Open Access Article
Open Access ArticleA cascade of a denitrification bioreactor and an aerobic biofilm reactor for heavy oil refinery wastewater treatment†
Honghong Dongad,
Xiaoyan Jiangb,
Shanshan Sun*a,
Li Fangb,
Wei Wanga,
Kai Cuia,
Tiantian Yaoa,
Heming Wanga,
Zhiyong Zhanga,
Ying Zhang d,
Zhongzhi Zhang
d,
Zhongzhi Zhang a and
Pengcheng Fuc
a and
Pengcheng Fuc
aState Key Laboratory of Heavy Oil Processing, China University of Petroleum, Beijing, 102249, P. R. China. E-mail: sssun33@163.com; Tel: +86-10-89734284
bChina National Petroleum Corporation Liaohe Petrochemical Company, Panjin 124000, P. R. China
cState Key Laboratory of Marine Resource Utilization in South China Sea, Hainan University, Hainan 570228, P. R. China
dCollege of Science, China University of Petroleum, Beijing, China
First published on 6th March 2019
Abstract
The performance of an efficient denitrification bioreactor–aerobic biofilm reactor cascade for heavy oil refinery wastewater treatment was investigated. Optimum operation parameters for denitrification were found as follows: (1) hydraulic retention time of 8 h; (2) C/NO3−–N molar ratio of 3.75 with acetate as the carbon source; (3) 20% (v/v) carrier filling ratio in the denitrification bioreactor. Under such optimal conditions, a volumetric removal of 0.82 kg N m−3 d−1 was obtained. As an alternative low-cost carbon source to acetate, secondary DAF effluent (COD/NO3−–N mass ratio of 5.4) was also detected and a stable activity of denitrification was achieved with adding 25% volume fraction of secondary DAF effluent. Effluent COD of the subsequent aerobic biofilm reactor further decreased satisfying the requirements of the current discharge standards. High-throughput sequencing results exhibited that Rhodocyclaceae and Comamonadaceae were the dominant denitrifiers in the denitrification reactor and Pseudomonas was the dominant microbe in the aerobic biofilm reactor.
1 Introduction
Heavy oil refinery wastewater comprises several kinds of aliphatic and aromatic petroleum hydrocarbons which are poorly soluble and biodegradable,1 resulting in a high concentration of total nitrogen (TN) in the effluent. Sewage with high TN may cause serious water pollution problems such as black-odor rivers, eutrophication, and red tides in coastal zones. With tightened regulations and discharge standards for wastewater, the need to treat oil refinery wastewater has increased significantly. The cyclic activated sludge technology (CAST) process was commonly used to remove TN in heavy oil refinery wastewater; however, the effluent from the CAST process still contains a high concentration of TN which is mainly composed of nitrate nitrogen.2 Current wastewater emission standards in China, i.e., GB 31570-2015 regulates that the TN should be less than 30 mg L−1. In addition, some provinces or local places require even more stringent standards for TN, i.e., Liaoning Province in Northeast China (DB21/1627-2008) regulates that the TN should be less than 15 mg L−1. To satisfy the current wastewater discharge regulations and standards, nitrate nitrogen must be properly removed from heavy oil refinery wastewater.Denitrification is a popular method because of its low cost, efficient removal rate, and specificity of denitrifier.3 The technologies of microbial immobilization have been widely applied to biological wastewater treatment for nitrate removal.4 Recently, the gel entrapment immobilization, which is regarded as the most common immobilization approach, has been used in municipal wastewater treatment.5 Most studies have focused on cell immobilization of pure denitrifying bacterial strains,4 but the degradation process of pollutants is usually completed by a microbial consortia rather than a single bacterial strain.4 However, few studies have investigated the immobilized cells by using mixed microbial culture from activated sludge.
A novel waterborne polyurethane (WPUR) has been successfully tested as immobilized material in our previous work.2 The WPUR was an ideal material for cell immobilization of single bacteria strain in wastewater treatment and showed excellent denitrification performance due to its mechanical and physical properties.6–8 However, it is still a challenge to immobilize activated sludge in such WPUR for heavy oil refinery wastewater treatment.
Nitrates was transformed to nitrogen gas by heterotrophic bacteria, with an organic carbon source as the electron donor.9 However, denitrification of heavy oil refinery wastewater needs an external organic carbon source, in order to achieve complete nitrate removal. Acetate is usually utilized as the additional carbon source to improve denitrification activity in organic carbon poverty wastewaters.10 Considering to the flammability, cost, and security of carbon sources for transportation, storage, and operation etc, the organic matter in raw wastewater is preferred as a carbon source for biological denitrification.11
In this study, a cascade of denitrification bioreactor combined aerobic biofilm reactor was used to process heavy oil refinery wastewater, with simultaneous removal of nitrate and COD. And the main differences between this research and our previous2 are summarized as follows: firstly, the types of treated wastewater are totally different. In this study, the treated wastewater was heavy oily refinery wastewater, which has been recognized as one type of the most refractory wastewaters because of its complicated chemical composition and low biodegradability. However, our previous treated wastewater was the acrylonitrile wastewater, which is another kind of industrial wastewater. Secondly, the applications of reactors are different. In our present research, a series of pilot-scale denitrification–aerobic reactors were tested. However, our previous paper research only employed one-type laboratory-scale denitrification bioreactor. Thirdly, the function of carbon sources, which was added in the reactor for denitrification, are different. In our present study, acetate and air flotation effluent were both used as carbon source for denitrification. But in our present study, acetate was only used as carbon source for denitrification. The last but not the least, the conclusions obtained in our study are different from those of our present research.
In the research, the efficient denitrification bioreactor was constructed with WPUR as a carrier to immobilize the activated sludge. The aerobic biofilm reactor followed the denitrification bioreactor was to further remove the residual COD. During the whole experiment, the effluent of the secondary dissolved air flotation (DAF) was tested as a low-cost carbon source comparing to acetate. The optimal hydraulic retention time (HRT), C/NO3−–N molar ratio with acetate as carbon source and the volume fraction (COD/NO3−–N mass ratio) with secondary DAF effluent as carbon source for denitrification were all studied. Furthermore, the Miseq pyrosequencing technology was employed to analyze the composition of the microbial community in the immobilized carrier and biofilm with low-cost secondary DAF effluent as carbon source comparing to acetate.
