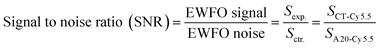Open Access Article
Open Access ArticleNucleic acid functionalized fiber optic probes for sensing in evanescent wave: optimization and application†
Xiyu Zhu‡
a,
Ruoyu Wang‡ a,
Kaidong Xiaab,
Xiaohong Zhou
a,
Kaidong Xiaab,
Xiaohong Zhou *a and
Hanchang Shia
*a and
Hanchang Shia
aState Key Joint Laboratory of ESPC, Research Centre of Environmental and Health Sensing Technology, Center for Sensor Technology of Environment and Health, School of Environment, Tsinghua University, 30 Shuangqing Road, Beijing 100084, China. E-mail: xhzhou@mail.tsinghua.edu.cn; Tel: +86-010-62796952
bNational Key Laboratory of Science and Technology on Micro/Nano Fabrication, Institute of Microelectronics, Peking University, Beijing 100871, China
First published on 18th January 2019
Abstract
Nucleic acid functionalized evanescent wave fiber optic (EWFO) biosensors have attracted much attention due to their remarkable advantages in both device configuration and sensing performance. One critical technique in EWFO biosensor fabrication is its surface modification, which requires (1) minimal nonspecific adsorption and (2) high-quality DNA immobilization to guarantee satisfactory sensing performances. Focusing on these two requirements, a series of optimizations have been conducted in this work to develop reliable DNA-functionalized EWFO probes. Firstly, the surface planeness of EWFO probes were found to be greatly improved by a novel HF/HNO3 mixture etching solution. Both atomic force microscopy (AFM) and X-ray photoelectron spectroscopy (XPS) were conducted to investigate the morphological structures and surface chemical compositions. Besides, EWFO sensing performances adopting moderate immobilization of irrelevant DNA were investigated for optimization purposes. Furthermore, a split aptamer based sandwich-type EWFO sensor was developed using adenosine (Ade) as the model target (LOD = 25 μM). To the best of our knowledge, this study is the first case to focus on the optimization of etching solution compositions in the fabrication of combination tapered fibers, which provides experimental basis for the understanding of the silica-etching mechanism using HF/HNO3 mixture solution and may further inspire related researches.
1. Introduction
Evanescent wave fluorescence biosensors have received much attention for decades because of their remarkable advantages such as robustness, immunity to electromagnetic interference, high sensitivity, high selectivity, high integration, and potential portability for in situ and online measurements.1,2 Optical fibers are cylindrical dielectric waveguides that serve as ideal candidates for evanescent wave transducers by transmitting light via the principle of total internal reflection (TIR). After removal of the cladding on the distal end of fiber, the evanescent field can be exposed, thus exciting fluorophores captured within the sensing region. Meanwhile, the fiber simultaneously serves as the collector of generated fluorescence, enabling the evanescent wave fiber optic (EWFO) biosensors easy to miniaturization.3–6 Therefore, the total fluorescent signal depends not only on fluorescence excited via the evanescent field on the fiber surface but also on the fraction of total emitted fluorescence coupling back into the detectable modes inside the fiber.2 Consequently, it is critical to optimize signal coupling and to reduce unwanted noises so that the relatively weak fluorescent signals can be detected.7Theoretically, removal of cladding results in V number mismatch between the cladded region and the sensing region of fiber immersed into a liquid medium. The V number, which determines the mode capacity of the fiber, is lower in the cladded region than that in the sensing region.8 Hence, the fluorescent signal excited by the evanescent wave in the unclad and immersed region of the fiber is lost when it enters the cladded region of the fiber because of a V number mismatch,7 which can be compensated by a properly designed taper in the fiber.9 Many previous researchers have reported that the adoption of a combination tapered probe instead of other tapered types could result in more significant signal enhancement.7,10,11 The abovementioned efforts greatly facilitated EWFO biosensors for applications in various fields, such as immunoassay based food safety evaluation,12 clinical diagnosis,13,14 and environmental monitoring.15–17 As a counterpart of antibody, nucleic acid based surface affinity recognition receives much attention in term of biosensing. Generally, surface immobilized nucleic acid was specifically designed to capture its complementary strand in solution.18–20 Other functional nucleic acid biomolecules (such as aptamers, DNAzymes) can also be used for detection of various targets, ranging from metal ions to whole cell, with lower price and less strict reaction/storage condition.21–23 Therefore, it is of great significance to develop EWFO sensing systems using nucleic acid as signal recognition element for the enrichment/improvement of biosensors.
One way to produce tapered silica fiber optic probes is to adopt a multi-step procedure that requires a high-precise, computer controlled stage to lower the silica fiber into concentrated hydrofluoric acid (HF) bath over a number of time intervals, which may be time-consuming and potentially non-reproducible.24,25 Another facile and widely accepted way is to produce tapered silica fiber optic probes via capillary motion by inserting the fibers into dilute HF solution for several hours (i.e. tube-etching method).26,27 In previous studies, HF solution was used as the only etchant with varied concentrations (10%, 30% or others). However, we found that facile HF etching might induce irreversible structural damage of fiber, resulting in a coarse surface with irregular grooves.28 Considering the small size, flexible chain structure and special charge distribution of single strand nucleic acid, great blocking challenges appear when facing the harsh interface conditions. Moreover, researches on improvement of surface induced restricted nucleic acid accessibility and hybridization efficiency have been extensively exploited with respect to gold electrode surface,29 silicon oxide substrate,30 flat silica slide,31 untreated distal end of silica fiber,32 and polystyrene microsphere.33 However, systematic investigations are still rare when it comes to nucleic acid immobilized tapered fiber surface.
