Open Access Article
Open Access ArticleHigh throughput metabolomics-proteomics investigation on metabolic phenotype changes in rats caused by Radix Scrophulariae using ultra-performance liquid chromatography with mass spectrometry†
Fang Lu‡
a,
Ning Zhang‡b,
Tao Ye‡ b,
Hongwei Zhaoa,
Mu Panga and
Shu-min Liu
b,
Hongwei Zhaoa,
Mu Panga and
Shu-min Liu *ac
*ac
aChinese Medicine Toxicological Laboratory, Heilongjiang University of Chinese Medicine, Harbin, P. R. China
bFist Affiliated Hospital of Guiyang University of Traditional Chinese Medicine, Guizhou, Guiyang, P. R. China
cDrug Safety Evaluation Center, Heilongjiang University of Chinese Medicine, He Ping Road 24, Harbin 150040, P. R. China. E-mail: keji-liu@163.com; Fax: +86 45182193278; Tel: +86 45182193278
First published on 6th June 2019
Abstract
Radix Scrophulariae, a traditional Chinese herb, is used to treat various diseases, including H2O2-induced apoptosis in cardiomyocytes, HaCaT cells, hyperuricaemia, and depression. This study screened metabolites, proteins and common pathways to better understand both the therapeutic effects and side effects of this herb. Methods: Untargeted metabolomics based on UPLC-TOF-MS, coupled with proteomics based on nano-UPLC-Q-Exactive-MS/MS, were used to investigate the effects of R. Scrophulariae in rats. Fifty-one identified metabolites in urine samples and 76 modulated proteins in liver tissue were potential biomarkers for R. Scrophulariae treatment. The biomarkers and common pathways involved were steroid hormone biosynthesis, drug metabolism-cytochrome p450, drug metabolism-other enzymes, pentose and glucuronate interconversions, and starch and sucrose metabolism. Some biomarkers were beneficial for treating diseases such as cancer, tuberculosis and isovaleric acidaemia, while other biomarkers caused side effects. Metabolomic and proteomic analyses of R. Scrophulariae-treated rats provided valuable information on the biological safety and efficacy of using R. Scrophulariae clinically.
1 Introduction
Radix Scrophulariae, the dried root of Scrophularia ningpoensis Hemsl., belongs to the Scrophulariaceae family and has been used in TCM for thousands of years. Per the Pharmacopoeia of the People's Republic of China (2015 Edition), the species' traditional functions include treating febrile diseases, constipation, hot eyes, pharyngalgia, diphtheria, and scrofula. Modern pharmacological research has shown that R. Scrophulariae inhibits ventricular remodelling,1–4 hypoxia-induced microglial activation and neurotoxicity,5 hypertension and attenuating arteriosclerosis,6 proliferation, apoptosis induction in cancer cell lines,7 and antioxidative activity.8 TCMs are administered orally; therefore, their metabolites and proteins are disturbed when the blood circulation contacts the target organs. TCMs are therapeutic but also have side effects. For clinical use, the safety and effectiveness of TCMs are most important. These fundamental rules will guide exploitation of the biological effects of R. Scrophulariae.The integrated metabolomics and proteomics based on mass spectrometry represents an innovative approach to characterize molecule fingerprints related to the function.9–13 Here, metabolomics coupled with iTRAQ-based proteome profile analysis of these biological effects were employed to screen the key metabolites from urine samples and liver proteins by UPLC-TOF-MS and nano-UPLC-Q-Exactive-MS/MS, respectively.
2 Materials and methods
2.1 Plant material and extract preparation
The root of Scrophularia ningpoensis Hemsl. is a natural medicine. R. Scrophulariae was acquired from the Heilongjiang Province Drug Company (Harbin, PR China). The voucher specimen (hlj-20120623012) for the herb was authenticated by Prof. Zhenyue Wang of the Department of Resources and Development of TCM at Heilongjiang University of Traditional Chinese Medicine, meeting the standards of the Pharmacopoeia of the People's Republic of China (2015 edition), page 117.Fatty oil of R. Scrophulariae was prepared with the 1 kg crude drug, which was extracted twice with 0.6 l petroleum ether for 12 h each. The two portions were mixed and concentrated into cream, then the drug residue was freeze-dried and extracted twice with 10 and 8 l distilled water (DW) for 1.5 h each, respectively. The two portions were mixed and concentrated into cream, comprising the aqueous extract of R. Scrophulariae. The eluates were freeze-dried to make extracts with a yield of 50.7%.
2.2 Rats and treatments
Healthy male Sprague-Dawley rats, weighing 200 ± 20 g each, were purchased from Liaoning Changsheng Biotechnology Co., Ltd. (PR China) (Animal Certificate No: SCXK [Liao] 2015-0001). Rats were fed a standard diet with free access to water and housed 1 per metabolic cage at a temperature of 21–23 °C and humidity at 40–50% in controlled rooms with a 12 h/12 h light/dark cycle. This study was conducted in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was subject to approval by the Committee on the Ethics of Animal Experiments of the College of Pharmacy of Heilongjiang University of Chinese Medicine.After acclimation for 1 week, 20 rats were randomly divided into two groups, the control group and the water decoction of R. Scrophulariae group (n = 10 per group). Rats in the R. Scrophulariae group received decoction of R. Scrophulariae (1350 mg crude drug per kg, i.g.) once daily for 15 consecutive days, and rats in the control group received the same volume of 0.9% saline once daily for 15 days.
2.3 Sample collection and preparation
For the metabolomic analysis, urine was collected from all rats in each group on days 0, 1, 3, 5, 7, 9, 11, 13 and 15. Urine was centrifuged twice at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 10 min at 4 °C to remove the solid residue. Supernatants were transferred to a 1.5 ml polypropylene tube and filtered through a syringe filter (0.22 μm). Two μl of the supernatant was injected into the UPLC/TOF-MS for analysis.
000 g for 10 min at 4 °C to remove the solid residue. Supernatants were transferred to a 1.5 ml polypropylene tube and filtered through a syringe filter (0.22 μm). Two μl of the supernatant was injected into the UPLC/TOF-MS for analysis.
For the proteomic analysis, rats were anaesthetized with 1% sodium pentobarbital anaesthesia (0.15 ml/100 g), and liver samples were obtained. Each group was analysed in triplicate (n = 10 per group) and then mixed into 3 mixed samples and stored at −80 °C. After thawing, 100 μl of STD buffer (4% SDS [161-0302, Bio-Rad], 1 mM DTT [161-0404 Bio-Rad], 150 mM Tris–HCl pH 8.0) per 20 μg sample was added and homogenized with a tissue homogenizer for 5 min in a boiling water bath. The mix was ultrasonicated (80 W) for 10 s and intermittently for 15 s ten times, then incubated in boiling water for 5 min and centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 10 min. The supernatant was removed and subjected to 12.5% SDS-PAGE electrophoresis.
