Open Access Article
Open Access ArticleUltrasensitive strips for the quadruple detection of nitrofuran metabolite residues†
Wei Wu a,
Ning Sun
a,
Ning Sun b,
Simin Zhuc,
Xiaomei Lib,
Jitao Chenb,
Chunzhao Liud,
Zhiyuan Fang*b and
Qingli Yang*a
b,
Simin Zhuc,
Xiaomei Lib,
Jitao Chenb,
Chunzhao Liud,
Zhiyuan Fang*b and
Qingli Yang*a
aCollege of Food Science and Engineering, Qingdao Agricultural University, No. 700 Changcheng Road, Qingdao, 266179, China. E-mail: rice407@163.com
bThe Fifth Affiliated Hospital of Guangzhou Medical University, No. 78, Hengzhigang Road, Guangzhou 510095, Guangdong Province, China. E-mail: fangjnu@126.com
cDepartment of Ophthalmology, Affiliated Hospital of Qingdao University, No. 1677 Wutaishan Road, Qingdao, China
dState Key Laboratory of Biochemical Engineering, Institute of Process Engineering, Qingdao University, Qingdao 266003, China
First published on 21st January 2019
Abstract
In this biosensor system, metabolite residues were derived by using a previous B-CBA synthesis method to label a biotin moiety for enrichment by streptavidin coated magnetic beads. Antibodies specific for derivatives were conjugated with carboxyl-modified barcode DNAs which were used as templates for strand displacement amplification (SDA). The assay can detect trace levels of 7.20 ppt of SEM, 11.58 ppt of AHD, 7.24 ppt of AOZ and 2.31 ppt of AMOZ, respectively.
Nitrofurans are broad-spectrum antibiotics used to treat animals with bacterial infections. Due to their mutagenicity and carcinogenicity,1–4 the use of these antibiotics has been banned in food-producing animals in European Union, the USA and China. The overuse of antibiotics in densely populated food-producing animals can cause high level of residues in the product and promote the emergence of new antibiotic-resistant bacteria, which pose a great threat to both animal and human health. Therefore, the routine monitoring of nitrofuran metabolites in food chain is the major way to ensure public health.
Nitrofuran antibiotics are metabolized rapidly in vivo with a very short half-life,5 making parent drug detection ineffective. Their metabolite residues are more stable and have been used as marker residues for the monitoring of nitrofuran abuse.6 Currently, the confirmative analytical methods for detection of these antibiotics are mainly based on chromatography-based methods.7–10 But expensive equipment and well trained operators are pre-requisite for accurate detection. Furthermore, the tedious derivatisation and solid-phase extraction are still required for the analysis of marker residues in the rapid methods, including enzyme-linked immunosorbent based assay,11–14 lateral flow immunoassay,15 impedimetric immunoassay16 and lateral flow biosensor assay.17,18 The organic extraction solvent can be involved in the hybridization, which may interfere with the accuracy and reproducibility of the assay. In a previous study, we synthesized a novel biotinylated aldehyde derivative (B-CBA) for the derivatisation of semicarbazide (SEM), which makes the detection of SEM free of extraction procedure.19 But the established assay doesn't have the multiplex ability to detect all four nitrofuran metabolite residues simultaneously. Furthermore, due to the complexity of the sample matrix and lack of maximum residue limits set, a highly sensitive and selective bioanalytical method is needed by regulatory agencies of many countries.