2 Materials and methods
2.1 Immobilized carriers
The seed-activated sludge was obtained from the Petro China Dalian Petrochemical Company, which is located in Liaoning Province of China. The detailed procedure of immobilization has been described by Dong et al.2 The mixed liquor volatile suspended solid (MLVSS) concentration of the sludge was 14.2 g L−1. Detailed protocols for immobilization were as follows: firstly, 1 L activated sludge, 600 mL WPUR (33.3%, w/v) solution, 60 g powdered activated carbon, and 60 g powdered iron oxide were mixed uniformly in a beaker. Then, polymerization was induced by adding 100 mL of 4% (w/v) potassium persulfate solution as an initiator and 100 mL of 1% (w/v) N,N,N,N′-tetramethylethylenediamine solution as catalyst. The mixture was allowed to stand for 5–10 min at 25–35 °C. The gel-immobilized activated sludge then formed a solid jelly-bean-shaped block of hydrosol.2.2 Wastewater samples
The actual wastewater samples were obtained from Liaohe Petrochemical Branch Company (Liaoning Province, China). The technological processes of Liaohe Petrochemical Branch Company include CAST and secondary DAF, with a full process flow (please see Fig. S1 in ESI†). The characteristic of original wastewater was summarized in Table S1.† The characteristic of CAST effluent is summarized in Table 1, which was used as the influent of the denitrification bioreactor. The effluent of the secondary DAF was used as a low-cost carbon source (Table 2) comparing to acetate.| Parameter | Concentration (mg L−1) |
|---|---|
| COD | 56 ± 3 |
| NH4+–N | 0.82 ± 0.04 |
| NO3−–N | 22.3 ± 1.2 |
| NO2−–N | 0.041 ± 0.002 |
| TN | 24.2 ± 1.1 |
| pH | 8.01 ± 0.2 |
| Parameter | Concentration (mg L−1) |
|---|---|
| COD | 400 ± 3 |
| NH4+–N | 34.3 ± 1.3 |
| NO3−–N | 3.78 ± 0.31 |
| NO2−–N | 0.052 ± 0.002 |
| TN | 37.5 ± 1.6 |
| Oil | 17.8 ± 2.1 |
| Volatile phenol | 21.2 ± 1.1 |
| Sulfide | 2.4 ± 0.1 |
| Volatile phenol | 27.3 ± 0.3 |
| pH | 8.07 ± 0.02 |
2.3 Experimental design
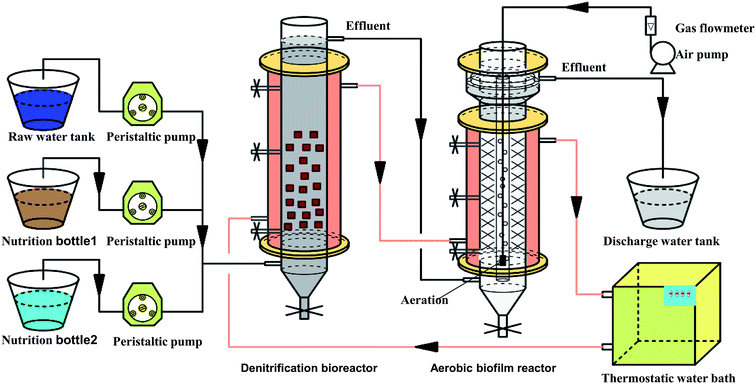
As shown in Fig. 1, a pilot-scale denitrification bioreactor and an aerobic biofilm reactor were connected in series for the treatment of CAST effluent. This experiment firstly last for 90 days with acetate added as electron donor and COD/NO3−–N ratio of 3.75, which was divided into 5 experimental stages (stages I–V) under different HRT conditions. From stages I to V, the HRT was changed from 24 h to 6 h. At the end of stage IV, samples were collected for high throughput analysis. Then, this experiment last for 118 days with DAF added as carbon source and HRT of 8 h, which was divided into 5 experimental stages (stages I–V) under different COD/NO3−–N conditions. From stages I to V, the COD/NO3−–N ratio was changed from 7 to 3.5. At the end of stage IV, samples were collected for further high throughput analysis. The detailed process flow was drawn in Fig. S2 of ESI.†The experiments were carried out in a cascade of plexiglass denitrification bioreactor and aerobic biofilm reactor (Fig. 1). The denitrification bioreactor contained 17 L in volume. The bioreactor inoculated with the immobilized carriers with 20% effective volume. Sodium acetate was employed as the carbon source to maintain C/NO3−–N molar ratios to 3.75 for 90 days denitrification testing. Then, DAF was added into the bioreactor as the carbon source with the volumetric fraction of DAF changed from 40%, 35%, 30%, and 25% to 20% corresponding to the COD/N = 7.0, 6.5, 6.0, 5.4, and 3.5 during denitrification of 118 days testing. The experimental temperature was set at 30 ± 0.5 °C. The pH value of wastewater was controlled at 7.6.
The aerobic biofilm reactor has an effective volume of 17 L. The structural features of aerobic biofilm reactor have been described by our team in 2016.3 Dissolved oxygen (DO) concentration was controlled at approximately 4 mg L−1. The pH value was 8.8, and the experiment temperature was set at 30 ± 0.5 °C.