In this work, reliable DNA-functionalized optic probes for EWFO sensing have been developed based a series of optimizations focusing on two important aspects: (1) smooth fiber surface with small nonspecific adsorption and (2) high-quality immobilization of capture DNA. Firstly, a new type of HF/HNO3 mixture etchant was proposed to circumvent unwanted coarse surface of tapered fiber and to enhance the sensitivity of EWFO system. Moreover, the influence of surface ssDNA immobilization density on EWFO sensing performances was also investigated via adding irrelevant DNA sequences into the complementary ones. Consequently, for the first time, a split aptamer based sandwich-type assay was developed using the optimized nucleic acid functionalized fiber optic probes for facile sensing in the home-made EWFO platform.
2. Experimental
2.1 Chemicals and instrumentation
Silica fibers (99.99% SiO2) with a length (l) of 8.5 cm and a diameter (2R) of 600 μm were purchased from Chunhui Science & Technology Industrial Co., China. The 5.0 cm claddings (h2 + h3) were stripped away from one distal end of the fibers. These fibers were then perpendicularly dipped in different etching solutions (see Table 1 for specific compositions) for about 3 h to form a combination tapered structure with a final diameter (2r) of 220 μm (Fig. S1†). The schematic illustration of etching solution based wet-process for the fabrication of combination tapered silica fiber is shown in Fig. S2.† The etched silica fibers were thoroughly washed with ultrapure water and dried in N2 stream for long-term storage. The instrumental details of the home-made EWFO biosensing platform was described in previous study.22 Flat silica chips (99.99% SiO2, 4 mm × 4 mm × 2 mm) for AFM and XPS characterization were bought from Weida Company, China.All DNA strands (HPLC purified) were synthesized by Sangon Biotech. Co. (Shanghai, China), please see Table 2 for their names and sequences. Basically, amino-modified C6-DNA is designed to be immobilized onto fibers via covalent amide bonds. The sequence of capture DNA is underlined in italic font. CT-Cy5.5 and A20-Cy5.5 were applied in EWFO sensing as complementary and non-complementary strands to the immobilized capture DNA, respectively. See ESI† for other chemicals, buffers and instrument used in this work.
| Name | Sequences (5′–3′) |
|---|---|
| C6-DNA | NH2-(CH2)6-AGAGAGAGAGGGAGAGAGAGAGGG |
| CT-Cy5.5 | Cy5.5-CCCCTCTCTCTCTCTCTCTCTCT |
| A20-Cy5.5 | Cy5.5-AAAAAAAAAAAAAAAAAAAA |
| A10 | NH2-(CH2)6-CCCAAAAAAAAAA |
| ASA1 | NH2-(CH2)6-ACCTGGGGGAGTAT |
| ASA2 | Cy5.5-TGCGGAGGAAGGT |
The diameters of etched fiber optic probes were monitored with a microelectrode polisher (model 2002-C, Inbio Life Science Instrument Co., Ltd., Hubei, China). The surface structures of the etched fiber optic probes were observed by scanning electron microscopy (SEM, JEOL, JSM-700AF). The surface morphologies of SiO2 chips treated by different etching solutions observed by atomic force microscopy (AFM, Bruker Dimension FastScan, German) and the surface chemical state and composition were determined by X-ray photoelectron spectroscopy ESCALAB 250Xi (XPS, USA). All the XPS spectra were calibrated by C 1s line at 284.8 eV and using Al Kα radiation.
2.2 Immobilization of ssDNA strands onto tapered fiber
First of all, aldehyde-functionalized tapered silica fibers were prepared prior to the immobilization of ssDNA strands. Briefly, above-mentioned tapered fibers (etched by solution c) were cleaned and hydroxyl-activated in hot piranha solution (v (H2SO4)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v (30% H2O2) = 3
v (30% H2O2) = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for 1 h at 120 °C. Then the fibers were washed with water and dried in N2 stream. Next, a layer of APTS molecules was covalently linked onto the fibers by immersing them in 1% v/v APTS toluene solution for 2 h at 37 °C. The APTS-activated fibers were washed in toluene and fixed at 200 °C for 1 h. After that, the fibers were immersed in 1% v/v glutaraldehyde aqueous solution overnight at 37 °C to be aldehyde-functionalized.
1) for 1 h at 120 °C. Then the fibers were washed with water and dried in N2 stream. Next, a layer of APTS molecules was covalently linked onto the fibers by immersing them in 1% v/v APTS toluene solution for 2 h at 37 °C. The APTS-activated fibers were washed in toluene and fixed at 200 °C for 1 h. After that, the fibers were immersed in 1% v/v glutaraldehyde aqueous solution overnight at 37 °C to be aldehyde-functionalized.
Since the immobilization quality of capture DNA largely determines the sensitivity and selectivity achieved in EWFO sensing, careful considerations should be made about the ssDNA immobilization strategy adopted. Up to date, three major immobilization techniques have been developed, including physical adsorption, affinity-based (streptavidin–biotin) immobilization, and covalent immobilization.34 Among these techniques, covalent immobilization is the most widely applied strategy owing to multiple advantages: good stability, high binding strength, and long storage term.35,36 In the specific case of covalent ssDNA immobilization, a linker segment is usually needed in order to fully expose capture DNA sequences for target recognition (Fig. S4†). Both linear aliphatic carbon chain and polyhedral nanostructures like tetra-DNA29 are able to serve as linker segments for ssDNA immobilization recognition. In this case, the 6-carbon linker was applied to immobilize ssDNA strands onto tapered fibers (referred to as “C6-DNA” in the following context).
The aldehyde-functionalized silica fibers were immersed in 300 nM of C6-DNA (dissolved in Tris buffer) for 6 h at room temperature with gentle shaking. In this way, ssDNA strands would be covalently immobilized onto fiber surfaces due to the formation Schiff bases, which resulted from the reaction between surface aldehydes and 5′-terminal amino groups of DNAs. Next, unreacted surface aldehyde sites were blocked by 20 mM glycine aqueous solutions for 1 h at room temperature. After that, the formed Schiff bases were further stabilized using 20 mg mL−1 of NaCNBH3 aqueous solution. Before usage in EWFO sensing, all fibers were blocked in 2 mg mL−1 isoelectric BSA solution to reduce nonspecific adsorption of non-target DNA.28
Irrelevant ssDNA (A10) was applied as a class of mixer strand in C6-DNA based ssDNA immobilization. First of all, aldehyde-functionalized silica fibers were obtained following the protocol described in 2.2. Then the fibers were immersed in Tris buffer containing 300 nM of C6-DNA and 100 nM of A10 for 6 h at room temperature with gentle shaking. Both A10 and C6-DNA would be immobilized onto the fiber surface. Other following steps using glycine, NaCNBH3, and isoelectric BSA were same as above.