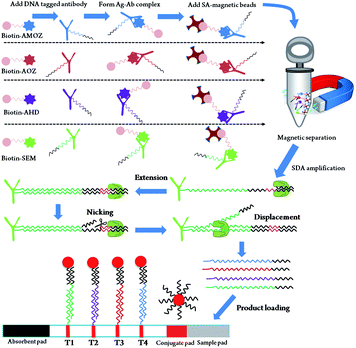
In this work, we report an isothermal strand displacement amplification (SDA) based method for ultrasensitive and quadruple detection of four nitrofuran metabolite residues. The abuse of furazolidone, nitrofurantoin, furaltadone, and nitrofurazone were analyzed by detecting 3-amino-2-oxazolidinone (AOZ), 1-aminohydantoin (AH), 3-amino-5-morpholinomethyl-1,3-oxazolidin-2-one (AMOZ), and semicarbazide (SEM), respectively. B-CBA synthesized in a previous work was used for the derivatisation,19 therefore each of the resulted residue derivatives was labeled with a biotin moiety (Fig. S1†). The metabolite residue derivatives were then captured by their specific DNA-tagged antibodies. The formed derivative–antibody complexes (De–Ab complexes) were further enriched by streptavidin coated magnetic beads (SAMB). Each De–Ab was tagged with barcode DNA of 10–20 nt, which was amplified by an isothermal strand displacement amplification. After amplification, the synthesized single strand DNA (ssDNA) was visually detected by a lateral flow biosensor. Four test lines that complementary to the four barcode DNA indicated the contamination of their respective nitrofuran antibiotics in sample (Scheme 1).
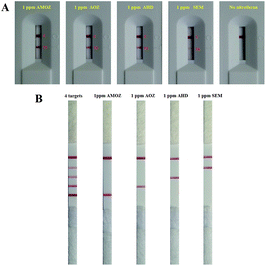
In order to verify the viability of designed methods for nitrofuran detection, we firstly performed a single test for the AOZ, AHD, AMOZ, and SEM, respectively. The designed nucleotides in Table S1† were used for AuNP coating and C-line/T-line (C/T line) preparation. The nitrofuran metabolisms of 1 ppm were derived by using biotin-CBA and formed biotin-CBA derivative. After SDA reaction, the product was subjected to LFB and the results were shown in Fig. 1A. The red T line was visual in the presence of nitrofuran metabolites and no positive T line was seen in the absence of target. These results indicated that the designed sequences were suitable for SDA reaction and LFB monitoring, which demonstrated that the designed method was usable in nitrofuran metabolite detection.
Based on the success of single test, we then challenged quadruplex detection of nitrofuran metabolites in a SDA reaction (Fig. 1B). Four test lines were observed in the presence of four targets. In case of only one type of nitrofuran metabolite, the products from multiplex SDA could result in one corresponding T line and no cross reaction was observed. These results suggested that the multiple SDA and quadruple LFB was workable in nitrofuran metabolite detection.
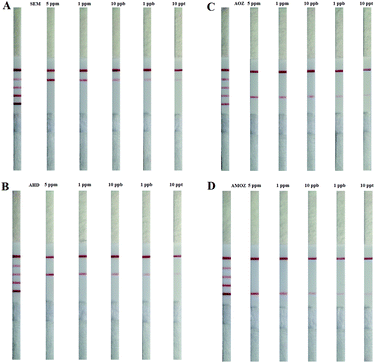
The sensitivity of LFB in nitrofuran metabolite detection was evaluated by serial dilution of different targets from 5 ppm to 10 ppt. 10 μL of the purified multiplex SDA product was diluted with 40 μL of PBS (pH 7.4) and pipetted onto the sample pad. The color of the test and control lines was visible to the naked eye after 5 min. Fig. 2A shows digital photographic images of LFB for the detection of SEM on multiplex SDA products, a simultaneous detection result of 4 targets as internal reference. With increasing SEM concentration, more De–Ab complexes were formed, and the color of the test lines increased proportionally. Furthermore, the color of the test lines was not visible to the naked eye below concentrations of 10 ppt. Therefore, 10 ppt were considered as the visual detection limit of the multi-SDA/LFB for the detection of SEM (Fig. 2A). The detection limit of the multi-SDA/LFB for the detection of AHD, AOZ, and AMOZ was all of 10 ppt (Fig. 2B–D).
