by which Aquilaria reacts to wounding, we applied a proteomics approach using liquid chromatography electrospray-ionization coupled with tandem mass spectrometry (LC-MS/MS) coupled with bioinformatics analysis and principal component analysis. Protein samples were extracted from wood tissues collected from drilled wounds on the stems of five-year old Aquilaria malaccensis. Samples were collected at different time-points of 0, 2, 6, 12, and 24 h after mechanical wounding for protein identification. Venn diagram analysis showed that 564 out of 2227 identified proteins were time-point specific proteins. GO analysis using the REViGO software (including functional proteins) supported these findings. In total, 20 wound-response proteins and one unknown protein were identified as having important roles in the signaling response to wounding, response to stress, activation of plant defense systems, and plant regeneration. The detected biological processes include brassinosteroid stimulus, polyamine catabolism, hypersensitive response, response to cadmium ions, response to oxidative stress, and malate metabolism, suggesting that the wounded trees must have undergone major plant cell damage. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis indicated that several wound-response proteins were involved in agarwood formation. Our proteomics data thus provide useful information for understanding the wound response mechanisms that trigger agarwood formation.
Introduction
Agarwood is a natural forest product that is rich in fragrance and medicinal compounds. It is obtained from a botanical resource of tropical origin, Aquilaria. Agarwood trade fetches a lucrative income in the international market and its demand overdrives its harvest from tree species such as Aquilaria malaccensis Lam. As a result, A. malaccensis was the first species placed under Appendix II of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES).1 At present, seven Aquilaria species are currently listed as ‘Vulnerable’ in the International Union for Conservation of Nature (IUCN) Red List.2 The practice of collecting from the wild is not a sustainable option for commercial agarwood production. Therefore, efforts have been made to cultivate Aquilaria trees in large-scale plantations.3Agarwood does not form on its own. Aquilaria requires severe induction to form the substances found in agarwood. Its formation is rare under natural conditions; it occurs when an injury is inflicted on the tree stem, such as from a falling branch, and from pest and disease infestation.4 Mechanical wounding techniques such as nailing, pruning, using a burning chisel, and hole-drilling have been conducted on planted Aquilaria trees to induce agarwood production.3 Considered as the tree's defensive response to abiotic and biotic stresses, agarwood resin acts as a barrier to prevent further damage to the tree, and from invasion by pathogens like fungi or bacteria.5 Microscopic observation revealed that agarwood formation is localized in wood tissues neighboring the wounded or pathogen-infected areas.6,7 Generally, it is agreed upon that wounding in Aquilaria stems is the primary damaging act that triggers the tree's defense signals, which are further transmitted to cells responsible for synthesizing the agarwood substances. Sesquiterpenes and phenylethyl chromone derivatives are two major compounds in agarwood.8 Both compounds have been reported to possess several medicinal properties, and have been used as analgesic, traditional sedative, and digestive medicines.9
For the past few years, biological studies have been conducted on model organisms, and for plants, most of the functional genomics studies have been conducted extensively on model plant species and other characterized species.10 While most plants of interest are essential as food, feed, or energy resources, the lack of genomic information from non-model plants makes it impossible to characterize their unique features and processes. As a result, valuable genomic information may be overlooked during the identification process, which hinders the work to reveal an accurate finding on the plant of interest. However, to complement transcriptomic studies, the proteomics approach was reported to be of potential importance in studying non-model species. Proteins are end products of gene expression, with sequences that are more conserved when compared to genes for high-throughput identification. Although proteome studies have been carried out in model plants such as Arabidopsis and rice,11,12 they still could not properly characterize proteins from woody plants with long life cycles and with special features that offer them a greater key to survival. This phenomenon applies to the mechanism of stress-induced biosynthesis in agarwood, which is actively being investigated using Aquilaria transcriptomes,13–15 but has been limited in proteomic approaches. A draft genome from A. agallocha (synonym A. malaccensis) originating from Myanmar has been reported,16 and a proposal on the Aquilaria terpenoid biosynthesis pathway and its possible regulators has greatly improved the available genetic information on Aquilaria. However, these efforts were only able to reflect fragments of the agarwood biosynthesis mechanism in Aquilaria.
In general, mechanical wounding is known to threaten plant life by affecting gaseous exchange, intercellular solute dynamics, photosynthesis rate, and creating opportunities for microbial invasion.6 However, wounding inflicted on a plant could provide a response toward modification of the damage sites, which helps it regulate its interaction with its surroundings. Such a process could lead to the release of intercellular fluid or guttation fluid around the wounds, accompanied with several extracellular defense proteins that act to prevent pathogen ingress.17 Similar to other plants with such defense characteristics, the Aquilaria tree produces agarwood resin when subjected to substantial wounding. Such stress-related changes in the proteome have been characterized in several plant species,18,19 and to date, 2-dimensional gel electrophoresis (2DE) and iTRAQ analysis are the only proteome analyses reported available for Aquilaria.20,21 The latter, reported on the mechanism of agarwood formation using chemical inducers as the trigger.21 Despite the few studies conducted to investigate the Aquilaria proteome, the available information is still sparse and inconclusive. Through this study, we aim to provide some insights on the early response mechanism to wounding that triggers agarwood formation in Aquilaria and contributes to a new body of information in plant proteomics research. To the best of our knowledge, this is the first report on protein response toward mechanical wounding on a time-frame basis in an agarwood producing tree using the LC-MS approach.