000 g for 10 min. The supernatant was removed and subjected to 12.5% SDS-PAGE electrophoresis.
2.4 Metabolic profiling platform
2.5 Proteomics analysis
The electrophoretic conditions were kept at a constant current for 90 min. The 300 μg samples were filter-aided for sample preparation (FASP). Eighty μg peptide samples from each group were labelled per the iTRAQ Reagent-8plex Multiplex Kit (AB SCIEX) instructions. Strong cation exchange (SCX) chromatography was performed on an AKTA Purifier 100 (GE Healthcare) coupled with a polysulfoethyl 4.6 × 100 mm column (5 μm, 200 Å) (PolyLC, Inc., Maryland, U.S.A.). Buffers were composed of buffer A (10 mM KH2PO4 pH 3.0, 25% ACN) and buffer B (10 mM KH2PO4 pH 3.0, 500 mM KCl, 25% ACN) with a flow rate of 1000 μl min−1. SCX gradient was set as follows: 0–25.01 min, 100% A; 25.01–32.00 min, 100–90% A; 32.01–42.00 min, 90–80% A; 42.01–47.00 min, 80–55% A; 47.01–52 min, 55–0% A; 52–60 min, 0% A; 60–60.01 min, 0–100% A; 60.01–75.00 min, 100% A.Samples were separated by an automated Easy-nLC system coupled with a Q-Exactive spectrometer (Thermo Finnigan, USA). Buffer was composed of solution A (water containing 0.1% FA) and solution B (84% ACN containing 0.1% FA). Protein samples were performed on a thermo scientific EASY C18 column (2 cm × 100 μm, 5 μm), and separated on a thermo scientific EASY C18 column (75 μm × 100 mm, 3 μm). The flow rate was 300 nL min−1. The gradient elution procedure was as follows: 0–55 min, 0–40% B; 55–58 min, 40–100% B; 58–60 min, 100% B. The scan range was set to 300–1800 m/z in positive ion mode. The AGC target was set to 3 × 106. The maximum injection time was 10 ms. The normalized collision energy was 30 eV. The underfill ratio was 0.1%. The mass resolution for full MS and dd-MS2 were 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 17
000 and 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, respectively.
500, respectively.
3 Results and discussion
3.1 Metabolomic profile analysis
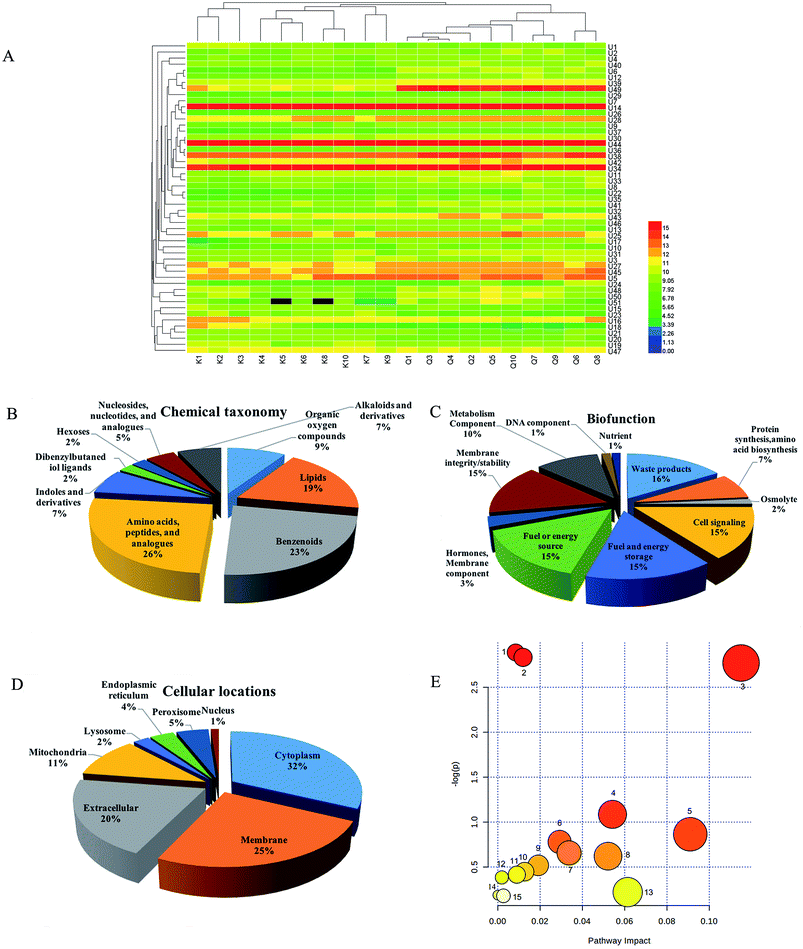
In OPLS-DA, urine samples from the R. Scrophulariae group were separated from the control group, revealing that the rats' metabolic profiles changed after R. Scrophulariae treatment (Fig. 1A). VIP-plot and S-plot for evaluating contribution degree are shown in Fig. 1B and C. Fifty-one differential metabolites were consistent with P < 0.05, VIP > 1, and maximum fold change > 1 after being retrieved and matched by Metlin, HMDB and KEGG (Fig. 2A and Table 1).Endogenous small metabolites were classified using the HMDB database, and 26% were classified as amino acids, peptides, and analogues, 23% were benzenoids, and 19% were lipids (Fig. 2B). For biofunction, 16% were subgrouped into waste products, and cell signalling, fuel and energy storage, fuel or energy source and membrane integrity/stability accounted for 15% each (Fig. 2C). These metabolites were primarily located in the cytoplasm, membrane, extracellular matrix and mitochondria (Fig. 2D).| No. | Rt-m/z | HMDB ID | Ions mode | KEGG | Formula | Metabolites | VIP value | Trends (Q/K) | Anova (p) | q value | Max fold change |
|---|---|---|---|---|---|---|---|---|---|---|---|
| a Compared with control group, *p < 0.05,**p < 0.01. U represents urine. K represents control group, Q represents R. Scrophulariae group. | |||||||||||
| U1 | 10.70_207.1034 | HMDB11603 | pos | C16453 | C10H13N3O2 | 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone | 1.1366 | Down* | 0.0171 | 0.0450 | 1.5678 |
| U2 | 4.45_202.1225 | HMDB00792 | pos | C08277 | C10H18O4 | Sebacic acid | 1.9215 | Up* | 0.0145 | 0.0402 | 1.4905 |
| U3 | 1.39_153.0816 | HMDB04825 | pos | C04227 | C8H11NO2 | p-Octopamine | 1.1147 | Up** | 0.0029 | 0.0123 | 1.2995 |
| U4 | 2.56_160.1227 | HMDB02038 | pos | C02728 | C7H16N2O2 | N(6)-Methyllysine | 1.0838 | Up** | 0.0000 | 0.0009 | 1.3617 |