2.3 EWFO based adenosine (Ade) detection
Based on the optimized nucleic acid functionalized fiber optic probe, surface based sandwich-type biosensor could be developed using split aptamers. In this work, one fragment of adenosine split aptamer (ASA1) was modified onto silica fiber using the method described in 2.2. In EWFO based adenosine (Ade), the ASA1-immobilized fiber was first rinsed in equilibrium buffer to obtain a stable baseline. Then different concentrations of Ade in Ade binding buffer containing 20 nM ASA2 were pumped into the flow cell of EWFO platform. ASA2 was labeled with a Cy5.5 fluorophore to generate fluorescence signals using this EWFO platform. The excitation and emission wavelengths of Cy5.5 fluorophore are 635 nm and 650 nm, respectively. Then the pump was set to stop for 3 min for surface based Ade recognition and fluorescence excitation. After the recognition procedure, unbound ASA2 and Ade were subsequently washed away using washing buffer. The generated EWFO signals were automatically collected by a built-in computer.3. Results and discussion
3.1 Improved etching effect using HF/HNO3 mixture solutions
Silica structures obtained after wet-process adopting HF etching solution often exhibit coarse surfaces with porous silicon layers.37 Although it has been reported the sensitivity of EWFO sensors toward small molecules (glucose) showed a volcanic response with increasing surface roughness,38 porous sensing surfaces may act as physical barriers against macromolecules (proteins, nucleic acids, etc.) in fluids based sensing and further lead to unwanted nonspecific adsorption.28 We attribute the coarse surface by adopting HF etching solution to the following reason. HF etching proceeds through several sequential steps to remove SiO2 as silicon tetrafluoride (SiF4) and fluorosilicic acid (H2SiF6). Eqn (1)–(4) describe the basic chemical reactions involved in HF-based silica fiber etching.37,39,40 H2SiF6 could easily decompose into SiF4 and HF (eqn (5)),40 and the hydrolysis of SiF4 would further produce colloidal silica (orthosilicic acid, H4SiO4, eqn (6)).41 The accumulation of H4SiO4 may lead to the formation of a precipitate layer near the fiber, hindering the further reaction between HF and SiO2.| 4HF + SiO2 → SiF4 + 2H2O | (1) |
| 2HF + SiF4 → H2SiF6 | (2) |
| 2H2O + 3SiF4 → 2H2SiF6 + SiO2 | (3) |
| 6HF + SiO2 → H2SiF6 + 2H2O | (4) |
| H2SiF6 → SiF4 + 2HF | (5) |
| SiF62− + H2O → H4SiO4 + 4HF + 2F− | (6) |
To meet this challenge, we assume that the addition of strong oxidizing nitric acid (HNO3) can promote the decomposition of H2SiF6 and the volatilization of SiF4, thus reducing the hydrolysis of SiF4 and enhancing the sectional flow of solution near the fiber to reduce H4SiO4 precipitate. Therefore, the original HF solution was doped with a certain proportion of HNO3 as new etching solutions (see Table 1 for specific formulations).
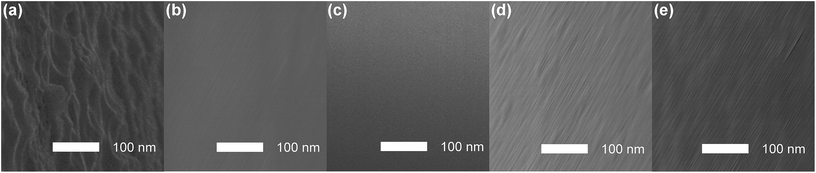
The effect of adopting new etching solutions was analyzed based on scanning electron microscopy (SEM) images (Fig. 1). As shown in Fig. 1a, after pure HF etching, coarse surface with irregular roughness structures could clearly be observed. On the fiber surfaces etched by solution (d) and (e), grooves with fuzzy boundaries could still be observed (Fig. 1d and e); Whereas in the cases using etching solution (b) and (c), such grooves became almost invisible (Fig. 1b and c). Overall, the surface flatness has been improved after adding HNO3 to the original HF etching solution (b–e). The etched fiber surfaces are observed to be smooth in the order of: (c) > (b) > (e) > (d). Therefore, fiber optic probes prepared under the optimal condition (etching solution c) were employed for further studies on morphologies and surface chemical compositions.