Materials and methods
Plant materials and mechanical wounding treatment
Five-year-old A. malaccensis trees were grown in polybags at a plant nursery belonging to the Faculty of Forestry, Universiti Putra Malaysia (UPM), Serdang, Selangor, Malaysia. The trees were mechanically wounded as described by Azzarina et al.22 A drill machine equipped with a 3 mm diameter drill bit was used to drill the stems to a depth of 1 to 2 cm, depending on the stem diameter, enabling the wound to reach the xylem. Wounding was carried out in a single straight line on the tree, from the base to the apex along sectors of the circumference with each wound spaced at approximately 11 cm apart. Twelve trees were wounded at the same time and three unwounded trees were used as controls representing 0 h. A total of 15 trees were utilized in the experiment. Harvesting was conducted at four different time-points post-wounding: 2, 6, 12, and 24 h. The sampled area (SA) was 1 mm from the drilled point and 5 cm in length away from the drilled point were pooled together. All tree stems were harvested in each group, and SAs were sorted out for total protein extraction. The samples were kept in zip-lock bags and stored at −80 °C prior to protein extraction.Total protein extraction
Total proteins of fresh wood samples from five different time-points (0, 2, 6, 12, and 24 h) and from three biological replicates were extracted using the ReadyPrep™ Protein Extraction Kit (Total Protein) (Bio-RAD, USA) following the manufacturer's protocol. In total, 1 g of wood sample was powdered in liquid nitrogen using a mortar and pestle. The sample was transferred into a 5 mL microcentrifuge tube containing 2 mL of 2-D rehydration/sample buffer I, 20 μL of ReadyPrep TBP reducing agent and 0.2% (w/v) of the final concentrate of ampholyte. Subsequent steps were carried out according to the manufacturer's protocol. Total protein concentration was determined using the Quick Start™ Bradford protein assay kit (Bio-Rad, USA) on a Nanophotometer™ (IMPLEN, Germany) and bovine serum albumin (Bio-Rad, USA) was used as a standard. Upon quantification, 5 mg of total protein was transferred into a new 1.5 mL microcentrifuge tube, precipitated using 2-D Clean Up Kit (GE Healthcare Life Science, UK), following the manufacturer's protocol, and stored at −20 °C prior to in-solution tryptic digestion.In-solution digestion (tryptic digestion)
Approximately 0.5 mg of total protein sample was resuspended, denatured, and reduced by addition of a mixture containing 25 μL of ammonium bicarbonate, 25 μL of trifluroethanol denaturation agent, and 1 μL of 200 mM DTT. The mixture was briefly vortexed, centrifuged, and incubated at 60 °C for 1 h. Alkylation was carried out by adding 4 μL of iodoacetamide (IAM) and incubating at room temperature in the dark for 1 h. Excess IAM was quenched by addition of 1 μL DTT. Dilution was carried out by adding 300 μL of distilled water and pH was raised by adding 100 μL of ammonium bicarbonate. Digestion was performed by adding 10 μL of freshly prepared trypsin (ThermoScientific, USA) stock solution (ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 by mass of enzyme–substrate) to the mixture, briefly vortexing and incubating overnight at 37 °C. At the end of the digestion step, 1 μL of formic acid was added into the tube to lower the pH and stop trypsin activity. The digested protein samples were kept at −20 °C prior to LC-MS/MS analysis.
20 by mass of enzyme–substrate) to the mixture, briefly vortexing and incubating overnight at 37 °C. At the end of the digestion step, 1 μL of formic acid was added into the tube to lower the pH and stop trypsin activity. The digested protein samples were kept at −20 °C prior to LC-MS/MS analysis.
Nanoflow liquid chromatography electrospray-ionization coupled with tandem mass spectrometry (nanoflow-ESI-LC-MS/MS)
LC-MS/MS analysis was performed using Agilent QTOF 6550 at the LC-MS/MS laboratory, Monash University Malaysia, Selangor, Malaysia. The digested fraction was loaded into an Agilent Large Capacity Chip (Agilent, USA) comprising a 160 mL enrichment column and a 75 μM × 150 mm analytical column, which was packed with 5 μM of Zorbax 300SB-C18. The solvent systems used to elute the peptides were Solvent A (0.1% formic acid in MilliQ water), and Solvent B (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of 0.1% formic acid in acetonitrile
1 ratio of 0.1% formic acid in acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MilliQ water). The gradient used was programmed as: 3–50% of solvent B for 30 min, 50–95% of solvent B for 2 min, 95% of solvent B for 7 min, and 95–3% of solvent B for 47 min. The quadrupole time-of-flight (Q-TOF) polarity was set at positive, the capillary and fragmentor voltages were set at 2050 V and 300 V, respectively, and the gas flow was set at 5 L min−1 and 325 °C. The peptide spectra were acquired using the Agilent MassHunter Workstation Data Acquisition software (Agilent Technologies, USA) in an auto MS/MS mode ranging from 110 to 3000 m/z for the MS scan, and from 50 to 3000 m/z for the MS/MS scan. The chromatograms were analyzed using the Agilent MassHunter Qualitative Analysis B.05.00 software (Agilent Technologies, USA).
MilliQ water). The gradient used was programmed as: 3–50% of solvent B for 30 min, 50–95% of solvent B for 2 min, 95% of solvent B for 7 min, and 95–3% of solvent B for 47 min. The quadrupole time-of-flight (Q-TOF) polarity was set at positive, the capillary and fragmentor voltages were set at 2050 V and 300 V, respectively, and the gas flow was set at 5 L min−1 and 325 °C. The peptide spectra were acquired using the Agilent MassHunter Workstation Data Acquisition software (Agilent Technologies, USA) in an auto MS/MS mode ranging from 110 to 3000 m/z for the MS scan, and from 50 to 3000 m/z for the MS/MS scan. The chromatograms were analyzed using the Agilent MassHunter Qualitative Analysis B.05.00 software (Agilent Technologies, USA).