| U5 | 2.56_143.0966 | HMDB04827 | pos | C10172 | C7H13NO2 | Proline betaine | 3.5748 | Up* | 0.0109 | 0.0325 | 1.3654 |
| U6 | 3.50_275.1305 | HMDB13209 | pos | C00806 | C14H17N3O3 | Alanyltryptophan | 2.5677 | Up** | 0.0000 | 0.0000 | 5.6666 |
| U7 | 3.96_286.1515 | HMDB00343 | pos | C05298 | C18H22O3 | 2-Hydroxyestrone | 1.0815 | Up** | 0.0028 | 0.0120 | 1.4051 |
| U8 | 0.93_172.0633 | HMDB01138 | pos | C00624 | C7H11NO5 | N-Acetylglutamic acid | 1.4061 | Up** | 0.0000 | 0.0007 | 1.5997 |
| U9 | 0.96_202.1108 | HMDB00216 | pos | C00547 | C8H11NO3 | Norepinephrine | 1.3643 | Up** | 0.0000 | 0.0005 | 1.5478 |
| U10 | 0.97_284.1007 | HMDB00472 | pos | C01017 | C11H12N2O3 | 5-Hydroxy-L-tryptophan | 1.1434 | Up** | 0.0016 | 0.0080 | 1.8401 |
| U11 | 1.81_267.1359 | HMDB05056 | pos | C18166 | C18H22O4 | Enterodiol | 1.7389 | Up** | 0.0004 | 0.0031 | 1.4417 |
| U12 | 1.84_144.0672 | HMDB01514 | pos | C00329 | C6H13NO5 | Glucosamine | 1.9440 | Up** | 0.0000 | 0.0000 | 2.5749 |
| U13 | 2.23_275.1260 | HMDB00273 | pos | C00214 | C10H14N2O5 | Thymidine | 1.0469 | Up** | 0.0022 | 0.0103 | 1.4222 |
| U14 | 2.31_297.1462 | HMDB06344 | pos | C04148 | C13H16N2O4 | Alpha-N-phenylacetyl-L-glutamine | 13.5022 | Up** | 0.0009 | 0.0055 | 1.4501 |
| U15 | 2.92_267.1004 | HMDB00933 | pos | C16308 | C12H20O4 | Traumatic acid | 1.2758 | Up** | 0.0000 | 0.0008 | 2.1394 |
| U16 | 4.24_254.1154 | HMDB41959 | pos | C11785 | C16H17NO3 | Normorphine | 2.1217 | Down* | 0.0388 | 0.0802 | 1.5050 |
| U17 | 10.65_190.0521 | HMDB01553 | pos | C01180 | C5H8O3S | 2-Oxo-4-methylthiobutanoic acid | 1.3511 | Up* | 0.0287 | 0.0664 | 1.7704 |
| U18 | 10.09_151.0469 | HMDB02091 | pos | C03033 | C11H18O8 | Isovalerylglucuronide | 2.2095 | Down** | 0.0001 | 0.0013 | 36.6655 |
| U19 | 6.49_316.1969 | HMDB00306 | pos | C00483 | C8H11NO | Tyramine | 1.0905 | Down* | 0.0350 | 0.0748 | 1.3971 |
| U20 | 5.54_246.1721 | HMDB02176 | pos | C18319 | C5H10O2 | Ethylmethylacetic acid | 1.1394 | Down** | 0.0009 | 0.0055 | 1.5188 |
| U21 | 5.35_278.1073 | HMDB10328 | pos | C03033 | C14H19NO7 | Tyramine glucuronide | 1.0563 | Down** | 0.0015 | 0.0080 | 1.4124 |
| U22 | 4.79_158.0630 | HMDB00821 | pos | C05598 | C10H11NO3 | Phenylacetylglycine | 1.2436 | Up** | 0.0000 | 0.0008 | 3.3653 |
| U23 | 4.33_348.1204 | HMDB01476 | pos | C00632 | C7H7NO3 | 3-Hydroxyanthranilic acid | 1.7225 | Down** | 0.0003 | 0.0027 | 3.1281 |
| U24 | 3.36_153.0933 | HMDB00784 | pos | C08261 | C9H16O4 | Azelaic acid | 1.1179 | Up* | 0.0142 | 0.0398 | 2.6331 |
| U25 | 2.73_245.1513 | HMDB00201 | pos | C02571 | C9H17NO4 | L-Acetylcarnitine | 2.8023 | Up** | 0.0090 | 0.0282 | 1.4032 |
| U26 | 2.62_197.0838 | HMDB02035 | pos | C00811 | C9H8O3 | 4-Hydroxycinnamic acid | 1.0570 | Up** | 0.0001 | 0.0012 | 1.6887 |
| U27 | 2.49_230.1422 | HMDB04063 | pos | C05588 | C10H15NO3 | Metanephrine | 1.8485 | Up* | 0.0144 | 0.0400 | 1.1881 |
| U28 | 2.45_243.1353 | HMDB13248 | pos | C03343 | C16H22O4 | Monoethylhexyl phthalic acid | 2.7083 | Up** | 0.0007 | 0.0048 | 1.3543 |
| U29 | 2.27_200.0756 | HMDB00462 | pos | C01551 | C4H6N4O3 | Allantoin | 1.1143 | Up** | 0.0000 | 0.0002 | 1.8271 |
| U30 | 2.18_261.0885 | HMDB00181 | pos | C00355 | C9H11NO4 | L-Dopa | 2.0865 | Up** | 0.0000 | 0.0002 | 1.5815 |
| U31 | 2.05_413.1250 | HMDB10334 | pos | C03033 | C22H22O9 | Ketoprofen glucuronide | 1.1382 | Up** | 0.0084 | 0.0268 | 1.3105 |
| U32 | 1.68_255.0778 | HMDB01858 | pos | C01468 | C7H8O | p-Cresol | 1.0839 | Up** | 0.0001 | 0.0011 | 1.5645 |
| U33 | 1.65_265.1570 | HMDB00010 | pos | C05299 | C19H24O3 | 2-Methoxyestrone | 1.3779 | Up** | 0.0006 | 0.0040 | 1.5384 |
| U34 | 1.40_204.1350 | HMDB00450 | pos | C16741 | C6H14N2O3 | 5-Hydroxylysine | 11.9554 | Up** | 0.0000 | 0.0003 | 1.5922 |
| U35 | 1.34_173.0461 | HMDB12710 | pos | C00944 | C7H10O6 | 3-Dehydroquinate | 1.3519 | Up** | 0.0012 | 0.0069 | 2.1674 |