3.2 Morphological structures and surface chemical compositions of fibers etched by HF and HF/HNO3 solutions
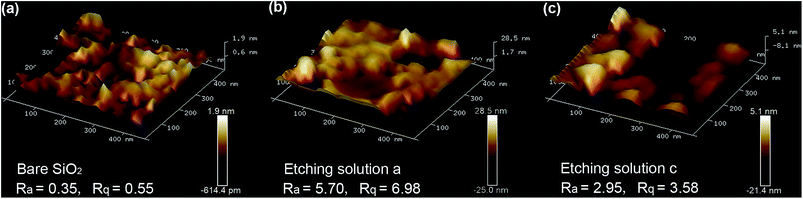
The morphological changes after HF and HF/HNO3 based etching procedures were further studied by means of AFM. Since columnar fiber optic probes may lead to unwanted deviations in AFM testing due to their arcuate surfaces, flat SiO2 chips of the same purity as the fibers were etched using HF (etching solution a) and the optimal HF/HNO3 mixture solution (etching solution c) were further used for AFM tests. As shown in Fig. 2, unetched SiO2 showed a relatively flat surface with a height range (δh) around 1.3 nm, which became larger in the cases based on both HF etching (δh ∼ 53.5 nm) and HF/HNO3 etching (δh ∼ 26.5 nm), indicating that HF/HNO3 mixture solution produced a smoother morphology than HF, although in both cases the roughness increased compared with unetched SiO2. Besides increased δh, two profile roughness parameters, Ra§ and Rq,¶ were also compared to evaluate the impact of different etching agents on the surface roughness. As labeled in Fig. 2, both Ra and Rq followed the order of HF > HF/HNO3 > unetched SiO2, which was consistent with the conclusion obtained after δh analysis.More-detailed chemical and electronic states of the as-prepared HF etching sample and HF/HNO3 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) etching sample were characterized by X-ray photoelectron spectroscopy (XPS) for further investigation of the surface information. The corresponding results are presented in Fig. 3. A full-survey-scan spectrum in Fig. 3a indicates the presence of Si, O, C and F elements after silica chip etched by HF solution. From the survey spectrum of after silica chip etched by HF/HNO3 mixture solution (Fig. 3b), the existence of elements Si, O, C, F and N are also confirmed. C 1s peak existed in both of the survey spectrum are attributed to the contaminated carbonaceous that accumulated during the storage of the sample from the environment.42 The Si 2p, F 1s and O 1s high-resolution spectrum of silica substrate surface etched by pure HF solution and HF/HNO3 (10
3) etching sample were characterized by X-ray photoelectron spectroscopy (XPS) for further investigation of the surface information. The corresponding results are presented in Fig. 3. A full-survey-scan spectrum in Fig. 3a indicates the presence of Si, O, C and F elements after silica chip etched by HF solution. From the survey spectrum of after silica chip etched by HF/HNO3 mixture solution (Fig. 3b), the existence of elements Si, O, C, F and N are also confirmed. C 1s peak existed in both of the survey spectrum are attributed to the contaminated carbonaceous that accumulated during the storage of the sample from the environment.42 The Si 2p, F 1s and O 1s high-resolution spectrum of silica substrate surface etched by pure HF solution and HF/HNO3 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) solution can clearly observed in Fig. 4c–e. As shown in Fig. 3c, a strong major peaks of Si 2p which located at binding energy of 103.3 eV can be detected corresponding to the HF/HNO3 etched sample (red line), it is agreeing well with the spin–orbit peaks of the SiO2 phase.43 The Si 2p spectrum of pure HF etched sample (blue line) can be divided into two fitting peaks and located at 103.5 eV and 104 eV, the first peak at 103.5 eV can also describe as SiO2 and the second peak at 104 eV can be almost entirely ascribed to the characteristic peak of H4SiO4 colloidal polymer configuration.44,45 The comparison of two spectra proved that the addition of HNO3 can promote the decomposition of H2SiF6 and the volatilization of SiF4 to prevent the formation of H4SiO4. According to the measurement, the Si 2p peak area ratio of HF/HNO3 etched sample is wider than that of pure HF etched ones, which can further indicate the assumption that HNO3 prevents the HF etching reaction on the silica surface. Fig. 3d depicts a weak visible peak of F 1s (located at 685.3 eV) in HF etched sample which is associated with residual F−.46 By contrast in the spectrum of HF/HNO3 etched sample cannot observe the peak of F 1s indicate the conclusion of HNO3 as a decomposition promoter. For O 1s spectrum (Fig. 3e), two samples with different etchant have similar binding energy of O 1s peak which located in 532.8 eV and 532.9 eV, respectively. Both of the spectra in accordance with previous data observed for SiO2.47,48 Fig. 3f represent a weak peak at 400.2 eV in the N 1s high-resolution spectrum, which can certify the residual NO3− ion on the silica surface after etched by HF/HNO3 mixture solution.49 Additionally, the Si 2p and O 1s high-resolution spectrums of SiO2 prior to etching are presented in Fig. S3,† the binding energy position of these characteristic peak (located at 532.4 eV in Si 2p, 103.1 eV in O 1s) indicate the high purity of SiO2 phase without any surface etching. Based on the above discussions, the mechanism of two different etchant caused different silica surface roughness can further confirmed.
3) solution can clearly observed in Fig. 4c–e. As shown in Fig. 3c, a strong major peaks of Si 2p which located at binding energy of 103.3 eV can be detected corresponding to the HF/HNO3 etched sample (red line), it is agreeing well with the spin–orbit peaks of the SiO2 phase.43 The Si 2p spectrum of pure HF etched sample (blue line) can be divided into two fitting peaks and located at 103.5 eV and 104 eV, the first peak at 103.5 eV can also describe as SiO2 and the second peak at 104 eV can be almost entirely ascribed to the characteristic peak of H4SiO4 colloidal polymer configuration.44,45 The comparison of two spectra proved that the addition of HNO3 can promote the decomposition of H2SiF6 and the volatilization of SiF4 to prevent the formation of H4SiO4. According to the measurement, the Si 2p peak area ratio of HF/HNO3 etched sample is wider than that of pure HF etched ones, which can further indicate the assumption that HNO3 prevents the HF etching reaction on the silica surface. Fig. 3d depicts a weak visible peak of F 1s (located at 685.3 eV) in HF etched sample which is associated with residual F−.46 By contrast in the spectrum of HF/HNO3 etched sample cannot observe the peak of F 1s indicate the conclusion of HNO3 as a decomposition promoter. For O 1s spectrum (Fig. 3e), two samples with different etchant have similar binding energy of O 1s peak which located in 532.8 eV and 532.9 eV, respectively. Both of the spectra in accordance with previous data observed for SiO2.47,48 Fig. 3f represent a weak peak at 400.2 eV in the N 1s high-resolution spectrum, which can certify the residual NO3− ion on the silica surface after etched by HF/HNO3 mixture solution.49 Additionally, the Si 2p and O 1s high-resolution spectrums of SiO2 prior to etching are presented in Fig. S3,† the binding energy position of these characteristic peak (located at 532.4 eV in Si 2p, 103.1 eV in O 1s) indicate the high purity of SiO2 phase without any surface etching. Based on the above discussions, the mechanism of two different etchant caused different silica surface roughness can further confirmed.