2.4 Materials characterization
2.4.2.1 Sample pretreatment. The immobilized pellets samples are named as IP-S, where IP represents immobilized pellets, and S for sodium acetate as electron donor for the denitrification. Similarly, for IP-W, W is for DAF utilized as carbon source for the denitrification. Sample IP-0 was used as a control, and 0 is for without any electron donor from the denitrification process. The DNA extraction of immobilized particles were detailed described in our previous study.2
Biofilm samples were taken from the aerobic biofilm reactor. In biofilm samples B-S and B-W, B stands for biofilm, S stands for sodium acetate addition as carbon source, and W stands for DAF as carbon source. For DNA extraction of biofilm samples, this method was referenced as described previously.2
2.4.2.2 DNA extraction and polymerase chain reaction (PCR) amplification. The total genomic DNA from five samples (IP-0, IP-S, IP-W, B-S, and B-W) was extracted using the Fast DNA Spin Kit for Soil. The primers F: (5′-AGAGTTTGATCMTGGCTCAG-3′) and R: (5′-CTACGGGNGGCWGCAG-3′) were applied to target the 16S rRNA gene at V1–V2 regions.2 To multiplex the samples during the sequencing process, barcodes were added to the 5′ termini of the forward primers. The PCR volume and reactions were played as described previously.2 PCR was performed with a 25 μL reaction volume containing 12.5 μL of the Taq PCR Master Mix (Tiangen Biotech Co., Ltd., Beijing, China), 1 μL of each primer (10 μM), 20 ng template DNA, and 8.5 μL PCR grade water. The PCR program was detailed described in our previous study.2 The amplification program consisted of the following steps: (1) initial denaturation at 95 °C for 10 min, (2) six cycles of denaturation at 98 °C for 45 s, (3) annealing at 50 °C for 30 s per cycle, (4) extension at 72 °C for 30 s, followed by 30 cycles of denaturation at 98 °C for 45 s, 5. annealing at 68 °C for 30 s, (6) extension at 72 °C for 30 s, and 7. final elongation for 9 min. Each PCR reaction was run in triplicate. After PCR amplification, 5 μL of PCR products were mixed with the same volume of 1× loading buffer (containing SYBR green). Then, electrophoresis was conducted on 1% agarose gel to detect positive amplified bands. Each PCR reaction was run in triplicate. PCR products were purified with a gel extraction kit (MinElute Gel Extraction Kit, Qiagen, Germany). Nucleic acids were quantified with the NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, USA). The mixed PCR products were then sent to the Beijing Institute of Genomics, Chinese Academy of Sciences for sequencing conducted on a Miseq platform.
2.4.2.3 Sequence data processing. The method of FLASH, which was described by Magoc and Salzberg in 2011, was used for joining and quality-filtering 16S rRNA sequence paired-end data.13 The QIIME (version 1.9.1) was employed to analyze the sequence data.14 Chimeric sequence was analyzed by usearch 61.15 The RDP classifier was used for assigning all operational taxonomic units (OUTs) from taxonomy.16 The whole detailed sequence data processing was described in our previous findings.2
3 Results and discussion
3.1 Performance of cascade denitrification reactor–aerobic biofilm reactor with sodium acetate as carbon source
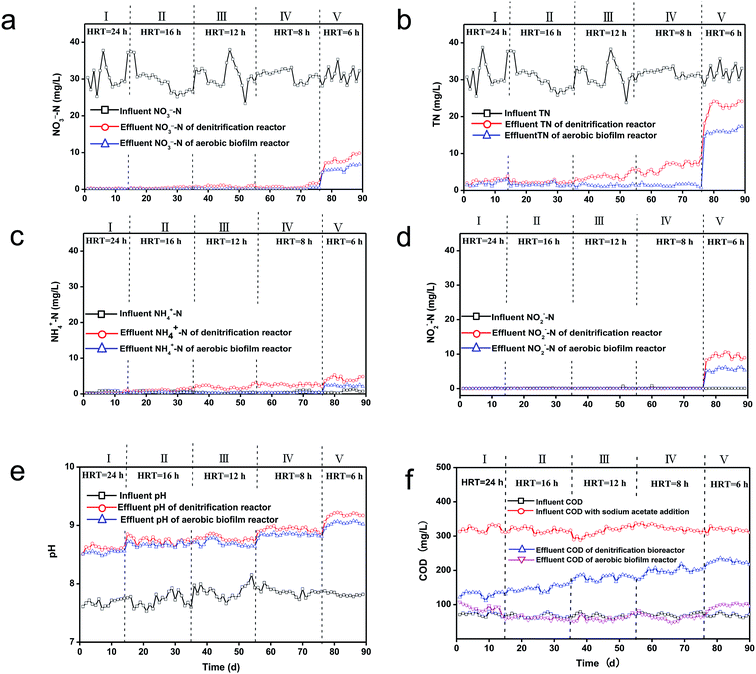
The performance of cascade denitrification reactor–aerobic biofilm reactor was investigated using a continuously flow operation mode (Fig. 2). As exhibited in Fig. S3,† without adding the carbon source of acetate, the average effluent COD was decreased from 65.8 mg L−1 to 48.21 mg L−1. However, the denitrification effect was low. The average effluent TN was decreased to 21.14 mg L−1, exceeding the wastewater standards of Liaoning Province of China (less than 15 mg L−1). Then with sodium acetate addition, the effect of different HRTs was examined and the C/NO3−–N molar ratio maintained a constant of 3.75. The average influent NO3−–N and COD concentrations were 30.5 and 73 mg L−1, respectively. With the reduction of wastewater HRT in the denitrification bioreactor from 24 h to 6 h from stages I to V, the molar ratio of C/NO3−–N was kept at 3.75.As shown in Fig. 2a and b, with HRT of 8 h in stage IV, the denitrification for both NO3−–N and TN achieved their highest value, i.e., 97.7% NO3−–N removal efficiency and residual 0.7 mg L−1 concentration of NO3−–N (Fig. 2a). The average concentration in effluent total nitrogen was 6.3 mg L−1 and the corresponding TN efficiency of removal was up to 81.5% (Fig. 2b), and the average volumetric TN rate of removal attained 0.82 kg N m−3 d−1. Furthermore, the average TN concentration in effluent decreased to 1.5 mg L−1 (Fig. 2b) after aerobic treatment, meeting the Liaoning Province (China) sewage discharge standards, i.e., TN < 15 mg L−1. In stage V, when HRT was decreased to 6 h, the average effluent NO3−–N and TN in cascade denitrification–aerobic biofilm reactor were increased to 5.7 mg L−1 and 16.1 mg L−1, respectively (Fig. 2a and b), exceeding the wastewater standards of Liaoning Province, China (less than 15 mg L−1).