Data analysis
Data processing and protein identification were performed using Spectrum Mill (Ver. B.06.00.201, Agilent Technologies, USA) against an organism-specific database (NCBI taxa: Malvales; as of October 2016). Carbamidomethylation was selected as the fixed modification parameter and trypsin as the digestion enzyme with the maximum number of missed cleavages equal to 2. The MS searches were auto-validated at a false discovery rate (FDR) of 1.2%, followed by the generation of protein–protein comparison files and data export to Mass Profiler Professional (MPP v 14.9.1, Agilent Technologies, USA) for further analysis. The data files were then processed by principle component analysis (PCA) using the MPP software.Protein annotation and analysis
Protein annotation was carried out using the tblastn (https://www.ncbi.nlm.nih.gov/) Ubuntu Linux platform, based on a probability-based scoring system applied against the NCBI non-redundant (NR) protein database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/; as of January 2017). The BLAST output, in .xml format, was imported into BLAST2GO software23 for Gene Ontology (GO) annotation. All identified proteins were combined to produce the Venn diagram. Data in the form of identified proteins from Blast analyses were submitted to the on-line ‘Create-A-Venn’ software (http://bioinformatics.psb.ugent.be/webtools/Venn/). Venn diagram calculated the intersections and produced a graphical output, which pointed to the time-point specific (TPS) proteins. All of the TPS proteins were selected based on the Blast results and Venn diagram analyses. Annotated sequences with the corresponding enzyme commission (EC) numbers were obtained and mapped into the Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathway database (http://www.genome.jp/kegg/).GO result visualization
GO enrichment analysis was carried out to identify the enriched functional categories in the plant UniProt database based on Fisher's exact test and a false discovery rate (FDR) < 0.05, using BLAST2GO software.23 Default parameters were used for all analyses. Results obtained from the enrichment analysis were visualized and summarized by removing the redundant GO terms using Reduce and Visualize Gene Ontology (REViGO; http://revigo.irb.hr/).24 Additional filtering steps based on several conditions such as the percentage of identity (PID), bit score, and e-value, were performed to obtain our targeted protein identities. The parameters were set at: PID ≥ 55%; bit score ≥30.0; and e-value ≤ 0, as the minimum criteria for significant protein identities.Results
LC-MS/MS analysis of wound-response proteins
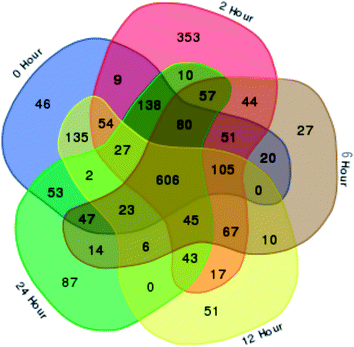
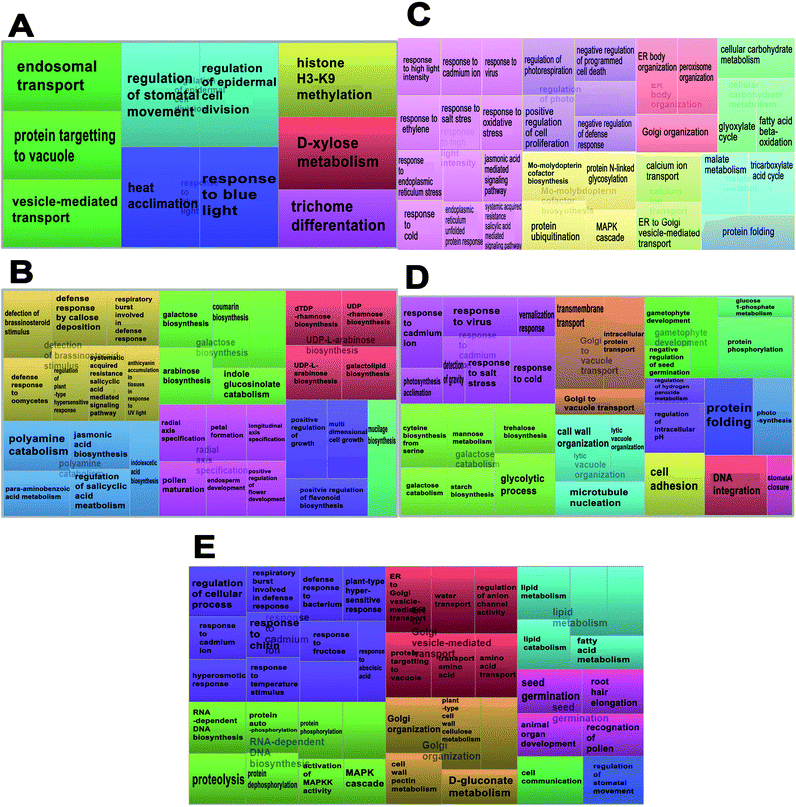
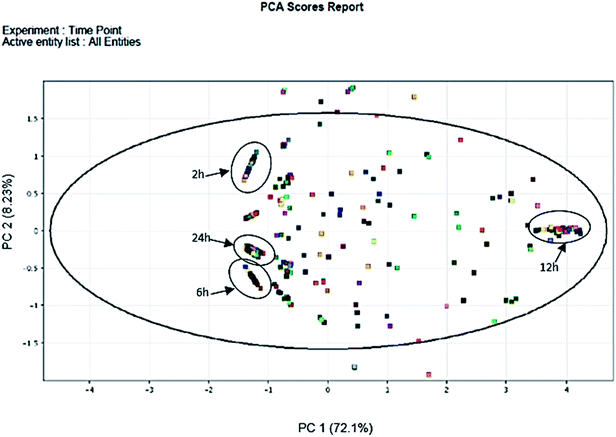
In total, 15 protein extracts representing three biological replicates of each control (unwounded, 0 h) and the mechanically wounded trees at 2, 6, 12, and 24 h post-treatment were prepared. All the protein samples had concentrations above 1000 μg mL−1. The samples were tryptic-digested and analyzed using LC-MS/MS. As a result, 4188 MS fragmented data were obtained. Searches against the NCBI database yielded a total of 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 884 proteins, however 93% were removed as they were common proteins over the five time-points. Approximately, 7% of the identified proteins (2227) were further analyzed using a Venn diagram, from which 564 were TPS proteins (Fig. 1). Fig. 1 illustrates the distribution of proteins present across the five different time-points as represented by five different colors: blue (0 h), red (2 h), brown (6 h), yellow (12 h), and green (24 h). Intersections between time-points represent numbers of overlapping proteins, while the value at the center represents the number of proteins present at all time-points. About 8% of the TPS proteins were specific to the unwounded sample (0 h), while the remaining TPS proteins were triggered by wounding (2–24 h). A rapid change in the number of TPS proteins (7.6-fold proteins) was observed at 2 h after wounding compared to that at 0 h (Fig. 1). However, a drastic decrease (<0.08-fold) was observed 6 h later and small increments in the number of TPS proteins were observed at 12 h and 24 h after wounding. The high number of TPS proteins present at 2 h can be explained by a tree's immediate response to wounding. A majority of these proteins were involved in the signaling response (Fig. 2B). TPS proteins identified at 6, 12 and 24 h appeared to be related to stress and defense response rather than signaling (Fig. 2C–E). All the identified TPS proteins when analyzed through PCA displayed four clusters (Fig. 3). PCA shows a clear separation between the four time-points (2 h, 6 h, 12 h, and 24 h). This supports findings from the Venn diagram (Fig. 1).