| U36 | 1.30_259.1671 | HMDB00824 | pos | C03017 | C10H19NO4 | Propionylcarnitine | 1.4708 | Up** | 0.0021 | 0.0100 | 1.5050 |
| U37 | 1.22_215.1049 | HMDB32049 | pos | C06354 | C13H10O | Benzophenone | 1.3260 | Up** | 0.0000 | 0.0003 | 2.9951 |
| U38 | 1.22_166.0736 | HMDB02303 | pos | C00580 | C2H6S | Dimethylsulfide | 5.9549 | Up** | 0.0000 | 0.0001 | 1.4267 |
| U39 | 1.20_158.0904 | HMDB00904 | pos | C00327 | C6H13N3O3 | Citrulline | 2.6437 | Up** | 0.0000 | 0.0001 | 1.7277 |
| U40 | 1.05_168.0684 | HMDB00742 | pos | C05330 | C4H9NO2S | Homocysteine | 1.0606 | Up** | 0.0028 | 0.0122 | 1.3436 |
| U41 | 0.95_261.1481 | HMDB13835 | pos | C15205 | C16H22O4 | Diisobutyl phthalate | 1.1423 | Up** | 0.0014 | 0.0076 | 1.2734 |
| U42 | 0.90_269.1267 | HMDB00014 | pos | C00881 | C9H13N3O4 | Deoxycytidine | 3.0487 | Up** | 0.0000 | 0.0001 | 1.5761 |
| U43 | 0.87_227.0455 | HMDB01890 | pos | C06809 | C5H9NO3S | Acetylcysteine | 3.1726 | Up** | 0.0001 | 0.0012 | 1.6787 |
| U44 | 0.80_212.1014 | HMDB41821 | pos | C07585 | C8H9N3O2 | Acetylisoniazid | 14.5218 | Up** | 0.0000 | 0.0003 | 1.6217 |
| U45 | 0.74_144.1041 | HMDB01010 | pos | C00745 | C10H13N2 | Nicotine imine | 2.8013 | Up** | 0.0048 | 0.0179 | 1.3663 |
| U46 | 0.68_176.0135 | HMDB00875 | pos | C01004 | C7H7NO2 | Trigonelline | 1.2566 | Up** | 0.0008 | 0.0049 | 1.4882 |
| U47 | 7.86_253.1075 | HMDB02004 | neg | C08309 | C13H18N2O | 5-Methoxydimethyltryptamine | 1.4738 | Down* | 0.0235 | 0.5348 | 1.3094 |
| U48 | 4.02_290.0707 | HMDB00855 | neg | C03150 | C11H15N2O5 | Nicotinamide riboside | 1.2816 | Up* | 0.0252 | 0.5368 | 1.6344 |
| U49 | 3.72_247.0284 | HMDB04983 | neg | C11142 | C2H6O2S | Dimethyl sulfone | 13.8303 | Up** | 0.0000 | 0.0000 | 12.5966 |
| U50 | 1.72_238.0757 | HMDB13318 | neg | C00977 | C11H13N3O | Tryptophanamide | 1.6212 | Up* | 0.0113 | 0.4388 | 1.8896 |
| U51 | 1.72_483.1785 | HMDB10317 | neg | C03033 | C24H32O8 | 17-Beta-estradiol glucuronide | 1.3788 | Up* | 0.0194 | 0.5261 | 2.7223 |
Pathway analysis was performed using MetaboAnalyst 3.0 software, revealing that endogenous small molecule metabolites were concentrated in the metabolisms of nicotinate and nicotinamide, phenylalanine, tyrosine and pyrimidine, and in phenylalanine, tyrosine and tryptophan biosynthesis (Fig. 2E).
3.2 Liver proteomic profile
Seventy-six significant proteins were selected by P < 0.05 and ratio >1.2 or <0.833 and are listed in Table 2. Compared with the control, expression levels of 31 proteins were down-regulated, while 45 were up-regulated. Seventy-six changed proteins were further enriched using the Bonferroni correction for multiple testing (P < 0.05) through GO analysis, and the GO terms for fold enrichment are shown in Fig. 3A. Seventy-six proteins participated in single-organism metabolic processes. These proteins were primarily involved in catalytic and electron carrier activities and were mainly located in the organelles. It suggested that the function of R. Scrophulariae is related to a complicated biological process. Twelve significant KEGG pathways were selected by −log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p value > 2 and are shown in Fig. 3B. The protein–protein interaction (PPI) network was analysed by the publicly available programme STRING (http://string-db.org/). STRING is a database of known and predicted protein interactions. PPI nodes such as COX2, ND2, Cyp17a1, Hsd17b2, Mgst3, Ugt2b, Cyp2c13, RT1-CE1, Mn1 and Psmb8 might play key roles in the functional mechanism of R. Scrophulariae (Fig. 3C).
p value > 2 and are shown in Fig. 3B. The protein–protein interaction (PPI) network was analysed by the publicly available programme STRING (http://string-db.org/). STRING is a database of known and predicted protein interactions. PPI nodes such as COX2, ND2, Cyp17a1, Hsd17b2, Mgst3, Ugt2b, Cyp2c13, RT1-CE1, Mn1 and Psmb8 might play key roles in the functional mechanism of R. Scrophulariae (Fig. 3C).