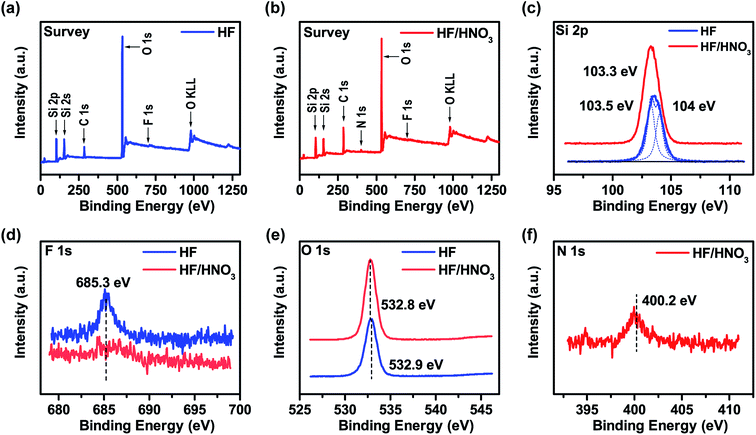 | ||
Fig. 3 XPS spectra of SiO2 etched by HF pure solution and HF/HNO3 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) mixture solution: (a and b) survey; (c) Si 2p; (d) F 1s; (e) O 1s; (f) N 1s. 3) mixture solution: (a and b) survey; (c) Si 2p; (d) F 1s; (e) O 1s; (f) N 1s. | ||
3.3 Comparison of HF and HF/HNO3 etched fiber optic probes with applications to EWFO sensing
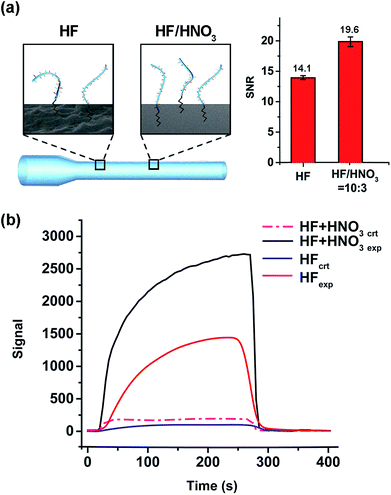
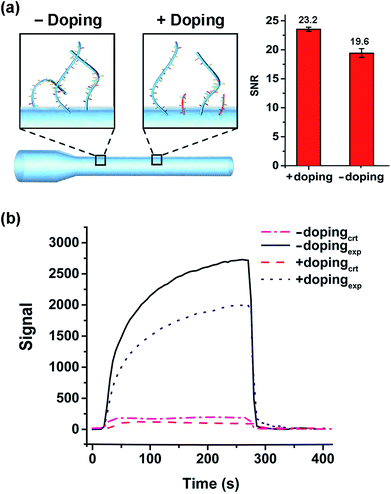
After immobilizing C6-DNA onto fibers (using the method described in 2.2), 10 nM of complementary (CT-Cy5.5) and non-complementary (A20-Cy5.5) DNAs were applied as experimental (exp.) and control (ctr.) groups in EWFO based tests. The control groups were used to represent the nonspecific adsorption on the surface. Based on exp. and ctr. signals generated on different fibers, signal to noise ratios (SNR, also known as S/N) could be calculated according to the following equation as a general indicator to evaluate sensing performances of different fibers.EWFO signals for fibers etched merely by HF and by HF/HNO3 mixture solution (c) were shown in Fig. 4b. The signal amplitude for CT-Cy5.5 has been enhanced in the case of HF/HNO3 mixture solution etched fiber, which may suggest for more effective exposure of capture probe. Besides, nonspecific signal induced by A20-Cy5.5 on both fibers were comparable. Overall, the SNRs are calculated to be 14.1 and 19.6 for fibers etched merely by HF and by HF/HNO3 mixture solution (c), respectively (Fig. 4a, right). The enhanced SNR quantitatively indicated the beneficial effect of the developed HF/HNO3 mixture etching solution. To the best of our knowledge, this study is the first case to focus on the optimization of etching solution compositions in the fabrication of combination tapered fibers, which provides an experimental basis for the understanding of silica-etching mechanism using HF/HNO3 mixture solution and may further inspire other related research.