As displayed in Fig. 2c, the effluent NH4+–N content remained steady during the experiment period, whereas a sudden increase was observed in residual nitrite (up to 9.4 mg L−1) at stage V. Meanwhile, the average NO2−–N concentration in effluent was reduced to 5.6 mg L−1 (Fig. 2d) after aerobic treatment.
Fig. 2e shows the effluent pH was increased in throughout the period of denitrification experiment. This may be due to the generation of OH−– in denitrification process and accumulated acetate in the wastewater.1 In the literature, an optimum pH of 7–8 for denitrification has been frequently proposed.17 In comparison, our study indicated that the effluent pH of the cascade denitrification–aerobic biofilm reactor was constantly above 8 form stage I to IV, but an effective denitrification performance was obtained. However, at stage V, the average effluent TN in cascade denitrification–aerobic biofilm reactor was increased in step to 16.1 mg L−1 (Fig. 2b), exceeding the wastewater discharge standards of TN. The average effluent pH was increased 9.2 at stage V.
Fig. 2f showed that the COD of raw influent to the treatment facility was relatively stable at 73 mg L−1. Its initial value at the inlet raised to 318 mg L−1 by adding the carbon of sodium acetate, but all reduced to 166 mg L−1 in the effluent of denitrification reactor. However, the effluent COD concentration of denitrification reactor was higher than that of the original wastewater. This consequence attributed to the production of biomass and amount of unconsumed acetate.2 The COD concentration would further decrease to 66 mg L−1 after treatment in the aerobic biofilm reactor, which was equivalent to the original influent without adding the carbon from stage I to IV. When HRT was further decreased to 6 h, the effluent COD from aerobic biofilm reactor was greater than that from the influent without adding the carbon. And this result was due to the increasing content of unconsumed acetate.9 The increase of COD could be induced by the erosion phenomenon on the biofilm surface because of the severe turbulence in liquid flow of aerobic biofilm reactor.18
3.2 Performance of cascade denitrification reactor–aerobic biofilm reactor with DAF as carbon source
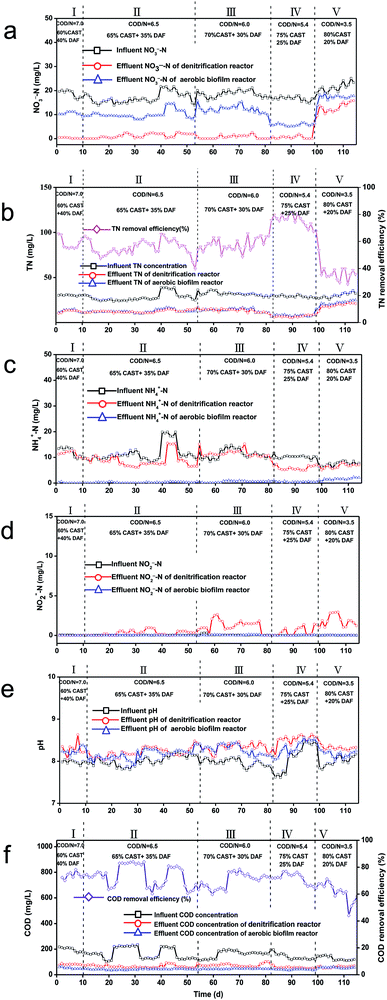
The performance of cascade denitrification reactor–aerobic biofilm reactor was further studied by employing a continuously flowing bioreactor system, where varying volumetric fractions of DAF (COD/N) was examined when the HRT maintained constant at 8 h. Fig. 3 shows that from stages I to V, the volumetric fraction of DAF added into the bioreactor was changed from 40%, 35%, 30%, and 25% to 20% corresponding to the COD/N = 7.0, 6.5, 6.0, 5.4, and 3.5. As shown in Fig. 3a and b, the average influent NO3−–N and TN concentrations were relatively stable at 18.1 mg L−1 and 29.7 mg L−1, respectively. When the COD/N was 5.4 (stage IV), the effects of denitrification for both TN and NO3−–N were the best. Almost complete denitrification was attained, i.e., at this stage, the removal efficiency of both effluent NO3−–N and NO3−–N concentration were up to 97.2% and of 0.5 mg L−1 (Fig. 3a), respectively. The effluent TN content was 7.7 mg L−1 in average and the corresponding TN removal rate was 76.3% (Fig. 3b). In addition, the volumetric denitrification rate in average came up to 0.66 kg N m−3 d−1. At the optimal stage IV, the effluent NO3−–N concentration of the aerobic biofilm reactor increased to 5.74 mg L−1 but the TN maintained at 6.8 mg L−1, meeting Liaoning Province (China) local sewage discharge standards, i.e., TN < 15 mg L−1.When the COD/N was further reduced to 3.5 (stage V), the denitrification efficiency decreased because of the lack of carbon source. The effluent NO3−–N and TN concentration increased to 13.2 and 21.9 mg L−1 (Fig. 2a and b). Meanwhile, the effluent NO3−–N content of the aerobic biofilm reactor was increased to 17.2 mg L−1, possibly attribute to the ammonia transform to nitrate in the aerobic biofilm reactor. The effluent TN content of aerobic biofilm reactor slightly decreased to 19.2, higher than the Liaoning Province's discharge standards.
Fig. 3c shows that the average NH4+–N concentrations in influent raised to 10.4 mg L−1 throughout the experiment period. This finding indicated that when DAF was used as carbon source in biological denitrification, it inevitably raised the proportion of ammonium nitrogen in the wastewater. NH4+–N content would decrease to 8.9 mg L−1 in the effluent of denitrification reactor and then further drop down to 0.5 mg L−1 after the treatment in aerobic biofilm reactor. This phenomenon was because of the NH4+–N reduction to NO3−–N by nitrifying bacteria.
Fig. 3d shows that the NO2−–N content of denitrification reactor raise up from trace level to over 2 mg L−1 at stage III. In comparison, the effluent NO2−–N content of aerobic biofilm reactor was stable throughout the experiment period. This result was achieved by the reduction of NO2−–N to NO3−–N by nitrifying bacteria.