884 proteins, however 93% were removed as they were common proteins over the five time-points. Approximately, 7% of the identified proteins (2227) were further analyzed using a Venn diagram, from which 564 were TPS proteins (Fig. 1). Fig. 1 illustrates the distribution of proteins present across the five different time-points as represented by five different colors: blue (0 h), red (2 h), brown (6 h), yellow (12 h), and green (24 h). Intersections between time-points represent numbers of overlapping proteins, while the value at the center represents the number of proteins present at all time-points. About 8% of the TPS proteins were specific to the unwounded sample (0 h), while the remaining TPS proteins were triggered by wounding (2–24 h). A rapid change in the number of TPS proteins (7.6-fold proteins) was observed at 2 h after wounding compared to that at 0 h (Fig. 1). However, a drastic decrease (<0.08-fold) was observed 6 h later and small increments in the number of TPS proteins were observed at 12 h and 24 h after wounding. The high number of TPS proteins present at 2 h can be explained by a tree's immediate response to wounding. A majority of these proteins were involved in the signaling response (Fig. 2B). TPS proteins identified at 6, 12 and 24 h appeared to be related to stress and defense response rather than signaling (Fig. 2C–E). All the identified TPS proteins when analyzed through PCA displayed four clusters (Fig. 3). PCA shows a clear separation between the four time-points (2 h, 6 h, 12 h, and 24 h). This supports findings from the Venn diagram (Fig. 1).
Functional categories of wound-response proteins
The 564 TPS proteins were categorized into different biological processes and pathways using REViGO and KEGG analysis. In total, 518 TPS proteins were triggered by wounding (2–24 h) (Fig. 1). These wound-response (WR) proteins were involved in 133 biological process categories (Fig. 2). In total, 21 WR proteins with high e-values and bit scores were identified, which have active biological processes in relation to wound and defense mechanisms.GO analysis showed that 37 GO terms were assigned to these WR proteins, of which 12, including the jasmonic acid biosynthesis pathway, terpenoid backbone biosynthesis pathway, phenylalanine metabolism pathway, linolenic acid metabolism, galactose metabolism, amino sugar and nucleotide sugar metabolism, biosynthesis of secondary metabolites, cysteine and methionine metabolism, pentose phosphate pathway, and glutathione metabolism, were assigned to KEGG pathways (Table 1).
| Time-point (hours) | Protein name | Biological process | PID% | Bit score | EC number |
|---|---|---|---|---|---|
| a Indicates protein identified through the KEGG pathway, which is associated to agarwood formation (Siah et al., 2016). | |||||
| 2 | (1) Brassinosteroid receptor | Detection of brassinosteroid stimulus (GO:0009729) | 70 | 32.1 | 2.7.11.25a |
| (2) UDP-d-xylose 4-epimerase | Defense response by callose deposition (GO:0052542) | 84.62 | 29.5 | 5.1.3.2 | |
| Defense response to oomycetes (GO:0052542) | |||||
| (3) Cinnamyl alcohol dehydrogenase | Regulation of plant-type hypersensitive response (GO:0010363) | 56 | 34.7 | 1.1.1.255 | |
| Salicylic acid mediated signaling pathway (GO:0009862) | |||||
| Para-aminobenzoic acid metabolism (GO:0046482) | |||||
| (4) Caffeoyl-3-o-methyltransferase | Polyamine catabolism (GO:0006598) | 85 | 38.9 | 2.1.1.104a | |
| (5) Unknown protein | Jasmonic acid biosynthesis (GO:0009695) | 66.67 | 30.1 | 1.13.11.12a | |
| (6) UDP-glucose 4-epimerase | Galactose biosynthesis (GO:0046369) | 5.1.3.5 | |||
| 6 | (7) Heat shock protein 70 | Response to high light intensity (GO:0009644) | 85.71 | 31.3 | |
| Response to cadmium ion (GO:0046686) | |||||
| Response to virus (GO:0009615) | |||||
| Response to endoplasmic reticulum stress (GO:0034976) | |||||
| (8) Green ripe-like 1 | Response to ethylene (GO:0009723) | 80 | 42.3 | ||
| (9) Protein transport protein | Response to salt stress (GO:0009651) | 70.59 | 32.7 | ||
| (10) Protein transport protein sec24-like at4g32640-like | Response to oxidative stress (GO:0006979) | 70.59 | 32.7 | ||
| (11) Calnexin 1 | Jasmonic acid mediated signaling pathway (GO:0009867) | 100 | 41 | ||
| (12) Malate dehydrogenase | Response to cold (GO:0009409) | 95.65 | 51 | 1.1.1.37a | |
| (13) Glyoxysomal malate dehydrogenase | Malate metabolism (GO:0080093) | 85.71 | 47.6 | ||
| Regulation of photorespiration (GO:0080093) | |||||
| 12 | (14) Heat shock protein | Response to cadmium ion (GO:0046686) | 81.48 | 42.4 | |
| Response to virus (GO:0009615) | |||||
| Response to cold (GO:0009409) | |||||
| Response to salt stress (GO:0009651) | |||||
| (15) Cysteine synthase | Photosynthetic acclimation (GO:0009643) | 100 | 31.2 | 2.5.1.47 | |
| (16) Cytosolic phosphoglucomutase | Trehalose biosynthesis (GO:0005992) | 92.31 | 30.6 | 5.4.2.2 | |
| Galactose catabolism (GO:0019388) | |||||
| Starch biosynthesis (GO:0019252) | |||||
| 24 | (17) 6-Phosphogluconate dehydrogenase | Response to cadmium ion (GO:0046686) | 100 | 39.7 | 1.1.1.44 |
| Response to abscisic acid (GO:0009737) | |||||
| Response to fructose (GO:0009750) | |||||
| (18) Cysteine-rich receptor-like protein kinase | Response to chitin (GO:0010200) | 72.73 | 32.7 | ||
| Defense response to bacterium (GO:0042742) | |||||
| (19) Cysteine-rich receptor-like protein kinase | Activation of MAPKK activity (GO:0000186) | 72.73 | 39.7 | 2.7.11.25a | |
| MAPK cascade (GO:0000165) | |||||
| (20) Protein phosphatase | Protein dephosphorylation (GO:0006470) | 82.35 | 34.7 | ||
| (21) Receptor-like serine threonine protein kinase | Protein autophosphorylation (GO:0046777) | 51.85 | 37.4 | ||
Hierarchical treemap and interactive graph analysis
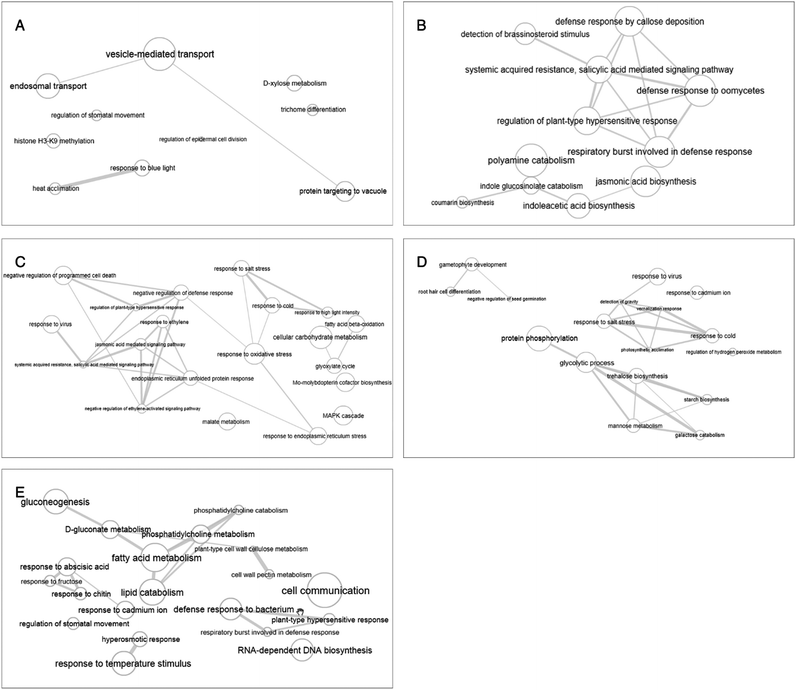