3.4 Effect of irrelevant ssDNA mixer added immobilization on EWFO hybridization signals
Besides etching technology, the immobilization chemistry, such as ssDNA immobilization density, also plays an important role in high-quality EWFO sensing:31 high surface density may be beneficial to obtain high hybridization signal,33 but it may result in mediocre hybridization efficiency due to steric/electrostatic hindrance; conversely, extremely low immobilization density may lead to undetectable signals hidden beneath background noises.50 One simple but practical method to adjust DNA immobilization density is to mix a certain amount of irrelevant short-chain DNA during the immobilization procedure. We tested the effect of mixing-based immobilization on EWFO hybridization signals using A10 as mixer agent (Fig. 5a, left). As shown in Fig. 5, although the signal amplitude decreased to some extent, the SNR increased by ∼20% simply by mixing A10 strands during immobilization. The influences of the specific type, length, GC content, and adding amount of mixers on EWFO sensing performances are under investigation. Within the scope of this work, A10 mixed immobilization was employed for further sensing applications. Besides, the quantification of functional groups or molecules on the surface is a key point regarding the quality of the sensing surface.51 However, considering the low specific surface area of fiber probes in this work, traditional quantification methods like ninhydrin tests are not sensitive enough to calculate the small amount of chemical groups modified on the surface. In this work, the successful modification can be proved by subsequential sensing performances in a qualitative way.3.5 EWFO based adenosine (Ade) detection
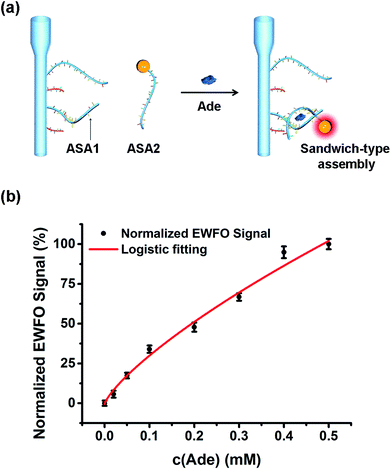
In the past decades, functional DNAs showing affinity toward various types of targets have been vastly developed and applied as sensors.52,53 As one type of functional DNAs, split aptamers are two nucleic acid fragments that can assemble together in the presence of specific target. This feature makes split aptamers particularly attractive in developing surface-based sandwich-type biosensor.54 By tethering one fragment of split aptamer onto fiber, the detection scope of EWFO system could be expanded to non-nucleic acid targets based on sandwich-type biosensing. A small molecule, adenosine (Ade), was used as the model target in this work. As shown in Fig. 6a, one fragment of the reported Ade split aptamer55 (ASA1) was immobilized onto the fiber using our optimized strategy to capture fluorescently labeled ASA2 via the formation of a sandwich-type ASA1/Ade/ASA2 assembly. Fluorophores labeled on ASA2 could be excited within the evanescent field around the fiber surface, generating EWFO signals that are positively correlated with the concentrations of Ade. The calibration curve of Ade was shown in Fig. 6b. After non-linear four-parameter logistic fitting, the LODAde was calculated to be 25 μM with a linear range from 50 μM to 3.5 mM. This sensitivity is appropriate to biomedical test applications and comparable with other fluorescence-based aptasensor,56 electrochemical method,57 and colorimetric method adopting the same split aptamer fragments.54 Additional amplification strategies could be integrated to further improve the sensing performance.544. Conclusions
In summary, reliable DNA-functionalized optic probes for sensing in evanescent wave have been developed based a series of optimizations on the etching solution and immobilization chemistry. Two major findings of this work include: (1) for the first time, HF/HNO3 mixture etching solution was adopted to prepare high-quality combination tapered fiber probes with enhanced sensing performances, and the composition of the etching solution was optimized to be v (HF)/v (HNO3) = 10/3; (2) moderate addition of irrelevant ssDNA onto tapered fiber was benefit to further improve the sensitivity of EWFO sensing. Under the optimized experimental conditions, a split aptamer based sandwich-type EWFO sensing system was developed using Ade molecule as the model target, reaching a LOD of 25 μM. The developed sandwich-type sensing system could serve as a universal platform for other targets of interest and act as a powerful analytical device in the fields of environmental science, personal health care product development, clinical diagnosis and others. Moreover, this work may provide useful information/inspirations for other related researches, especially for silica surface based sensors using nucleic acids.Conflicts of interest
None.Acknowledgements
This research is supported by special fund of State Key Joint Laboratory of Environment Simulation and Pollution Control (16Y04ESPCT), National Nature Science Foundation (21677082) and Shanghai Tongji Gao Tingyao Environmental Science & Technology Development Foundation in China.References
- C. R. Taitt, G. P. Anderson and F. S. Ligler, Evanescent wave fluorescence biosensors, Biosens. Bioelectron., 2005, 20, 2470–2487 CrossRef CAS PubMed.
- C. R. Taitt, G. P. Anderson and F. S. Ligler, Evanescent wave fluorescence biosensors: advances of the last decade, Biosens. Bioelectron., 2016, 76, 103–112 CrossRef CAS PubMed.
- R. Orghici, U. Willer, M. Gierszewska, S. Waldvogel and W. Schade, Fiber optic evanescent field sensor for detection of explosives and CO2 dissolved in water, Appl. Phys. B: Lasers Opt., 2008, 90, 355–360 CrossRef CAS.
- A. Leung, P. M. Shankar and R. Mutharasan, A review of fiber-optic biosensors, Sens. Actuators, B, 2007, 125, 688–703 CrossRef CAS.
- X. D. Wang and O. S. Wolfbeis, Fiber-optic chemical sensors and biosensors (2008-2012), Anal. Chem., 2013, 85, 487–508 CrossRef CAS PubMed.
- Y. Xiong, J. Tan, C. Wang, Y. Zhu, S. Fang and Q. Wang, et al., A miniaturized oxygen sensor integrated on fiber surface based on evanescent-wave induced fluorescence quenching, J. Lumin., 2016, 179, 581–587 CrossRef CAS.
- J. P. Golden, L. C. Shriver-Lake, G. P. Anderson, R. B. Thompson and F. S. Ligler, Fluorometer and tapered fiber optic probes for sensing in the evanescent wave, Opt. Eng., 1992, 31, 1458–1462 CrossRef.
- N. Nidhi and A. Sneh, Evanescent wave fiber optic fluorosensor: effect of tapering configuration on the signal acquisition, Opt. Eng., 1998, 37, 220–228 CrossRef.
- T. R. Glass, S. Lackie and T. Hirschfeld, Effect of numerical aperture on signal level in cylindrical waveguide evanescent fluorosensors, Appl. Opt., 1987, 26, 2181–2187 CrossRef CAS PubMed.
- G. P. Anderson, J. P. Golden and F. S. Ligler, An evanescent wave biosensor. I. Fluorescent signal acquisition from step-etched fiber optic probes, IEEE Trans. Biomed. Eng., 1994, 41, 578–584 CrossRef CAS PubMed.
- J. P. Golden, G. P. Anderson, S. Rabbany and F. Ligler, An evanescent wave biosensor. II. Fluorescent signal acquisition from tapered fiber optic probes, IEEE Trans. Biomed. Eng., 1994, 41, 585–591 CrossRef CAS PubMed.
- S. H. Ohk and A. K. Bhunia, Multiplex fiber optic biosensor for detection of Listeria monocytogenes, Escherichia coli O157: H7 and Salmonella enterica from ready-to-eat meat samples, Food Microbiol., 2013, 33, 166–171 CrossRef CAS PubMed.