As shown in Fig. 3e, the influent pH was 8.01. The effluent pH of the denitrification reactor raised to 8.30, which was higher than that of influent. This finding was in agreement with the literature that denitrification may lead to the increase of pH.2 However, the effluent pH from the aerobic biofilm reactor slightly decreased to 8.22. As shown in Fig. 4f, the influent COD was fluctuant from stages I to V with DAF addition. The average effluent COD was 40 mg L−1 for denitrification, whereas the average effluent COD of aerobic biofilm reactor was 54 mg L−1.
Joel et al.19 found that the volumetric denitrification rate was 0.17 kg NO3−–N per m3 per d with fixed-bed column reactor during sewage treatment from real ground water in their work. Researchers reported the maximum removal rate of volumetric denitrification was up to 0.209 kg N m−3 d−1 with an anaerobic continuous stirred tank reactor, which was derived from petroleum wastewater.20 In all the reported denitrification treatment methods, the utilization of free cells in a continuous reactor results in a decrease in biomass with each washout of the column reactor.21 However, immobilized cell system in our study have a major advantage of retaining a high cell concentration. The reactor capacity can be improved by increasing the biomass retention time rather than the liquid retention time using an immobilized cell system. The volumetric TN removal rate in our work came up to 0.82 kg N m−3 d−1 and it was greatly higher than the above reported conclusions. Moreover, the cascade denitrification aerobic biofilm reactor in our research, in addition to saving the carbon source cost, is simultaneous removal of nitrate and COD, which met wastewater discharge standard in China. The immobilization protocol and the approach to use lower cost wastewater as carbon source in this research can be further scaled up for wide scale application. Moreover, the operational parameters reported in this paper, for example, the COD/N mass ratio, and HRT could be checked up in a up-scaling bioreactor before applying in real industry.
3.3 The characterization of microbial communities in denitrification bioreactor and aerobic biofilm reactor
As displayed in Table 3, after sequence data processing, we found that there have 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 682–52
682–52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 effective reads in the DNA samples from microbial analysis results. Good's coverage ranged from 91.61% to 96.98%, indicating that the variety of microorganism from the bioreactors were well analyzed via the construction of libraries sequence. The index of Chao 1 displayed a decrease from activated sludge to IP-W, suggesting that the quantity of denitrifying bacterial species decreased on the surface of carrier immobilizing activated sludge. This result was in accordance with a previous research, which studied by employing a system of activated sludge, showing that the quantity of microbial species richness in digestion sludge was less than inoculating sludge.22 Compared with IP-S sample, IP-W sample showed comparative more denitrifying bacteria richness. In the sample of activated sludge, the highest diversity (Shannon index was 8.22) was detected and it was confirmed by associated Simpson index. These results suggested that a highly complex and highest diversity microbial communities was contained in activated sludge. Nevertheless, overall diversity decreased because the denitrifying microbial communities gradually became the dominant, as discovered in previous report.23 Shannon indices of IP-S and IP-W samples in the acclimation phase were 7.96 and 8.08, respectively, which were similar with Simpson index. These obtained results suggested that the difference about microbial diversity was not important. However, the B-W sample had a higher diversity than that of the B-S sample. Bacterial communities with high Shannon index are displayed to be potentially resistant against the environmental stress and the growth of intrusive bacterial species.24
250 effective reads in the DNA samples from microbial analysis results. Good's coverage ranged from 91.61% to 96.98%, indicating that the variety of microorganism from the bioreactors were well analyzed via the construction of libraries sequence. The index of Chao 1 displayed a decrease from activated sludge to IP-W, suggesting that the quantity of denitrifying bacterial species decreased on the surface of carrier immobilizing activated sludge. This result was in accordance with a previous research, which studied by employing a system of activated sludge, showing that the quantity of microbial species richness in digestion sludge was less than inoculating sludge.22 Compared with IP-S sample, IP-W sample showed comparative more denitrifying bacteria richness. In the sample of activated sludge, the highest diversity (Shannon index was 8.22) was detected and it was confirmed by associated Simpson index. These results suggested that a highly complex and highest diversity microbial communities was contained in activated sludge. Nevertheless, overall diversity decreased because the denitrifying microbial communities gradually became the dominant, as discovered in previous report.23 Shannon indices of IP-S and IP-W samples in the acclimation phase were 7.96 and 8.08, respectively, which were similar with Simpson index. These obtained results suggested that the difference about microbial diversity was not important. However, the B-W sample had a higher diversity than that of the B-S sample. Bacterial communities with high Shannon index are displayed to be potentially resistant against the environmental stress and the growth of intrusive bacterial species.24
| Sample | Number of effective sequences | OTUs | Chao1 index | Good's coverage (%) | Shannon index | Simpson index |
|---|---|---|---|---|---|---|
| Activated sludge | 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 250 |
3862 | 5704 | 96.98 | 8.22 | 0.9866 |
| IP-0 | 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 250 |
3856 | 5669 | 96.67 | 8.18 | 0.9857 |
| IP-S | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 682 682 |
2621 | 4575 | 91.61 | 7.96 | 0.9780 |
| IP-W | 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 250 |
3847 | 5406 | 96.83 | 8.08 | 0.9819 |
| B-S | 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 250 |
2993 | 4630 | 97.20 | 4.71 | 0.7534 |
| B-W | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 906 906 |
2320 | 4150 | 93.89 | 7.69 | 0.9594 |
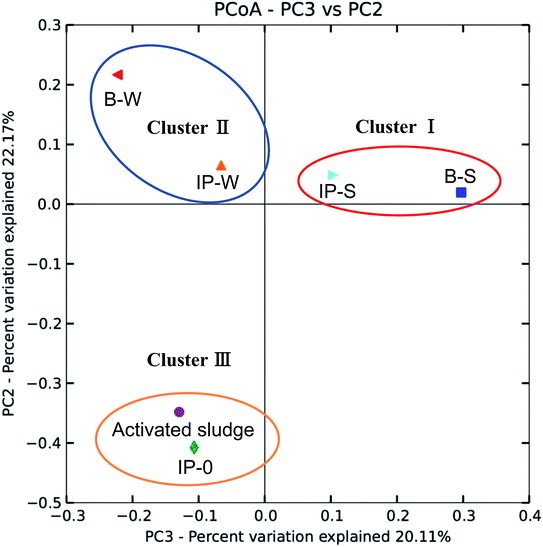
In Fig. 4, the variation of overall patterns among the bacterial communities in the seven samples by PCoA is presented. Samples IP-S and B-S clustered into Cluster I, which indicated that samples of acetate as carbon resource utilizers identified in denitrification reactor and aerobic biofilm reactor were similar. Samples IP-W and B-W clustered into Cluster II, indicating that DAF as carbon resource utilizers identified in denitrification reactor and aerobic biofilm reactor showed similar behavior. Moreover, the difference between the Clusters I and II was due to the difference in carbon resource provided during denitrification. However, samples of activated sludge and IP-0 (Cluster III) were an outliner, signifying that the communities in the carrier of un-acclimated activated sludge was vary from these samples that were in acclimated immobilized carrier and biofilm. With acclimation time increasing, bacterial community of immobilized carrier samples has changed dramatically.