The enrichment of the biological processes in this study was performed at all time-points, thereby providing an aggregate picture of the TPS proteins interactome. Using all 518 TPS proteins identified in this study, a draft map of biological processes enriched in the signaling and wound response is presented in Fig. 2. In this study, the most enriched biological processes are shown as larger components/boxes at each time-point and were found displaying different primary biological processes: “protein targeting to vacuole” at 0 h (Fig. 2A, green box) is a common biological process, which is important in the maintenance of cell organization; “detection of brassinosteroid stimulus” at 2 h (Fig. 2B, gold box) is one of the plant hormones that plays an important role in signaling response; “response to high light intensity” at 6 h (Fig. 2C, pink box) is a biological process, which monitors changes in the activity or state of a cell; “response to cadmium ion” at 12 h (Fig. 2D, pink box) is a biological process that responds to over exposure of toxic metals; and “response to cadmium ion” at 24 h (Fig. 2E, purple box) is a continued response to the over exposure of toxic metals. While the treemap only indicates the biological processes and the GO terms involved at each time-point, an interactive graph was constructed to show the relationship and functional network between each GO term.To reduce the redundancy of functional categories between enriched GO terms, a semantic similarity approach was used. The gene ontology interactive graph-based network encompasses functional homology between genes present at specific time-points (Fig. 4). At 0 h, the network comprised 10 nodes and 3 edges, whereas the remaining time-points (2 h, 6 h, 12 h, and 24 h) showed more than 10 nodes and 3 edges. The differences between 0 h and 2 h showed the presence of increased interaction after wounding treatment. The network interaction at 2 h showed that the detection of brassinosteroid stimulus (GO:0009729) was correlated with five nodes with GO terms related to signaling response after wounding treatment (GO:0052542, GO:0009862, GO:0010363 and GO:0002679). The GO:0009729 category includes a large number of genes that play a role in signaling response to wounding at 2 h (Fig. 4B). The network interactions in Fig. 4C (17 nodes and 24 edges), 4D (14 nodes and 20 edges), and 4E (24 nodes and 22 edges) were more complex and they showed that most of the GO terms are referred as defense responses.
Discussion
Studies on plant defense systems, specifically on early wound-response proteins, are essential to provide valuable information on agarwood formation in Aquilaria trees. Based on general observations from previous studies, agarwood is never produced in sound trees, and agarwood formation is correlated with tissue wounding and invasion by biotic agents.25 Mechanical wounding could induce early response in WR genes during the first 3 h.26 In fact, mechanical wounding has been acknowledged as being the primary cause for the formation of agarwood.27 Damage by mechanical wounding causes the A. malaccensis tree to weaken and become vulnerable to threats by biotic and abiotic agents. Known to be a component in the Aquilaria plant defense system, agarwood acts to close the wound and protect the tree from further pathogen or fungal invasion.14,28 In this study, protein changes in A. malaccensis upon wounding treatment and the underlying process, complement and validate the existing knowledge on agarwood formation. Additionally, we applied PCA to serve as an exploratory data analysis method in identifying proteins that are highly relevant to the treatment and also to substantiate that they were not randomly picked. As such 21 WR proteins, such as the brassinosteroid receptor, cinnamyl alcohol dehydrogenase, caffeoyl-3-o-methyltransferase, heat shock protein 70, calnexin 1, malate dehydrogenase, glyoxysomal malate dehydrogenase, heat shock protein, cysteine synthase, and 6-phosphogluconate dehydrogenase (Table 1), were selected for KEGG and REViGO analyses. Although a handful of agarwood formation-related genes from Aquilaria have been identified through transcriptomic analysis,13 the findings from this study are deemed valuable as this is the first report on the identification of WR protein expression in A. malaccensis within 24 h of mechanical wounding through a proteomics approach.Proteins responsive to wounding signals
In the early hours after mechanical wounding, a total of 564 TPS proteins and several biological processes involved in countering plant stress were identified in A. malaccensis. Wounding stress undergone by A. malaccensis activated brassinosteroid stimulus and polyamine catabolism at 2 h (Fig. 2B). The former includes a series of events in cells receiving the brassinosteroid stimulus, and further converting it to a molecular signal, whereas the latter involves chemical reactions and pathways that result in the breakdown of polyamines or any organic compound containing two or more amino groups.29 As a class of essential plant hormones, brassinosteroids monitor a wide range of plant growth and development processes. They interact with the signaling components of other phytohormones, such as cytokinins, auxins, gibberellins, jasmonic acid, salicylic acid, and polyamine, at several levels.30 Both jasmonic acid and salicylic acid have been reported as crucial signal transducers in Aquilaria agarwood formation.31 These phytohormones are also recognized to play an important role in regulating the defensive signaling network and are involved in agarwood sesquiterpene production.28,32 Jasmonic acid and salicylic acid are known to help plants such as potato, tomato, and bean to counter heat stresses.32,33 The presence of jasmonic acid in Aquilaria sinensis has been shown to cause upregulation of agarwood sesquiterpene genes in the tree as early as 30 min post-wounding treatment,32 thus complementing our study. In Aquilaria, wounding triggers formation of the main compounds in agarwood, specifically sesquiterpenoids via two pathways, the methylerythritol phosphate pathway (MEP) and mevalonic acid pathway (MEV).34 Although there is no specific study on brassinosteroid stimulus in Aquilaria, however, its interaction with other phytohormones such as jasmonic acid has been associated with agarwood formation.27,30 Based on the KEGG pathway analysis, a protein with potential role in linolenic acid metabolism has been identified. Linolenic acid is the precursor for lipoxygenase (LOX), which initiates the lipoxygenase and octadecanoate pathways responsible for the formation of jasmonic acid.33 Polyamine catabolism has also been proven essential for development and growth in all living organisms35 and is associated with plant defense.36 Based on the filtering parameters for GO analysis, six proteins with significant protein identities were identified at 2 h (Table 1). Three of the identified proteins matched those previously reported in KEGG pathways,14 namely, the terpenoid backbone biosynthesis and phenylalanine metabolic pathways, which are involved in the synthesis of sesquiterpenes,32,37 and the linolenic acid metabolic pathway, which is related to aroma production and jasmonate formation.38Activation of plant defense systems