Using a LFB, it was possible to achieve quantitative results by recording the optical intensity of the T-line, as gold nanoparticles had characteristic peaks. Quantitative analysis of was performed by monitoring the optical peak intensity of test lines. The optical intensity of the corresponding test lines was shown, and the intensity increased with increasing concentrations of targets. The resulting standard curve of optical intensity versus the logarithm of target concentration was displayed in Fig. 3. The linear curves have correlation coefficients of determination for SEM, AHD, AOZ and AMOZ of 0.977, 0.975, 0.984 and 0.980, respectively. The limit of detection was defined as the optical signals intensity was 3-fold higher (S/N > 3) than the average signal of negative control. At the concentrations of 7.20 ppt of SEM, 11.58 ppt of AHD, 7.24 ppt of AOZ and 2.31 ppt of AMOZ, the optical intensity of corresponding test line was 70.3, 68.2, 61.7 and 60.9, respectively, 3 folds higher than the negative control (23.4, 22.7, 20.6 and 20.3 for SEM, AHD, AOZ and AMOZ, respectively). Therefore, the detection limit of multi-SDA/LFB was 7.20 ppt of SEM, 11.58 ppt of AHD, 7.24 ppt of AOZ and 2.31 ppt of AMOZ.
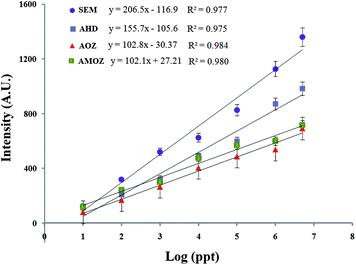 | ||
| Fig. 3 Standard curve of the assay. Optical densities are obtained from serially diluted standard solutions from 5 ppm to 10 ppt. | ||
Nitrofuran metabolites (AMOZ, AOZ, AHD and SEM) were added into meat samples from local market in Qingdao. Only meat samples that confirmed as negative for nitrofuran metabolites according to HPLC method were spiked. The samples were spiked with different concentrations of AMOZ, AOZ, AHD and SEM. As shown in Fig. S2,† the spiked nitrofuran metabolites can be detected in the meat homogenate by using our established LFB. The optical intensity of T-lines for 5 ppm (Fig. S2A†) was significantly stronger than those for 100 ppt (Fig. S2B†), which was similar to the results in pure PBS. Then, we calculated the recovery rate in food samples (Table S2†). The founded concentration was calculated by substituting the optical intensity of LFBs as y to the equation in Fig. 3. The recovery rate was equal to the ratio of founded target concentration and added target concentration. The recoveries of AMOZ, AOZ, AHD and SEM were 91.83–112.34%, 92.85–106.77%, 97.14–122.64%, and 95.35–104.76%, respectively. The LFB had a detection limit of AMOZ, AOZ, AHD and SEM in meat of 21.2, 23.8, 18.92 and 15.72 ppt, respectively. The detection limit of nitrofuran metabolites in food samples was slightly higher than that in a pure PBS solution. The result showed that this method was applicable for the detection of AMOZ, AOZ, AHD and SEM in meat samples.