Fig. 5 displays the population of bacterial in five samples at both genus and family levels by using phylogenetic characterization. In all, in the immobilized carrier, the denitrifying bacteria's population increased with the process of denitrification. In addition, the structure of microbial community are also transformed.
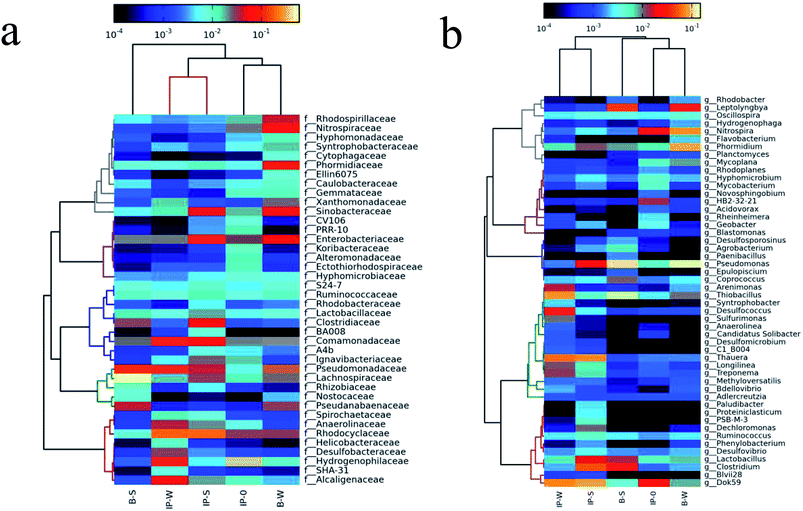 | ||
| Fig. 5 Phylogenetic analysis of bacterial populations in immobilized carrier and biofilm samples with different carbon source addition at family (a) and genus levels (b). | ||
Ahead of bioreactor operation (IP-0), the Hydrogenophilaceae (23.1%) family makes up bacterial community. The number of Hydrogenophilaceae members decreased to a minor component in other samples (Fig. 5a), and most of them were chemolithotrophic nitrite-oxidizing bacterium, reducing NO3−–N via employing a broad scope of inorganic electron donors, for example, hydrogen or the reduced sulfuric compounds.25 Due to lack of such substrates, the population of these taxa probably decreases during denitrification process.
By adding sodium acetate, the dominant microbes in the denitrification bioreactor (IP-S) were Rhodocyclaceae, Enterobacteriaceae, Comamonadaceae, Clostridiaceae, Sinobacteraceae, Pseudomonadaceae, and Lachnospiraceae, accounting for 19.2%, 9.0%, 7.4%, 5.2%, 5.1%, 4.0%, and 3.1% in the bacterial community at the stage IV, respectively. In the denitrification reactor with the DAF (IP-W) addition, the dominant bacterial families were shifted to strongly denitrifying species and enriched with Rhodocyclaceae (18.7%), Hydrogenophilaceae (6.1%), Alcaligenaceae (5.9%), Anaerolinaceae (3.8%), Desulfobacteraceae (2.9%), Comamonadaceae (6.9%), Enterobacteriaceae (2.3%), and Pseudomonadaceae (2.0%). The relative abundance of Rhodocyclaceae showed an increasing trend after acclimation, ranging from 3.1% (IP-0) to about 20% (IP-W and IP-S). This condition is capable of denitrification26 and played a vital role in immobilized carrier (IP-W and IP-S). The relative abundance of Comamonadaceae raised from 2.3% (IP-0) to about 7.0% (IP-S and IP-W), which is a well-known denitrifier and exhibited a denitrification performance in immobilized carrier.26 The relative abundance of Hydrogenophilaceae decreased from 23.1% (IP-0) to 6.1% (IP-W), demonstrating that DAF contained a certain amount of reduced sulfuric compounds as inorganic electron donors for denitrification. However, Hydrogenophilaceae members can't be detected due to the too low content in IP-S sample at the IV stage, reducing NO3−–N by utilizing an extensive range of inorganic electron donors, for example hydrogen or reduced sulfuric compound.25 Because of insufficient such substrates during the process of denitrification, we might observe the significant decline in the population of these taxa. A difference was the higher relative abundance of Enterobacteriaceae family in IP-S sample (6.04%) compared with the IP-W sample (2.3%). The Enterobacteriaceae family were responsible for the first reduction from nitrate to nitrite under denitrifying conditions.27
The aerobic biofilm reactor was located in the downstream of the denitrification reactor, and the most dominant families with the addition of sodium acetate (B-S) were Lachnospiraceae (26.9%), Pseudomonadaceae (11.1%), Clostridiaceae (3.0%), Enterobacteriaceae (2.2%), Rhodocyclaceae (2.1%), and Comamonadaceae (2.0%). With DAF (B-W) addition, the most dominant families were Pseudomonadaceae, Phormidiaceae, Nitrospiraceae, Sinobacteraceae, Enterobacteriaceae, Rhodospirillaceae, Pseudanabaenaceae, Lachnospiraceae, Xanthomonadaceae, Rhodocyclaceae, and Comamonadaceae, accounting for 15.6%, 8.5%, 7.4%, 6.7%, 5.8%, 4.2%, 3.1%, and 2.5%, at the stage IV. Pseudomonadaceae family was dominant both in B-S and B-W. Rhodocyclaceae, Comamonadaceae, and Enterobacteriaceae were also present in biofilm samples (B-S and B-W), indicating that it could be introduced by influent, and exhibited a denitrification performance. Compared with the IP-W sample (5.8%), the high relative abundance of Lachnospiraceae family was found in B-S sample (26.9%), which was butyrate-producing bacteria.28 The Phormidiaceae and Sinobacteraceae families, which were capable of denitrification, were only present in B-W sample.29 In addition, the Nitrospiraceae was dominant only in B-W sample responsible for nitrite nitrogen degradation.30
From Fig. 5b, we can get the conclusion that in the baseline sample (IP-0) the Thiobacillus composed the bacterial community from the genera level. And the relative abundance of Thiobacillus in IP-0 samples reduced from 23.1% to a minimal value of the other four samples. It was reported that Thiobacillus was capable to utilize S2O32− and electron donors i.e., other reduced sulfur compounds during denitrification process.25 The relative abundance of Thiobacillus decreases and the limited substrate availability was possibly the main reason for forming this phenomenon.