| No. | Accession | Gene name | Description | Average ratio Q/K | Trends Q/K | P value |
|---|---|---|---|---|---|---|
| a Compared with control group, *p < 0.05,**p < 0.01. L represents liver. K represents control group, Q represents R. Scrophulariae group. | ||||||
| L1 | Q6MG32 | RT1-CE12 | RT1 class I, CE12 | 0.4685 | Down** | 0.0002 |
| L2 | Q63042 | Gfer | FAD-linked sulfhydryl oxidase ALR | 0.5479 | Down** | 0.0022 |
| L3 | A0A0G2JSK1 | Serpina3c | Protein Serpina3c | 0.5502 | Down** | 0.0001 |
| L4 | P35286 | Rab13 | Ras-related protein Rab-13 | 0.6139 | Down* | 0.0458 |
| L5 | M0R9Q1 | Rbm14 | Protein Rbm14 | 0.6297 | Down** | 0.0005 |
| L6 | Q4VBH1 | Ighg | Ighg protein | 0.6415 | Down** | 0.0004 |
| L7 | A0A023IKI3 | Psmb8 | Proteasome subunit beta type | 0.6454 | Down** | 0.0047 |
| L8 | A0A0G2JX10 | Anks3 | Ankyrin repeat and SAM domain-containing protein 3 | 0.6764 | Down** | 0.0022 |
| L9 | P70473 | Amacr | Alpha-methylacyl-CoA racemase | 0.6981 | Down** | 0.0003 |
| L10 | M0RAJ5 | Prr14l | Protein Prr14l | 0.7129 | Down** | 0.0069 |
| L11 | Q6P756 | Necap2 | Adaptin ear-binding coat-associated protein 2 | 0.7407 | Down* | 0.0151 |
| L12 | Q80W92 | Vac14 | Protein VAC14 homolog | 0.7429 | Down* | 0.0338 |
| L13 | A0A097PE04 | COX2 | Cytochrome c oxidase subunit 2 | 0.7470 | Down** | 0.0005 |
| L14 | Q5RK24 | Pmvk | Phosphomevalonate kinase | 0.7552 | Down** | 0.0019 |
| L15 | Q99MS0 | Sec14l2 | SEC14-like protein 2 | 0.7571 | Down** | 0.0009 |
| L16 | E9PU17 | Abca17 | ATP-binding cassette sub-family A member 17 | 0.7599 | Down* | 0.0271 |
| L17 | F1LM99 | Phf12 | PHD finger protein 12 | 0.7733 | Down** | 0.0045 |
| L18 | A0A0G2JVQ0 | Rnf111 | Protein Rnf111 | 0.7744 | Down* | 0.0444 |
| L19 | D3ZTW7 | Atpaf2 | ATP synthase mitochondrial F1 complex assembly factor 2 (predicted), isoform CRA_c | 0.7886 | Down** | 0.0012 |
| L20 | P43424 | Galt | Galactose-1-phosphate uridylyltransferase | 0.7907 | Down** | 0.0076 |
| L21 | A0A0G2JV37 | LOC100910040 | Carboxylic ester hydrolase | 0.7914 | Down* | 0.0305 |
| L22 | P49889 | Ste | Estrogen sulfotransferase, isoform 3 | 0.7941 | Down** | 0.0050 |
| L23 | Q6AYW2 | Pah | Phenylalanine hydroxylase | 0.7994 | Down** | 0.0088 |
| L24 | P55006 | Rdh7 | Retinol dehydrogenase 7 | 0.8006 | Down** | 0.0001 |
| L25 | P0C5E9 | Crygs | Beta-crystallin S | 0.8026 | Down** | 0.0075 |
| L26 | A0A0G2KA12 | Kif1b | Kinesin-like protein KIF1B | 0.8111 | Down* | 0.0244 |
| L27 | F1LRB8 | Mat2a | S-Adenosylmethionine synthase | 0.8113 | Down** | 0.0057 |
| L28 | B2GV29 | Trmt13 | Ccdc76 protein | 0.8139 | Down** | 0.0016 |
| L29 | Q4QR81 | Rbms2 | Protein Rbms2 | 0.8203 | Down* | 0.0224 |
| L30 | D4AAP6 | Mn1 | Protein Mn1 | 0.8215 | Down** | 0.0016 |
| L31 | D4AB73 | Sprtn | Putative uncharacterized protein RGD1559496_predicted | 0.8298 | Down* | 0.0177 |
| L32 | Q5XHZ8 | Cog3 | Component of oligomeric golgi complex 3 | 1.2021 | Up** | 0.0011 |
| L33 | P00502 | Gsta1 | Glutathione S-transferase alpha-1 | 1.2046 | Up** | 0.0000 |
| L34 | P19488 | Ugt2b37 | UDP-glucuronosyltransferase 2B37 | 1.2057 | Up** | 0.0045 |
| L35 | Q6AXQ0 | Sae1 | SUMO-activating enzyme subunit 1 | 1.2057 | Up* | 0.0347 |
| L36 | F1LNM4 | LOC103689965 | Complement C4 (fragment) | 1.2075 | Up** | 0.0039 |
| L37 | F1LU27 | Focad | Protein Focad | 1.2138 | Up* | 0.0489 |
| L38 | Q32PY9 | Idnk | Probable gluconokinase | 1.2167 | Up** | 0.0068 |
| L39 | G3V647 | Pdxk | Pyridoxal kinase | 1.2222 | Up** | 0.0010 |
| L40 | P05545 | Serpina3k | Serine protease inhibitor A3K | 1.2300 | Up** | 0.0100 |
| L41 | G3V9N9 | Man1a1 | Alpha-1,2-mannosidase | 1.2303 | Up** | 0.0040 |
| L42 | Q566C7 | Nudt3 | Diphosphoinositol polyphosphate phosphohydrolase 1 | 1.2349 | Up** | 0.0001 |
| L43 | F1LN59 | Eif4g2 | Protein Eif4g2 | 1.2408 | Up** | 0.0084 |
| L44 | D4A284 | Nell1 | NEL-like 1 (chicken), isoform CRA_a | 1.2417 | Up* | 0.0180 |
| L45 | D3ZNJ5 | Inmt | Protein Inmt | 1.2494 | Up** | 0.0027 |
| L46 | A0A0G2JU41 | Dyrk4 | Protein Dyrk4 | 1.2503 | Up** | 0.0015 |
| L47 | D4ADS4 | Mgst3 | Protein Mgst3 | 1.2523 | Up** | 0.0007 |
| L48 | A0A0G2JSR8 | Cyp17a1 | Cytochrome P450, family 17, subfamily a, polypeptide 1 | 1.2576 | Up** | 0.0077 |
| L49 | A2VCW9 | Aass | Alpha-aminoadipic semialdehyde synthase, mitochondrial | 1.2637 | Up** | 0.0000 |
| L50 | F1M7N8 | Ugt2b37 | UDP-glucuronosyltransferase | 1.2647 | Up** | 0.0010 |
| L51 | P38659 | Pdia4 | Protein disulfide-isomerase A4 | 1.2670 | Up** | 0.0018 |
| L52 | Q6AXR4 | Hexb | Beta-hexosaminidase subunit beta | 1.2712 | Up** | 0.0021 |
| L53 | D4A3E8 | Mrps27 | Mitochondrial ribosomal protein S27 (predicted), isoform CRA_b | 1.2733 | Up* | 0.0453 |
| L54 | D3ZES7 | Plxna4 | Protein Plxna4 | 1.2853 | Up** | 0.0001 |
| L55 | P05183 | Cyp3a2 | Cytochrome P450 3A2 | 1.3085 | Up** | 0.0002 |
| L56 | Q5XIG0 | Nudt9 | ADP-ribose pyrophosphatase, mitochondrial | 1.3121 | Up** | 0.0001 |
| L57 | Q920L7 | Elovl5 | Elongation of very long chain fatty acids protein 5 | 1.3239 | Up** | 0.0021 |
| L58 | P08290 | Asgr2 | Asialoglycoprotein receptor 2 | 1.3334 | Up* | 0.0127 |
| L59 | A0A023IM45 | Psmb8 | Proteasome subunit beta type | 1.3358 | Up** | 0.0020 |
| L60 | Q62730 | Hsd17b2 | Estradiol 17-beta-dehydrogenase 2 | 1.3607 | Up** | 0.0000 |
| L61 | Q31256 | N/A | MHC class I RT1.Au heavy chain | 1.3778 | Up** | 0.0003 |
| L62 | A0A0A1G491 | ND2 | NADH-ubiquinone oxidoreductase chain 2 | 1.3851 | Up* | 0.0195 |