At 6 h post-wounding, signaling responses due to abiotic stresses were detected. Mechanical stress applied on the tree can induce abiotic and biotic stresses that can disrupt its biological processes at this time-point.39 Proteins identified at this time-point included signaling proteins toward irregular nutrient uptake, microorganisms, and unfavorable surroundings. Malate metabolism was present at 6 h after wounding treatment and is believed responsible toward the activation of carbohydrates metabolism (Fig. 2C). In Aquilaria, carbohydrate metabolism contributes to the production of agarwood resin as it was reported that brownish droplets consisting of phenolic compound substances were observed after the disappearance of starch grains in parenchyma cells.27 The event of agarwood formation is further reinforced upon the detection of calnexin 1 protein (Table 1), which is involved in the jasmonic acid-mediated signaling pathway, displaying a high possibility that agarwood formation transducing signals were sent off during this time point, which in return may trigger plant defense mechanisms to produce agarwood resin. Based on the REViGO analysis, all identified GO terms assigned to the 21 WR proteins were responsive to wounding, for example, brassinosteroid stimulus (GO:0009729) and polyamine catabolism (GO:0006598) (involved in the wound signaling response), defense response to oomycetes (GO:0052542), galactose biosynthesis (GO:0046369), response to cadmium ion (GO:0046686), response to virus (GO:0009615), response to salt stress (GO:0009651), response to oxidative stress (GO:0006979), jasmonic acid mediated signaling (GO:0009867), response to cold (GO:0009409), malate metabolism (GO:0080093), defense response to bacteria (GO:0042742), and activation of MAPKK activity (GO:0000186), whereby they are all associated with the plant defense systems.Accumulation of reactive oxygen species (ROS)
Heat shock protein 70 in A. malaccensis was expressed consistently from 6 h to 12 h, which is detected in the response to cadmium ions, viruses, cold, and salt stress. Based on treemap visualization (Fig. 2D), the presence of responses to cadmium ions, viruses, cold, and salt stress is highly over-represented/more significant compared to others. Cadmium ions are indirectly connected to the generation of reactive oxygen species (ROS; especially H2O2) and jasmonate,11,40 which both of them were reportedly involved in agarwood production.13,41 By highlighting the protein response to cadmium ions, this signal is transmitted by polyamines and plants hormones such as jasmonic acid, salicylic, ethylene, and abscisic acid.42,43 Within the cytoplasm, response to this heavy metal is at least in part, mediated by mitogen-activated protein kinases (MAPKs), which are stimulated by cadmium ions at transcriptional and post-translation levels (Fig. 2C).41,44 The MAPK response is recorded as early as 6 h post-wounding treatment and may thus prove the presence of oxidative stress like ROS, which is responsible for cell death, and this oxidative stress actually activates the MAPK signaling pathway. As physical wounding is believed to induce ROS accumulation in Aquilaria trees, cell death has been observed during agarwood formation.31 Hydrogen peroxide and superoxide involved in hydrogen peroxide metabolism (Fig. 2D) emerged as two key ROS that promote programmed cell death and salicylic acid accumulation upon agarwood formation.31 Cell death in Aquilaria is often accompanied by vacuolar collapse,31 which explains the presence of a lytic vacuole organization process that also involves cell wall organization and microtubule nucleation (Fig. 2D).Cell recovery and regeneration
Wood discoloration was reported to occur around the wounded region on A. malaccensis trees, possibly due to localized chemical accumulation (ROS and PCD) in the injured wood.5,41 Therefore, wounding treatment is probably the first and main trigger in the activation of agarwood-related metabolism, followed by microbial attack on the exposed wood, which further elevates gene expression leading to continuous agarwood induction.5 The biological response to chitin and defense response to bacterium were observed at 24 h post-wounding treatment, which explains the presence of microbial activities in A. malaccensis. The 6-phosphogluconate dehydrogenase protein identified at 24 h after wounding treatment is believed to be one of the contributing proteins responsible for wood discoloration in Aquilaria. During agarwood formation, 6-phosphogluconate dehydrogenase is involved in converting starch to glucose and results in the presence of a brownish color after the disappearance of starch grains.7 Additionally, fructose is produced to create a hard barrier along the wounded area, probably to prevent microorganism invasion.45 In response to the continuous and persistent pressure in abiotic stress, MAPKK activities are activated, indicating the tree to be undergoing cell recovery and regeneration processes.Conclusions
We demonstrated that wounding is a trigger to Aquilaria defense response. Although we could only verify a few WR proteins in this study, there is a possibility that the remaining identified proteins may also be related to wound responses. Limited availability of useful databases may hinder the identification of proteins in A. malaccensis. Nevertheless, our findings undoubtedly provide a valuable reference for future studies on agarwood proteomes to better understand agarwood formation. A sound understanding of the process will favor the development of effective agarwood induction techniques, and the selection of a high-quality breed.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work received financial support from the Ministry of Science, Technology and Innovation of Malaysia (Science fund Project No. 02-01-04-SF2102). The authors would like to acknowledge Zulfawwaz Mohmad from Halal Products Research Institute, Universiti Putra Malaysia, for assisting in PCA.References
- UNEP-WCMC (Comps.), The Checklist of CITES Species Website, CITES Secretariat, Geneva, Switzerland, UNEP-WCMC, Cambridge, UK, http://checklist.cites.org, accessed July 17, 2017 Search PubMed.