- R. Xiao, Z. Rong, S. Chen, W. Chen and S. Wang, Optic fiber-based immunosensor for the rapid and sensitive detection of hepatitis C virus in serum, RSC Adv., 2014, 4, 36125–36130 RSC.
- R. Kapoor and C. W. Wang, Highly specific detection of interleukin-6 (IL-6) protein using combination tapered fiber-optic biosensor dip-probe, Biosens. Bioelectron., 2009, 24, 2696–2701 CrossRef CAS PubMed.
- H. Q. Yin, R. Xiao, Z. Rong, P. P. Jin, C. F. Ji and J. G. Zhang, Establishment of evanescent wave fiber-optic immunosensor method for detection bluetongue virus, Methods, 2015, 90, 65–67 CrossRef CAS PubMed.
- K. D. King, J. M. Vanniere, J. L. LeBlanc, K. E. Bullock and G. P. Anderson, Automated fiber optic biosensor for multiplexed immunoassays, Environ. Sci. Technol., 2000, 34, 2845–2850 CrossRef CAS.
- Y. Lin, D. Li, S. Zeng and M. He, Changes of microbial composition during wastewater reclamation and distribution systems revealed by high-throughput sequencing analyses, Front. Environ. Sci. Eng., 2016, 10, 539–547 CrossRef CAS.
- X. Zhu, R. Wang, X. Zhou and H. Shi, Free-energy-driven lock/open assembly-based optical DNA sensor for cancer-related microRNA detection with a shortened time-to-result, ACS Appl. Mater. Interfaces, 2017, 9, 25789–25795 CrossRef CAS PubMed.
- A. V. Hine, X. Chen, M. D. Hughes, K. Zhou, E. Davies and K. Sugden, et al., Optical fibre-based detection of DNA hybridization, Biochem. Soc. Trans., 2009, 37, 445–449 CrossRef CAS PubMed.
- F. Long, S. Wu, M. He, T. Tong and H. Shi, Ultrasensitive quantum dots-based DNA detection and hybridization kinetics analysis with evanescent wave biosensing platform, Biosens. Bioelectron., 2011, 26, 2390–2395 CrossRef CAS PubMed.
- R. Wang, Y. Xiang, X. Zhou, L.-h. Liu and H. Shi, A reusable aptamer-based evanescent wave all-fiber biosensor for highly sensitive detection of Ochratoxin A, Biosens. Bioelectron., 2015, 66, 11–18 CrossRef CAS PubMed.
- R. Wang, X. Zhou and H. Shi, Triple functional DNA–protein conjugates: signal probes for Pb2+ using evanescent wave-induced emission, Biosens. Bioelectron., 2015, 74, 78–84 CrossRef CAS PubMed.
- R. Wang, X. Zhou, H. Shi and Y. Luo, T–T mismatch-driven biosensor using triple functional DNA-protein conjugates for facile detection of Hg2+, Biosens. Bioelectron., 2016, 78, 418–422 CrossRef CAS PubMed.
- S. Pilevar, C. C. Davis, A. J. Fielding and F. Portugal, Optical fiber evanescent field excited fluorosensor and method of manufacture, US Pat., US6558958B1, 2003 Search PubMed.
- K. Zhou, X. Chen, L. Zhang and I. Bennion, Implementation of optical chemsensors based on HF-etched fibre Bragg grating structures, Meas. Sci. Technol., 2006, 17, 1140–1145 CrossRef CAS.
- F. Long, M. He, H. Shi and A. Zhu, Development of evanescent wave all-fiber immunosensor for environmental water analysis, Biosens. Bioelectron., 2008, 23, 952–958 CrossRef CAS PubMed.
- L. H. Liu, X. H. Zhou and H. C. Shi, Portable optical aptasensor for rapid detection of mycotoxin with a reversible ligand-grafted biosensing surface, Biosens. Bioelectron., 2015, 72, 300–305 CrossRef CAS PubMed.
- R. Wang, X. Zhou, X. Zhu, C. Yang, L. Liu and H. Shi, Isoelectric bovine serum albumin: robust blocking agent for enhanced performance in optical-fiber based DNA sensing, ACS Sens., 2017, 2, 257–262 CrossRef CAS PubMed.
- Y. Wen, H. Pei, Y. Wan, Y. Su, Q. Huang and S. Song, et al., DNA nanostructure-decorated surfaces for enhanced aptamer-target binding and electrochemical cocaine sensors, Anal. Chem., 2011, 83, 7418–7423 CrossRef CAS PubMed.
- V. Dugas, G. Depret, Y. Chevalier, X. Nesme and É. Souteyrand, Immobilization of single-stranded DNA fragments to solid surfaces and their repeatable specific hybridization: covalent binding or adsorption?, Sens. Actuators, B, 2004, 101, 112–121 CrossRef CAS.
- E. Arslan and I. J. Laurenzi, An efficient algorithm for the stochastic simulation of the hybridization of DNA to microarrays, BMC Bioinf., 2009, 10, 411–427 CrossRef PubMed.
- J. A. Ferguson, T. C. Boles, C. P. Adams and D. R. Walt, A fiber-optic DNA biosensor microarray for the analysis of gene expression, Nat. Biotechnol., 1996, 14, 1681–1684 CrossRef CAS PubMed.
- J. A. Ferguson, F. J. Steemers and D. R. Walt, High-density fiber-optic DNA random microsphere array, Anal. Chem., 2000, 72, 5618–5624 CrossRef CAS PubMed.
- S. B. Nimse, K. Song, M. D. Sonawane, D. R. Sayyed and T. Kim, Immobilization techniques for microarray: challenges and applications, Sensors, 2014, 14, 22208–22229 CrossRef PubMed.
- T. Y. Lee and Y. B. Shim, Direct DNA hybridization detection based on the oligonucleotide-functionalized conductive polymer, Anal. Chem., 2001, 73, 5629–5632 CrossRef CAS PubMed.