| L63 | P20814 | Cyp2c13 | Cytochrome P450 2C13, male-specific | 1.4111 | Up** | 0.0000 |
| L64 | F1LMF4 | Fat3 | Protocadherin fat 3 | 1.4253 | Up** | 0.0060 |
| L65 | Q4V797 | RGD1309362 | Interferon-gamma-inducible GTPase Ifgga1 protein | 1.4272 | Up** | 0.0000 |
| L66 | P50169 | Rdh3 | Retinol dehydrogenase 3 | 1.4563 | Up** | 0.0000 |
| L67 | A0A0G2K222 | N/A | Uncharacterized protein | 1.5162 | Up | 0.0272 |
| L68 | Q5UAJ6 | COX2 | Cytochrome c oxidase subunit 2 | 1.5316 | Up** | 0.0001 |
| L69 | M0RC39 | Olr796 | Olfactory receptor | 1.5880 | Up* | 0.0484 |
| L70 | D3ZMQ0 | Mga | Protein Mga | 1.6374 | Up* | 0.0312 |
| L71 | Q6T5E9 | Ugt1a6 | UDP-glucuronosyltransferase | 1.7279 | Up** | 0.0003 |
| L72 | A1XF83 | Ugt2b | UDP-glucuronosyltransferase | 1.8256 | Up** | 0.0001 |
| L73 | D3ZXC8 | Ebpl | Emopamil binding protein-like (predicted), isoform CRA_a | 1.8324 | Up** | 0.0018 |
| L74 | F1LM22 | Ugt2b | UDP-glucuronosyltransferase | 1.8894 | Up** | 0.0000 |
| L75 | Q63002 | Igf2r | Mannose 6-phosphate/insulin-like growth factor II receptor | 2.1130 | Up** | 0.0080 |
| L76 | Q5BK88 | Amacr | Alpha-methylacyl-CoA racemase | 2.5845 | Up** | 0.0001 |
 | ||
| Fig. 3 Analysis of enriched gene ontology (A), KEGG pathway (B) and protein–protein interaction (C). | ||
3.3 Common pathways analysis
The common KEGG pathways between proteins and metabolism were steroid hormone biosynthesis, drug metabolism – cytochrome p450, drug metabolism – other enzymes, pentose and glucuronate interconversions, and starch and sucrose metabolism. Among them, the proteins engaged in steroid hormone biosynthesis were Ste, Ugt2b37, Cyp17a1, Cyp3a2, Hsd17b2, Cyp2c13, Ugt1a6 and Ugt2b, and the metabolites were 2-hydroxyestrone and 2-methoxyestrone. The proteins related to drug metabolism – cytochrome p450 were Gsta1, Ugt2b37, mgst3, Ugt1a6 and Ugt2b, and the metabolite was normorphine. The proteins participating in drug metabolism – other enzymes were carboxylic ester hydrolase, Ugt2b37, Ugt1a6 and Ugt2b. The proteins associated with the two pathways were Ugt2b37, Ugt1a6 and Ugt2b, and the metabolites were isovalerylglucuronide and tyramine glucuronide.R. Scrophulariae enhanced 2-hydroxyestrone and 2-methoxyestrone expression in the urine. The direct precursor of 2-methoxyestrone is 2-hydroxyestrone, while the direct precursor of the latter is estrone. Ste levels decreased in the liver. Ste catalyses the transfer reaction from estrone to estrone sulfate and adenosine 3′,5′-diphosphate (PAP).14 PAP accumulation is toxic to several cellular systems.15 In addition, R. Scrophulariae enhanced the levels of Ugt2b37, Ugt1a6, Ugt2b, Cyp3a2 and Cyp2c13 in liver tissue. UDPGT16 is important in the conjugation and subsequent elimination of potentially toxic xenobiotics and endogenous compounds, which catalyse the transfer of glucuronic acid from uridine diphosphoglucuronic acid to a variety of substrates, including steroid hormones. Ugt2b37 participates in the glucuronidation of testosterone and dihydrotestosterone, Ugt1a6 transforms small lipophilic molecules into water-soluble and excretable metabolites, Ugt2b conjugates lipophilic aglycon substrates with glucuronic acid;17 thus, R. Scrophulariae may detoxify liver tissue. Cyp3a2 and Cyp2c13 are important drug metabolic enzymes in rat livers. Cyp3a2 activity was suppressed and appeared in cases of acute formaldehyde poisoning,18 5 week-old Zucker fatty diabetic rats,19 and human immunodeficiency virus-infected rats.20 One previous study has shown that CYP2C13 was absent in male hyperlipidaemic Sprague-Dawley rats.21 However, R. Scrophulariae enhanced CYP2C13 levels in liver tissue, indicating that R. Scrophulariae may have protective effects.
However, down-regulation of carboxylic ester hydrolase and tyramine glucuronide by R. Scrophulariae may be toxic. Carboxylic ester hydrolase participates in phase I metabolism of xenobiotics such as toxins or drugs, and the resulting carboxylates are conjugated by other enzymes to increase solubility and are eventually excreted.22 Tyramine glucuronide is a natural body metabolite of tyramine generated in the liver by UDP glucanosyltransferase.23 Glucuronidation assists in excreting toxic substances, drugs and other substances that cannot be used as an energy source.24 Glucuronic acid25 attaches to the substance via a glycosidic bond, and the resulting glucuronide, which has a higher water solubility than the original substance, is eventually excreted by the kidneys. Therefore, further studies should be conducted on R. Scrophulariae toxicity.
R. Scrophulariae enhanced the level of Hsd17b2, which promotes the interconversion of estrone and oestradiol and regulates the biological activity of sex hormones.26 Oestradiol is essential for reproductive and sexual functioning in women, and it also affects other organs including bones.27 Thus, R. Scrophulariae may generate an oestrogen-like effect by raising Hsd17b2 levels. In addition, oestrogen assimilates protein in the liver and can also impact the male reproductive system, including androgen levels, causing testicular tissue structural changes and testicular cancer, reducing sperm counts, developing male breasts and leading to endocrine disorders.28 Therefore, we must administer R. Scrophulariae appropriately to take advantage of its assimilation rather than its side effects.