In the present study, a SDA-based lateral flow strip composed of 4 test lines and 1 control line was developed for simultaneous and multiplex detection of nitrofurans metabolite residues (AOZ, AHD, AMOZ and SEM). Different from traditional methods, this assay does not capture and analyze target molecules directly. Nitrofurans metabolite residues were used as hooks that were able to capture DNA signals immobilized on their specific antibodies, which were further amplified by SDA. It renders this LFB with high sensitivity. To the best of our knowledge, the present assay has the best sensitivity in the analysis of nitrofurans metabolite residues.20–22 In the last decades, mainstream methods for nitrofuran metabolisms were HPLC, LC/MS, and LC-MS/MS, which were sensitive but time-consuming, expensive, and requirement of sophisticated equipment. Then, various rapid detection methods have been reported. Kun et al. developed an efficient fluorescent biosensor based on fluorescence quenching of pyoverdine by furazolidone to detect the AMOZ. This fluorescent biosensor owned outstanding specificity and sensitivity in furazolidone detection within a LOD of 0.5 μM.23 Besides, immunochromatographic assay (ICA) has been gaining increasing attention, which was similar to our LFB and the readout was using a paper strip. Shuqun Li et al. proposed an ICA for simple, sensitive, rapid, convenient and semi-quantitative detection of AMOZ in meat and feed samples on site. The intensity of red color in the test line is inversely related to the analyte concentration and the visual detection limit was found to be 10 ng mL−1. The performance of this assay was simple and convenient because the tedious and time-consuming derivatization step was avoided. The ICA detection was completed within 10 min.24 Wang et al. established multiplex ICA using gold nanoparticles for the rapid and simultaneous detection of four nitrofuran metabolites in fish samples. The multi-ICA detection was accomplished in less than 15 min by the naked eye. The cutoff values for the strip test were 0.5 ng mL−1 for AOZ and 0.75 ng mL−1 for AHD, SEM, and AMOZ.25 Compared to the previous methods, our proposed SDA-based lateral flow strip is more sensitive and easy-to-operate. The real samples analysis also demonstrated its high recovery rate, which was reliable in routine applications. This method was successfully used for the detection of the four nitrofuran metabolites in meat and can potentially be used in other types of food or targets. The newly developed SDA-based LFB represents a high-throughput and improved option for the rapid detection of these four analytes in food and will substantially advance research requiring on-site detection, allowing fast initial screening. The proposed assay can also be popularized to environmental monitoring or disease diagnosis, in case of multiple toxins or multi-SNP sites needed for determination.26 This can be used as an initial screening to indicate presence or absence of toxins or SNPs, which reduces the number of samples to be analyzed by confirmatory analytical methods.27 Furthermore, the signal amplification strategy in this study is inspiring toward its applications in other small molecule detection.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support was provided by National Science Foundation of China (81703228 and 31671933), Applied Basic Research Fund of Qingdao (No. 16-5-1-77-jch), National Science Foundation of Shandong Province (ZR2016CQ10) and Guangdong Science and Technology Foundation (2014B02021011).Notes and references
- F. W. Kari, J. E. Huff, J. Leininger, J. K. Haseman and S. L. Eustis, Food Chem. Toxicol., 1989, 27, 129–137 CrossRef CAS PubMed.
- Y. Hiraku, A. Sekine, H. Nabeshi, K. Midorikawa, M. Murata, Y. Kumagai and S. Kawanishi, Cancer Lett., 2004, 215, 141–150 CrossRef CAS PubMed.
- K. Takegawa, K. Mitsumori, K. Yasuhara, M. Moriyasu, M. Sakamori, H. Onodera, M. Hirose and T. Nomura, Toxicol. Pathol., 2000, 28, 649–655 CrossRef CAS.
- M. Kobierska-Szeliga and H. Czeczot, Acta Biochim. Pol., 1994, 41, 1–5 CAS.
- R. J. McCracken, W. J. Blanchflower, C. Rowan, M. A. McCoy and D. G. Kennedy, Analyst, 1995, 120, 2347–2351 RSC.
- K. M. Cooper, P. P. Mulder, J. A. van Rhijn, L. Kovacsics, R. J. McCracken, P. B. Young and D. G. Kennedy, Food Addit. Contam., 2005, 22, 406–414 CrossRef CAS PubMed.
- A. Leitner, P. Zollner and W. Lindner, J. Chromatogr. A, 2001, 939, 49–58 CrossRef CAS PubMed.
- S. Effkemann and F. Feldhusen, Anal. Bioanal. Chem., 2004, 378, 842–844 CrossRef CAS PubMed.
- P. Edder, S. Vargas, D. Ortelli and C. Corvi, Clin. Chem. Lab. Med., 2003, 41, 1608–1614 CAS.