A few anaerobic genera or anoxic denitrifiers, for example Thauera, Dok59, Clostridium, and Pseudomonas, were enriched in IP-S sample, accounting for 6.6%, 9.0%, 6.9%, 4.9%, and 2.0% at the stage V, respectively. The Thiobacillus, Thauera, Dok59, Desulfococcus, and Pseudomonas were enriched in IP-W sample, accounting for 6.0%, 4.8%, 6.9%, 2.9%, and 2.2% at the stage V, respectively. Thauera, Dok59, and Pseudomonas were common dominant denitrifiers between IP-S and IP-W samples after acclimation. Thauera is the common denitrifying genus.31,32 The Pseudomonas apparently functioned as the heterotrophic denitrifiers.33 A difference seems to exist for the higher relative abundance of Clostridium in IP-S sample (4.9%) compared with the IP-W sample (0.2%). The Clostridium has shown to be capable of reducing nitrate.34 Meanwhile, the relative abundance of Thiobacillus and Desulfococcus in IP-W samples was much greater than that of IP-S sample, indicating that the DAF introduction contained a certain amount of reduced sulfuric compounds as inorganic electron donors for denitrification. It was reported that Thiobacillus and Desulfococcus were able to utilize S2O32− and other sulfur-reducing substances can be regarded as electron donors in denitrification system.25,34,35
In the aerobic reactor, the most dominant genera in the B-S sample were Pseudomonas (10.8%), Leptolyngbya (4.0%), Rhodobacter (2.84%), and Clostridium (2.79%). With DAF addition, the result showed that the dominant family of B-W sample was obviously different from those found in the B-S sample, i.e., Pseudomonas (14.87%), Phormidium (8.51%), Nitrospira (7.37%), and Leptolyngbya (3.05%) at the stage V. The Pseudomonas was dominant both in B-S and B-W samples. Pseudomonas, as a petroleum-degrading bacteria with high abundance in the biofilm, was confirmed to be the main driver for the high COD removal rate in the aerobic biofilm reactor.36 In addition, Pseudomonas is reported to be the heterotrophic nitrifier, which aerobically oxidized ammonia to hydroxylamine, nitrite, and nitrate.37,38 Nitrospira, as a nitrite-oxidizing bacteria,39 is only dominant in the B-W sample. Pseudomonas and Nitrospira existence was the possible explanations for the NH4+–N removal introduced by DAF in the aerobic biofilm reactor.
4 Conclusions
In this study, an efficient cascade denitrification bioreactor-aerobic biofilm reactor system was utilized for treating heavy oil refinery wastewater, with simultaneous removal of nitrate and COD which met wastewater the discharge standard of China (DB21/1627-2008). The activity of the denitrification bioreactor, which contains waterborne polyurethane carrier and immobilized activated sludge, was evaluated with low-cost secondary DAF effluent as carbon source comparing to acetate. COD/NO3−–N mass ratio of 5.4 was also detected and stable activity of denitrification was achieved with adding 25% volume fraction of secondary DAF effluent.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (No. 51634008), Science and Technology Major Project (No. 2017ZX05009-004).References
- Y. Wang, Q. Wang, M. Li, Y. Yang, W. He, G. Yan and S. Guo, Biochem. Eng. J., 2016, 105, 44–51 CrossRef CAS.
- H. H. Dong, W. Wang, Z. Song, J. Wang, S. Sun, Z. Zhang, M. Ke, W. M. Wu, G. Zhang and J. Ma, Bioresour. Technol., 2017, 239, 472–481 CrossRef CAS PubMed.
- S. Y. Oh, Y. D. Seo, B. Kim, I. Y. Kim and D. K. Cha, Bioresour. Technol., 2016, 200, 891–896 CrossRef CAS PubMed.
- F. Ma, Y. Sun, A. Li, X. Zhang and J. Yang, Bioresour. Technol., 2015, 187, 30–36 CrossRef CAS PubMed.
- K. Isaka, Y. Date, T. Sumino and S. Tsuneda, Appl. Microbiol. Biotechnol., 2007, 76, 1457–1465 CrossRef CAS PubMed.
- Y. Dong, Z. Zhang, Y. Jin, J. Lu, X. Cheng, J. Li, Y. Y. Deng, Y. N. Feng and D. Chen, J. Environ. Sci., 2012, 24, 999–1005 CrossRef CAS.
- Y. M. Dong, Z. J. Zhang, Y. W. Jin, Z. R. Li and J. A. Lu, J. Environ. Sci., 2011, 23, 366–371 CrossRef CAS.
- S. Tabassum, Y. Wang, X. Zhang and Z. Zhang, RSC Adv., 2015, 5, 88692–88702 RSC.