- The IUCN Red List of Threatened Species, version 2017-2, accessed November 11, 2017, www.iucnredlist.org.
- P. D. Azren, S. Y. Lee, D. Emang and R. Mohamed, History and perspectives of induction technology for agarwood production from cultivated Aquilaria in Asia: a review, J. For. Res., 2018, 1–11 Search PubMed.
- R. Mohamed, P. L. Jong and M. S. Zali, Fungal diversity in wounded stems of Aquilaria malaccensis, Fungal Divers., 2010, 43(1), 67–74 CrossRef.
- R. Mohamed, P. L. Jong and I. N. Irdayu, Succession patterns of fungi associated to wound induced agarwood in wild Aquilaria malaccensis revealed from quantitative PCR assay, World J. Microbiol. Biotechnol., 2014, 30(9), 2427–2436 CrossRef CAS PubMed.
- T. Nobuchi and S. A. Siripatanadilok, The formation of wood in tropical forest trees, in Cytological observations of Aquilaria crassna wood associated with the formation of aloeswood, ed T. Nobuchi and S. Mohd Hamami, UPM Press, Serdang, 2008, pp. 147–160 Search PubMed.
- R. Mohamed, M. T. Wong and R. Halis, Pertanika J. Trop. Agric. Sci., 2013, 36(1), 43–50 Search PubMed.
- R. Naef, The volatile and semi-volatile constituents of agarwood, the infected heartwood of Aquilaria species: a review, Flavour Fragrance J., 2011, 26(2), 73–87 CrossRef CAS.
- H. F. Dai, J. Liu, Y. B. Zeng, Z. Han, H. Wang and W. L. Mei, A new 2-(2-phenylethyl) chromone from Chinese eaglewood, Molecules, 2009, 14(12), 5165–5168 CrossRef CAS PubMed.
- A. S. Edison, R. D. Hall, C. Junot, P. D. Karp, I. J. Kurland, R. Mistrik, L. K. Reed, K. Saito, R. M. Salek, C. Steinbeck and L. W. Sumner, The time is right to focus on model organism metabolomes, Metabolites, 2016, 6(1), 8 CrossRef PubMed.
- J. Zou, A. Liu, X. Chen, X. Zhou, G. Gao, W. Wang and X. Zhang, Expression analysis of nine rice heat shock protein genes under abiotic stresses and ABA treatment, J. Plant Physiol., 2009, 166(8), 851–861 CrossRef CAS PubMed.
- S. Franz, B. Ehlert, A. Liese, J. Kurth, A. C. Cazalé and T. Romeis, Calcium-dependent protein kinase CPK21 functions in abiotic stress response in Arabidopsis thaliana, Mol. Plant, 2011, 4(1), 83–96 CrossRef CAS PubMed.
- Y. Xu, Z. Zhang, M. Wang, J. Wei, H. Chen, Z. Gao, C. Sui, H. Luo, X. Zhang, Y. Yang and H. Meng, Identification of genes related to agarwood formation: transcriptome analysis of healthy and wounded tissues of Aquilaria sinensis, BMC Genomics, 2013, 14(1), 227 CrossRef CAS PubMed.
- C. H. Siah, P. Namasivayam and R. Mohamed, Transcriptome reveals senescing callus tissue of Aquilaria malaccensis, an endangered tropical tree, triggers similar response as wounding with respect to terpenoid biosynthesis, Tree Genet. Genomes, 2016, 12(2), 33 CrossRef.
- X. Wang, B. Gao, X. Liu, X. Dong, Z. Zhang, H. Fan, L. Zhang, J. Wang, S. Shi and P. Tu., Salinity stress induces the production of 2-(2-phenylethyl) chromones and regulates novel classes of responsive genes involved in signal transduction in Aquilaria sinensis calli, BMC Plant Biol., 2016, 16(1), 119 CrossRef PubMed.
- G. Chen, T. C. Kuo, M. Yang, T. Chien, M. Chu, L. Huang, C. Chen, H. Lo, S. Jeng and O. L. Chen, Identification of cucurbitacins and assembly of a draft genome for Aquilaria agallocha, BMC Genomics, 2014, 15(1), 578 CrossRef PubMed.
- H. Chen, B. C. McCaig, M. Melotto, S. Y. He and G. A. Howe, Regulation of plant arginase by wounding, jasmonate, and the phytotoxin coronatine, J. Biol. Chem., 2004, 279(44), 45998–46007 CrossRef CAS PubMed.
- J. Zou, A. Liu, X. Chen, X. Zhou, G. Gao, W. Wang and X. Zhang, Expression analysis of nine rice heat shock protein genes under abiotic stresses and ABA treatment, J. Plant Physiol., 2009, 166(8), 851–861 CrossRef CAS PubMed.
- R. Bari and J. D. Jones, Role of plant hormones in plant defence responses, Plant Mol. Biol., 2009, 69(4), 473–488 CrossRef CAS PubMed.
- S. Y. Lee, S. A. Syazwan, D. U. Lamasudin and R. Mohamed, Differentially Expressed Woundresponse-related Proteins from a Major Agarwood-producing Tree, Aquilaria malaccensis Lam. Identified via 2-D Electrophoresis, Curr. Proteomics, 2018, 15(4), 291–298 CrossRef CAS.