In this study, Cyp17a1 and Gsta1 levels were increased by R. Scrophulariae. Cyp17a1 is a prominent inhibitory target in treating prostate cancer because it produces the androgen required for tumour cell growth.29 Studies found that Gsta1 was involved in metabolizing carcinogenic compounds.30 These results may suggest that R. Scrophulariae has potential anti-cancer effects. In urine samples, R. Scrophulariae inhibited normorphine expression, a major metabolite of morphine. It acts directly on the central nervous system (CNS) to diminish sensations of pain.31,32 The analgesic effect from R. Scrophulariae was minimal and likely related to the dosage.
R. Scrophulariae enhanced Gsta1 and Mgst3 expression in liver tissues. Gsta1 exhibits glutathione peroxidase activity, thereby protecting cells from reactive oxygen species and peroxidation products.33 Mgst3 (microsomal glutathione s-transferase 3) is involved in the producing leukotrienes and prostaglandin E, important mediators of inflammation, and it demonstrates glutathione-dependent peroxidase activity towards lipid hydroperoxides.34 Thus, R. Scrophulariae may produce antioxidant effects. However, it is worth noting that increases in serum and urinary Gsta1 have been found associated with hepatocyte and renal proximal tubular necrosis, respectively, and show potential for monitoring injury to these tissues.35 R. Scrophulariae reduced isovalerylglucuronide expression. Elevated isovalerylglucuronide was reported in isovaleric acidaemia,36 indicating that R. Scrophulariae may be used to treat isovaleric acidaemia by decreasing isovalerylglucuronide.
4 Conclusions
Untargeted urine metabolomics were performed by UPLC-TOF-MS, and proteomic liver profiling of R. Scrophulariae-treated rats was detected by nano-UPLC-Q- Exactive-MS/MS. We found that 5 common pathways were targeted by R. Scrophulariae, including steroid hormone biosynthesis, drug metabolism – cytochrome p450, drug metabolism – other enzymes, pentose and glucuronate interconversions, and starch and sucrose metabolism. These results show therapeutic effects as well as side effects. When administering R. Scrophulariae treatment, we should focus on its side effects. Curative effects of R. Scrophulariae included detoxification, anti-cancer, antioxidant and isovaleric acidaemia treatment. Since R. Scrophulariae was characterized by multiple targets and multiple pathways, finding the appropriate basis for its specific pharmacological effects is vital, as this process lays the foundation for clinically accurate and safe medication.Conflicts of interest
There are no conflicts to declare.References
- C. C. Zhang, W. L. Gu, X. M. Wu, Y. M. Li, C. X. Chen and X. Y. Huang, Active components from Radix Scrophulariae inhibits the ventricular remodeling induced by hypertension in rats, SpringerPlus, 2016, 5, 358, DOI:10.1186/s40064-016-1985-z.
- W. L. Gu, C. X. Chen, X. Y. Huang and J. P. Gao, The effect of angoroside C on pressure overload-induced ventricular remodeling in rats, Phytomedicine, 2015, 22(7–8), 705–712, DOI:10.1016/j.phymed.2015.05.002.
- W. L. Gu, C. X. Chen, Q. Wu, J. Lu, Y. Liu and S. J. Zhang, Effects of Chinese herb medicine Radix Scrophulariae on ventricular remodeling, Pharmazie, 2010, 65(10), 770–775 CAS.
- X. Y. Huang, C. X. Chen, X. M. Zhang, Y. Liu, X. M. Wu and Y. M. Li, Effects of ethanolic extract from Radix Scrophulariae on ventricular remodeling in rats, Phytomedicine, 2012, 19(3–4), 193–205, DOI:10.1016/j.phymed.2011.09.079.
- S. Y. Sheu, Y. W. Hong, J. S. Sun, M. H. Liu, C. Y. Chen and C. J. Ke, Radix Scrophulariae extracts (harpagoside) suppresses hypoxia-induced microglial activation and neurotoxicity, BMC Complementary Altern. Med., 2015, 15, 324, DOI:10.1186/s12906-015-0842-x.
- C. Chen, C. X. Chen, X. M. Wu, R. Wang and Y. M. Li, Effects of extracts of Radix Scrophulariae on blood pressure in spontaneously hypertensive rats and the underlying mechanisms, Zhongxiyi Jiehe Xuebao, 2012, 10(9), 1009–1017 Search PubMed.
- X. Shen, T. Eichhorn, H. J. Greten and T. Efferth, Effects of Scrophularia ningpoensis Hemsl. on Inhibition of Proliferation, Apoptosis Induction and NF-kappaB Signaling of Immortalized and Cancer Cell Lines, Pharmaceuticals, 2012, 5(2), 189–208, DOI:10.3390/ph5020189.
- Y. M. Li, Z. H. Han, S. H. Jiang, Y. Jiang, S. D. Yao and D. Y. Zhu, Fast repairing of oxidized OH radical adducts of dAMP and dGMP by phenylpropanoid glycosides from Scrophularia ningpoensis Hemsl, Acta Pharmacol. Sin., 2000, 21(12), 1125–1128 CAS.
- X. Wang, A. Zhang and P. Wang, et al., Metabolomics coupled with proteomics advancing drug discovery toward more agile development of targeted combination therapies, Mol. Cell. Proteomics, 2013, 12(5), 1226–1238 CrossRef CAS PubMed.
- S. Qiu, A. Zhang and T. Zhang, et al., Dissect new mechanistic insights for geniposide efficacy on the hepatoprotection using multiomics approach, Oncotarget, 2017, 8(65), 108760–108770 Search PubMed.
- A. Zhang, X. Zhou and H. Zhao, et al., Metabolomics and proteomics technologies to explore the herbal preparation affecting metabolic disorders using high resolution mass spectrometry, Mol. BioSyst., 2017, 13(2), 320–329 RSC.
- H. Cao, A. Zhang and H. Sun, et al., Metabolomics-proteomics profiles delineate metabolic changes in kidney fibrosis disease, Proteomics, 2015, 15(21), 3699–3710 CrossRef CAS PubMed.
- X. Wang, A. Zhang and G. Yan, et al., Metabolomics and proteomics annotate therapeutic properties of geniposide: targeting and regulating multiple perturbed pathways, PLoS One, 2013, 8(8), e71403 CrossRef CAS PubMed.
- M. P. Thomas and B. V. Potter, The structural biology of oestrogen metabolism, J. Steroid Biochem. Mol. Biol., 2013, 137, 27–49, DOI:10.1016/j.jsbmb.2012.12.014.