- S. P. Khong, E. Gremaud, J. Richoz, T. Delatour, P. A. Guy, R. H. Stadler and P. Mottier, J. Agric. Food Chem., 2004, 52, 5309–5315 CrossRef CAS PubMed.
- S. Y. Sheu, Y. T. Tai, W. R. Li, Y. C. Lei, K. H. Hsieh, C. Y. Lin, C. C. Cheng, T. H. Chang and T. F. Kuo, Vet. Med. Sci., 2012, 74, 1439–1446 CrossRef CAS.
- Z. L. Xu, Y. D. Shen, Y. M. Sun, K. Campbell, Y. X. Tian, S. W. Zhang, H. T. Lei and Y. M. Jiang, Talanta, 2016, 103, 306–313 CrossRef PubMed.
- Y. C. Liu, W. Jiang, Y. J. Chen, Y. Xiao, J. L. Shi, Y. B. Qiao, H. J. Zhang, T. Li and Q. Wang, J. Immunol. Methods, 2013, 395, 29–36 CrossRef CAS PubMed.
- Z. L. Xu, Y. D. Shen, Y. M. Sun, K. Campbell, Y. X. Tian, S. W. Zhang, H. T. Lei and Y. M. Jiang, Talanta, 2013, 103, 306–313 CrossRef CAS PubMed.
- Y. Tang, X. Xu, X. Liu, X. Huang, Y. Chen, W. Wang and J. Xiang, J. Food Sci., 2011, 76, T138–T143 CrossRef CAS PubMed.
- G. Yang, W. Jin, L. Wu, Q. Wang, H. Shao, A. Qin, B. Yu, D. Li and B. Cai, Anal. Chim. Acta, 2011, 706, 120–127 CrossRef CAS PubMed.
- X. Lu, X. Liang, J. Dong, Z. Fang and L. Zeng, Anal. Bioanal. Chem., 2016, 408, 6703–6709 CrossRef CAS.
- W. Wu, Z. H. Jing, X. Q. Yu, Q. L. Yang, J. X. Sun, C. Z. Liu, W. J. Zhang, H. He and L. Zeng, Sens. Actuators, B, 2019, 111, 239–251 CAS.
- Z. Fang, B. Jiang, W. Wu, Z. Xiang, C. Ouyang, T. Huang, J. Chen and L. Zeng, Chem. Commun., 2013, 49, 6164–6166 RSC.
- Q. Wang, Y. Liu, M. Wang, Y. Chen and W. Jiang, Anal. Bioanal. Chem., 2018, 410, 223–233 CrossRef CAS PubMed.
- F. Aldeek, K. C. Hsieh, O. N. Ugochukwu, G. Gerard and W. Hammack, J. Agric. Food Chem., 2017, 58, 167–172 Search PubMed.
- M. S. Park, K. T. Kim and J. S. Kang, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2017, 1046, 172–176 CrossRef CAS PubMed.
- K. Yin, W. Zhang and L. Chen, Biosens. Bioelectron., 2014, 51, 90–96 CrossRef CAS PubMed.
- S. Li, J. Song, H. Yang, B. Cao, H. F. Chang and A. P. Deng, J. Sci. Food Agric., 2014, 94, 760–767 CrossRef CAS.
- Q. Wang, Y. C. Liu, M. Y. Wang, Y. J. Chen and W. Jiang, Anal. Bioanal. Chem., 2018, 410, 223–233 CrossRef CAS.
- Y. S. Lee, A. M. Weakley, C. C. Nieman, J. Malvick and G. C. Lanzaro, J. Visualized Exp., 2015, 96, 52385–52392 Search PubMed.
- L. Rodriguez, M. Fraga, A. Alfonso, D. Guillebault, L. Medlin and L. M. Botana, Environ. Toxicol. Chem., 2017, 36, 645–654 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details and supplementary data. See DOI: 10.1039/c8ra10589h |
| This journal is © The Royal Society of Chemistry 2019 |