- V. Pavlov, Y. Xiao, R. Gill, A. Dishon, M. Kotler and I. Willner, Amplified chemiluminescence surface detection of DNA and telomerase activity using catalytic nucleic acid labels, Anal. Chem., 2004, 76, 2152–2156 CrossRef CAS PubMed.
- T. Unagami, Formation mechanism of porous silicon layer by anodization in HF solution, J. Electrochem. Soc., 1980, 127, 476–483 CrossRef CAS.
- N. Zhong, X. Zhu, Q. Liao, Y. Wang, R. Chen and Y. Sun, Effects of surface roughness on optical properties and sensitivity of fiber-optic evanescent wave sensors, Appl. Opt., 2013, 52, 3937–3945 CrossRef PubMed.
- J. K. Kang and C. B. Musgrave, The mechanism of HF/H2O chemical etching of SiO2, J. Chem. Phys., 2002, 116, 275–280 CrossRef CAS.
- Y. M. Hsu, C. Y. Wu, D. A. Lundgren, J. W. Nall and B. K. Birky, Chemical characteristics of aerosol mists in phosphate fertilizer manufacturing facilities, J. Occup. Environ. Hyg., 2007, 4, 17–25 CrossRef CAS PubMed.
- C. Crowe, J. Masmonteil and R. Thomas, Trends in matrix acidizing, Oilfield Rev., 1992, 4, 22–40 Search PubMed.
- L. Li, J. Xu, J. Lei, J. Zhang, F. McLarnon and Z. Wei, et al., A one-step, cost-effective green method to in situ fabricate Ni(OH)2 hexagonal platelets on Ni foam as binder-free supercapacitor electrode materials, J. Mater. Chem. A, 2015, 3, 1953–1960 RSC.
- M. Sivakumar, K. Venkatakrishnan and B. Tan, Characterization of MHz pulse repetition rate femtosecond laser-irradiated gold-coated silicon surfaces, Nanoscale Res. Lett., 2011, 6, 78 CrossRef PubMed.
- E. Brinley, S. Seal, R. Folks, E. Braunstein, L. Kramer and S. Seal, High efficiency SiO2–TiO2 hybrid sol-gel antireflective coating for infrared applications, J. Vac. Sci. Technol., A, 2006, 24, 1141–1146 CrossRef CAS.
- J. A. Taylor, G. M. Lancaster, A. Ignatiev and J. W. Rabalais, Interactions of ion beams with surfaces. Reactions of nitrogen with silicon and its oxides, J. Chem. Phys., 1978, 68, 1776–1784 CrossRef CAS.
- R. Miao, J. Yang, Z. Xu, J. Wang, Y. Nuli and L. Sun, A new ether-based electrolyte for dendrite-free lithium-metal based rechargeable batteries, Sci. Rep., 2016, 6, 21771 CrossRef CAS PubMed.
- T. V. Larina, L. S. Dovlitova, V. V. Kaichev, V. V. Malakhov, T. S. Glazneva and E. A. Paukshtis, et al., Influence of the surface layer of hydrated silicon on the stabilization of Co2+ cations in Zr–Si fiberglass materials according to XPS, UV-Vis DRS, and differential dissolution phase analysis, RSC Adv., 2015, 5, 79898–79905 RSC.
- A. Zatsepin, D. Zatsepin, D. Boukhvalov, N. Gavrilov, V. Y. Shur and A. Esin, The MRO-accompanied modes of Re-implantation into SiO2-host matrix: XPS and DFT based scenarios, J. Alloys Compd., 2017, 728, 759–766 CrossRef CAS.
- A. Kormányos, M. S. Hossain, F. W. Foss, C. Janáky and K. Rajeshwar, Electrocatalytic behavior of freely-diffusing and immobilized synthetic flavins in aqueous media, Catal. Sci. Technol., 2016, 6, 8441–8448 RSC.
- A. W. Peterson, R. J. Heaton and R. M. Georgiadis, The effect of surface probe density on DNA hybridization, Nucleic Acids Res., 2001, 29, 5163–5168 CrossRef CAS PubMed.
- Y. Vida, M. I. Montanez and D. Collado, Dendrimeric antigen–silica particle composites: an innovative approach for IgE quantification, J. Mater. Chem. B, 2013, 1, 3044–3050 RSC.
- Y. Lu and J. Liu, Functional DNA nanotechnology: emerging applications of DNAzymes and aptamers, Curr. Opin. Biotechnol., 2006, 17, 580–588 CrossRef CAS PubMed.
- J. Liu, Z. Cao and Y. Lu, Functional nucleic acid sensors, Chem. Rev., 2009, 109, 1948–1998 CrossRef CAS PubMed.
- A. Chen, M. Yan and S. Yang, Split aptamers and their applications in sandwich aptasensors, TrAC, Trends Anal. Chem., 2016, 80, 581–593 CrossRef CAS.
- F. Li, J. Zhang, X. Cao, L. Wang, D. Li and S. Song, et al., Adenosine detection by using gold nanoparticles and designed aptamer sequences, Analyst, 2009, 134, 1355–1360 RSC.
- Y. Bai, F. Feng, L. Zhao, Z. Chen, H. Wang and Y. Duan, A turn-on fluorescent aptasensor for adenosine detection based on split aptamers and graphene oxide, Analyst, 2014, 139, 1843–1846 RSC.
- X. Zuo, Y. Xiao and K. W. Plaxco, High specificity, electrochemical sandwich assays based on single aptamer sequences and suitable for the direct detection of small-molecule targets in blood and other complex matrices, J. Am. Chem. Soc., 2009, 131, 6944–6945 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra10125f |
| ‡ X. Z. and R. W. contributed equally to this work and should be considered as co first authors. |
| § Ra is the average of the arithmetic absolute values of the profile heights over the evaluation length. |
| ¶ Rq is the root mean square average of the profile heights over the evaluation length. |
| This journal is © The Royal Society of Chemistry 2019 |