- J. Park, R. Craggs and J. Sukias, Bioresour. Technol., 2009, 100, 3175–3179 CrossRef CAS PubMed.
- J. Shen, R. He, W. Han, X. Sun, J. Li and L. Wang, J. Hazard. Mater., 2009, 172, 595–600 CrossRef CAS PubMed.
- P. M. Sutton, B. E. Jank and B. A. Monaghan, Single sludge nitrogen removal systems. EPS, 1979, p. 164 Search PubMed.
- APHA, AWWA and WEF, Standard Methods for examination of water and wastewater, American Public Health Association (APHA), Washington, DC, USA, 22nd edn, 2012, p. 1360 Search PubMed.
- T. Magoc and S. L. Salzberg, Bioinformatics, 2011, 27, 2957–2963 CrossRef CAS PubMed.
- J. G. Caporaso, C. L. Lauber, W. A. Walters, D. Berg-Lyons, J. Huntley, N. Fierer, S. M Owens, J. Betley, L. Fraser, M. Bauer, N. Gormley, J. A. Gilbert, G. Smith and R. Knight, ISME J., 2012, 6, 1621–1624 CrossRef CAS PubMed.
- R. C. Edgar, Bioinformatics, 2010, 26, 2460–2461 CrossRef CAS.
- J. R. Cole, Q. Wang, E. Cardenas, J. Fish, B. Chai, R. J. Farris, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, T. Marsh and G. M. Garrity, Nucleic Acids Res., 2009, 37, D141–D145 CrossRef CAS PubMed.
- C. Hwang, W. M. Wu, T. Gentry, J. Carley, S. Carroll, C. Schadt, D. Watson, P. Jardine, J. Zhou and R. Hickey, Appl. Microbiol. Biotechnol., 2006, 71, 748–760 CrossRef CAS PubMed.
- K. Joshi, R. Rajan, G. Srinikethan and M. Saidutta, International Journal of Current Engineering and Technology, 2014, 4, 3304–3309 Search PubMed.
- J. M. Flere and T. C. Zhang, J. Environ. Eng., 1999, 125, 721–729 CrossRef CAS.
- J. Reyes-Avila, E. Razo-Flores and J. Gomez, Water Res., 2004, 38, 3313–3321 CrossRef CAS PubMed.
- R. R. Nair and F. D. Stanislaus, J. Ind. Microbiol. Biotechnol., 2012, 39, 1637–1643 CrossRef CAS PubMed.
- L. Ye and T. Zhang, Appl. Microbiol. Biotechnol., 2013, 97, 2681–2690 CrossRef CAS PubMed.
- B. Xie, B. Liu, Y. Yi, L. Yang, D. Liang, Y. Zhu and H. Liu, Bioresour. Technol., 2016, 207, 109–117 CrossRef CAS PubMed.
- E. I. Prest, F. Hammes, M. C. Loosdrecht and J. S. Vrouwenvelder, Front. Microbiol., 2016, 7, 45 Search PubMed.
- Q. Guo, H. Y. Hu, Z. J. Shi, C. C. Yang, P. Li, M. Huang, W. M. Ni, M. L. Shi and R. C. Jin, Chem. Eng. J., 2016, 297, 207–216 CrossRef CAS.
- Y. Song, L. Xiao, I. Jayamani, Z. He and A. M. Cupples, J. Microbiol. Methods, 2015, 108, 4–11 CrossRef CAS PubMed.
- X. H. Yi, J. Wan, Y. Ma and Y. Wang, Biochem. Eng. J., 2016, 107, 66–74 CrossRef CAS.
- M. Jaquet, I. Rochat, J. Moulin, C. Cavin and R. Bibiloni, Int. J. Food Microbiol., 2009, 130, 117–121 CrossRef CAS PubMed.
- H. L. Tian, J. Y. Zhao, H. Y. Zhang, C. Q. Chi, B. A. Li and X. L Wu, Appl. Microbiol. Biotechnol., 2015, 99, 3279–3290 CrossRef CAS PubMed.
- Z. Chen, X. Wang, Y. Yang, M. M. Mirino and Y. Yuan, Bioresour. Technol., 2016, 218, 580–588 CrossRef CAS PubMed.
- T. T. Fida, C. Chen and G. Okpala, Appl. Environ. Microbiol., 2016, 82, 4190 CrossRef CAS PubMed.
- C. M. Callbeck, A. Agrawal and G. Voordouw, Appl. Environ. Microbiol., 2013, 179, 059–5068 Search PubMed.
- C. Chen, N. Ren, A. Wang, Z. Yu and D. J. Lee, Appl. Microbiol. Biotechnol., 2008, 78, 1057–1063 CrossRef CAS PubMed.
- X. J. Xu, C. Chen, X. Guan, Y. Yuan, A. J. Wang, D. J. Lee, Z. F. Zhang, J. Zhang, Y. J. Zhong and N. Q. Ren, Chem. Eng. J., 2017, 330, 63–70 CrossRef CAS.
- A. A. Grigoryan, S. L. Cornish, B. Buziak, S. Lin, A. Cavallaro, J. J. Arensdorf and G. Voordouw, Appl. Environ. Microbiol., 2008, 74, 4324–4335 CrossRef CAS PubMed.
- H. H. Dong, H. Dong, Z. Zhang, S. Sun, W. Wang, M. Ke, Z. Song, Z. Zhang, J. Wang and W. M. Wu, RSC Adv., 2016, 6, 107442–107451 RSC.
- H. Bothe, G. Jost, M. Schloter, B. B. Ward and K. P. Witzel, FEMS Microbiol. Rev., 2000, 24, 673–690 CrossRef CAS PubMed.
- M. Daum, W. Zimmer, H. Papen, K. Kloos, K. Nawrath and H. Bothe, Curr. Microbiol., 1998, 37, 281–288 CrossRef CAS PubMed.
- Y. Chen, Z. Zhao, Y. Peng, J. Li, L. Xiao and L. Yang, Bioresour. Technol., 2016, 220, 225–232 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additional analytical results including Fig. S1 and S2 were added. See DOI: 10.1039/c8ra10510c |
| This journal is © The Royal Society of Chemistry 2019 |