- W. Ye, W. Zhang, T. Liu, M. Zhu, S. Li, H. Li, Z. Huang and X. Gao, iTRAQ-Based Quantitative Proteomic Analysis of Chemically Induced Aquilaria sinensis Provides Insights into Agarwood Formation Mechanism, Proteomics, 2018, 18(20), 1800023 CrossRef PubMed.
- A. B. Azzarina, R. Mohamed, S. Y. Lee and M. Nazre, Temporal and spatial expression of terpene synthase genes associated with agarwood formation in Aquilaria malaccensis, N. Z. J. For. Sci., 2016, 46(1), 1–13 CrossRef.
- A. Conesa, S. Götz, J. M. García-Gómez, J. Terol, M. Talón and M. Robles, Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research, Bioinformatics, 2005, 21(18), 3674–3676 CrossRef CAS PubMed.
- F. Supek, M. Bošnjak, N. Škunca and T. Šmuc, REViGO summarizes and visualizes long lists of gene ontology terms, PLoS One, 2011, 6(7), e21800 CrossRef CAS PubMed.
- R. Mohamed, P. L. Jong and A. K. Kamziah, Fungal inoculation induces agarwood in young Aquilaria malaccensis trees in the nursery, J. For. Res., 2014, 25(1), 201–204 CrossRef CAS.
- M. T. Wong, C. H. Siah, Q. Z. Faridah and R. Mohamed, Characterization of wound responsive genes in Aquilaria malaccensis, J. Plant Biochem. Biotechnol., 2013, 22(2), 168–175 CrossRef CAS.
- S. Rasool and R. Mohamed, Understanding agarwood formation and its challenges, in Agarwood, ed. R. Mohamed, Springer, Singapore, 2016, pp. 39–56 Search PubMed.
- J. Zhao, L. C. Davis and R. Verpoorte, Elicitor signal transduction leading to production of plant secondary metabolites, Biotechnol. Adv., 2005, 23(4), 283–333 CrossRef CAS PubMed.
- K. Gupta, A. Dey and B. Gupta, Plant polyamines in abiotic stress responses, Acta Physiol. Plant., 2013, 35(7), 2015–2036 CrossRef CAS.
- S. Saini, I. Sharma and P. K. Pati, Versatile roles of brassinosteroid in plants in the context of its homoeostasis, signaling and crosstalks, Front. Plant Sci., 2015, 6, 90 Search PubMed.
- Y. Okudera and M. Ito, Production of agarwood fragrant constituents in Aquilaria calli and cell suspension cultures, Plant Biotechnol. J., 2009, 26(3), 307–315 CrossRef CAS.
- Y. H. Xu, Y. C. Liao, Z. Zhang, J. Liu, P. W. Sun, Z. H. Gao, C. Sui and J. H. Wei, Jasmonic acid is a crucial signal transducer in heat shock induced sesquiterpene formation in Aquilaria sinensis, Sci. Rep., 2016, 6, 21843 CrossRef CAS PubMed.
- E. Horváth, G. Szalai and T. Janda, Induction of abiotic stress tolerance by salicylic acid signaling, J. Plant Growth Regul., 2007, 26(3), 290–300 CrossRef.
- F. Chen, D. Tholl, J. Bohlmann and E. Pichersky, The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom, Plant J., 2011, 66(1), 212–229 CrossRef CAS PubMed.
- A. Scala, S. Allmann, R. Mirabella, M. A. Haring and R. C. Schuurink, Green leaf volatiles: a plant's multifunctional weapon against herbivores and pathogens, Int. J. Mol. Sci., 2013, 14(9), 17781–17811 CrossRef CAS PubMed.
- S. S. Cohen, Guide to the Polyamines, Oxford University Press, 1998 Search PubMed.
- W. Ye, H. Wu, X. He, L. Wang, W. Zhang, H. Li, Y. Fan, G. Tan, T. Liu and X. Gao, Transcriptome sequencing of chemically induced Aquilaria sinensis to identify genes related to agarwood formation, PLoS One, 2016, 11(5), e0155505 CrossRef PubMed.
- Y. Kumeta and M. Ito, Characterization of α-humulene synthases responsible for the production of sesquiterpenes induced by methyl jasmonate in Aquilaria, J. Nat. Med., 2016, 70(3), 452–459 CrossRef CAS PubMed.
- J. W. Walley, S. Coughlan, M. E. Hudson, M. F. Covington, R. Kaspi, G. Banu, S. L. Harmer and K. Dehesh, Mechanical stress induces biotic and abiotic stress responses via a novel cis-element, PLoS Genet., 2007, 3(10), 1800–1812 CrossRef CAS PubMed.
- M. Kumar, A. J. Bijo, R. S. Baghel, C. R. K. Reddy and B. Jha, Selenium and spermine alleviate cadmium induced toxicity in the red seaweed Gracilaria dura by regulating antioxidants and DNA methylation, Plant Physiol. Biochem., 2012, 51, 129–138 CrossRef CAS PubMed.
- J. Liu, Y. Xu, Z. Zhang and J. Wei, Hydrogen peroxide promotes programmed cell death and salicylic acid accumulation during the induced production of sesquiterpenes in cultured cell suspensions of Aquilaria sinensis, Funct. Plant Biol., 2015, 42(4), 337–346 CrossRef CAS.
- A. K. Sinha, M. Jaggi, B. Raghuram and N. Tuteja, Mitogen-activated protein kinase signaling in plants under abiotic stress, Plant Signaling Behav., 2011, 6(2), 196–203 CrossRef CAS PubMed.
- J. Chmielowska-Bąk, J. Gzyl, R. Rucińska-Sobkowiak, M. Arasimowicz-Jelonek and J. Deckert, The new insights into cadmium sensing, Front. Plant Sci., 2014, 5, 245 Search PubMed.
- D. Marino, C. Dunand, A. Puppo and N. Pauly, A burst of plant NADPH oxidases, Trends Plant Sci., 2012, 17(1), 9–15 CrossRef CAS PubMed.
- I. Morkunas and L. Ratajczak, The role of sugar signaling in plant defense responses against fungal pathogens, Acta Physiol. Plant., 2014, 36(7), 1607–1619 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra10616a |
| This journal is © The Royal Society of Chemistry 2019 |