- L. Yenush, J. M. Belles, J. M. Lopez-Coronado, R. Gil-Mascarell, R. Serrano and P. L. Rodriguez, A novel target of lithium therapy, FEBS Lett., 2000, 467(2–3), 321–325 CrossRef CAS PubMed.
- S. R. Salinas, A. A. Petruk, N. G. Brukman, M. I. Bianco, M. Jacobs and M. A. Marti, et al., Binding of the substrate UDP-glucuronic acid induces conformational changes in the xanthan gum glucuronosyltransferase, Protein Eng., Des. Sel., 2016, 29(6), 197–207, DOI:10.1093/protein/gzw007.
- T. Izukawa, M. Nakajima, R. Fujiwara, H. Yamanaka, T. Fukami and M. Takamiya, et al., Quantitative analysis of UDP-glucuronosyltransferase (UGT) 1A and UGT2B expression levels in human livers, Drug Metab. Dispos., 2009, 37(8), 1759–1768, DOI:10.1124/dmd.109.027227.
- M. Xu, H. Tang, Q. Rong, Y. Zhang, Y. Li and L. Zhao, et al., The Effects of Formaldehyde on Cytochrome P450 Isoform Activity in Rats, BioMed Res. Int., 2017, 2017, 6525474, DOI:10.1155/2017/6525474.
- S. Y. Park, C. H. Kim, J. Y. Lee, J. S. Jeon, M. J. Kim and S. H. Chae, et al., Hepatic expression of cytochrome P450 in Zucker diabetic fatty rats, Food Chem. Toxicol., 2016, 96, 244–253, DOI:10.1016/j.fct.2016.08.010.
- R. H. Ghoneim and M. Piquette-Miller, Endotoxin-Mediated Downregulation of Hepatic Drug Transporters in HIV-1 Transgenic Rats, Drug Metab. Dispos., 2016, 44(5), 709–719, DOI:10.1124/dmd.115.067827.
- P. J. Brassil, K. Debri, H. Nakura, S. Kobayashi, D. S. Davies and R. J. Edwards, Reduced hepatic expression of CYP7A1 and CYP2C13 in rats with spontaneous hyperlipidaemia, Biochem. Pharmacol., 1998, 56(2), 253–257 CrossRef CAS.
- Y. Chen, D. S. Black and P. J. Reilly, Carboxylic ester hydrolases: classification and database derived from their primary, secondary, and tertiary structures, Protein Sci., 2016, 25(11), 1942–1953, DOI:10.1002/pro.3016.
- K. P. Wong, The biosynthesis of tyramine glucuronide by liver microsomal fractions, Biochem. J., 1977, 164(3), 529–531 CrossRef CAS PubMed.
- H. Kaeferstein, Forensic relevance of glucuronidation in phase-II-metabolism of alcohols and drugs, Leg. Med., 2009, 11(suppl. 1), S22–S26, DOI:10.1016/j.legalmed.2009.01.037.
- S. S. Lewis, M. R. Hutchinson, Y. Zhang, D. K. Hund, S. F. Maier and K. C. Rice, et al., Glucuronic acid and the ethanol metabolite ethyl-glucuronide cause toll-like receptor 4 activation and enhanced pain, Brain, Behav., Immun., 2013, 30, 24–32, DOI:10.1016/j.bbi.2013.01.005.
- E. Hilborn, O. Stal and A. Jansson, Estrogen and androgen-converting enzymes 17beta-hydroxysteroid dehydrogenase and their involvement in cancer: with a special focus on 17beta-hydroxysteroid dehydrogenase type 1, 2, and breast cancer, Oncotarget, 2017, 8(18), 30552–30562, DOI:10.18632/oncotarget.15547.
- L. Tang, Z. Xia, Z. Luo, H. Long, Y. Zhu and S. Zhao, Low plasma PDGF-BB levels are associated with estradiol in postmenopausal osteoporosis, J. Int. Med. Res., 2017, 300060517706630, DOI:10.1177/0300060517706630.
- K. M. Lau and K. F. To, Importance of Estrogenic Signaling and Its Mediated Receptors in Prostate Cancer, Int. J. Mol. Sci., 2016, 17(9), 1434 CrossRef PubMed.
- S. N. Maity, M. A. Titus, R. Gyftaki, G. Wu, J. F. Lu and S. Ramachandran, et al., Targeting of CYP17A1 Lyase by VT-464 Inhibits Adrenal and Intratumoral Androgen Biosynthesis and Tumor Growth of Castration Resistant Prostate Cancer, Sci. Rep., 2016, 6, 35354, DOI:10.1038/srep35354.
- C. Martinez-Guzman, P. Cortes-Reynosa, E. Perez-Salazar and G. Elizondo, Dexamethasone induces human glutathione S transferase alpha 1 (hGSTA1) expression through the activation of glucocorticoid receptor (hGR), Toxicology, 2017, 385, 59–66, DOI:10.1016/j.tox.2017.05.002.
- P. A. Glare, T. D. Walsh and C. E. Pippenger, Normorphine, a neurotoxic metabolite?, Lancet, 1990, 335(8691), 725–726 CrossRef CAS.
- H. Li, R. Tao, J. Wang and L. Xia, Upregulation of miR-375 level ameliorates morphine analgesic tolerance in mouse dorsal root ganglia by inhibiting the JAK2/STAT3 pathway, J. Pain Res., 2017, 10, 1279–1287, DOI:10.2147/jpr.s125264.
- L. G. Higgins and J. D. Hayes, Mechanisms of induction of cytosolic and microsomal glutathione transferase (GST) genes by xenobiotics and pro-inflammatory agents, Drug Metab. Rev., 2011, 43(2), 92–137, DOI:10.3109/03602532.2011.567391.
- F. P. Liu, X. Ma, M. M. Li, Z. Li, Q. Han and R. Li, et al., Hepatoprotective effects of Solanum nigrum against ethanol-induced injury in primary hepatocytes and mice with analysis of glutathione S-transferase A1, J. Chin. Med. Assoc., 2016, 79(2), 65–71, DOI:10.1016/j.jcma.2015.08.013.
- V. Amlabu, C. Mulligan, N. Jele, A. Evans, D. Gray and H. J. Zar, et al., Isoniazid/acetylisoniazid urine concentrations: markers of adherence to isoniazid preventive therapy in children, Int. J. Tuberc. Lung Dis., 2014, 18(5), 528–530, DOI:10.5588/ijtld.13.0730.
- L. Dorland, M. Duran, S. K. Wadman, A. Niederwieser, L. Bruinvis and D. Ketting, Isovalerylglucuronide, a new urinary metabolite in isovaleric acidemia. Identification problems due to rearrangement reactions, Clin. Chim. Acta; international journal of clinical chemistry, 1983, 134(1–2), 77–83 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra10443c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |