Open Access Article
Open Access ArticleEfficient synthesis of Ibrutinib chiral intermediate in high space-time yield by recombinant E. coli co-expressing alcohol dehydrogenase and glucose dehydrogenase†
Yitong Chen,
Baodi Ma *,
Songshuang Cao,
Xiaomei Wu and
Yi Xu*
*,
Songshuang Cao,
Xiaomei Wu and
Yi Xu*
School of Chemical and Environmental Engineering, Shanghai Institute of Technology, 100 Haiquan Road, Shanghai 201418, China. E-mail: xuyi@sit.edu.cn; mabaodi2005@163.com
First published on 18th January 2019
Abstract
The production of (S)-N-boc-3-hydroxy piperidine (NBHP) via asymmetric bioreduction of 1-boc-3-piperidinone with reductase is impeded by the need for expensive coenzymes NAD(P)H. In order to regenerate the coenzyme in situ, the gene of alcohol dehydrogenase from Thermoanaerobacter brockii and glucose dehydrogenase from Bacillus subtilis were ligated into the multiple cloning sites of pRSFDuet-1 plasmid to construct the recombinant Escherichia BL21 (DE3) that co-expressing alcohol dehydrogenase and glucose dehydrogenase. Different culture conditions including the medium composition, inducer and pH etc were systematically investigated to improve the enzyme production. The enzyme activity was increased more than 11-fold under optimal culture condition, from 12.7 to 139.8 U L−1. In the further work, the asymmetric reduction of 1-boc-3-piperidinone by whole cells of recombinant E. coli was systematic optimized to increase the substrate concentration and reaction efficiency. At last, S-NBHP (>99% ee) was prepared at 500 mM substrate concentration without external addition of cofactors. The conversion of S-NBHP reached 96.2% within merely 3 h, corresponding a high space-time yield around 774 g L−1 d−1. All these results demonstrated the potential of recombinant E. coli BL21 (DE3) coupled expressing alcohol dehydrogenase and glucose dehydrogenase for efficient synthesis of S-NBHP.
1. Introduction
Chiral hydroxyl piperidines and their derivatives are of great importance in the pharmaceutical industry and many of them have pharmaceutical activity, such as anti-tumor agents, anti-biotics and anti-senile dementia agents.1 Many bioactive molecules and active pharmaceutical ingredients (API) contain one or more piperidine rings.2,3 Ibrutinib is a targeted anti-cancer drug that inhibits Bruton tyrosine kinase for the treatment of mantle cell lymphoma. (S)-N-boc-3-hydroxy piperidine (NBHP) is a key intermediate in the synthesis of Ibrutinib.Currently, several approaches have been developed for the synthesis of chiral alcohol, including classic diastereomeric resolution,4 asymmetric synthesis,5 and asymmetric reduction.6 Of these methods, asymmetric bioreduction has attracted extensive attention due to the high stereoselectivity, mild condition and 100% theoretical yield. Asymmetric bioreduction has been successfully applied in the preparation of (S)-(4-chlorophenyl)-(pyridin-2-yl)methanol,7 (R)-1-phenyl-1,2-ethanediol8 and ethyl (R)-4-chloro-3-hydroxybutanoate9 etc. However, there were few reports about the synthesis of (S)-N-boc-3-hydroxy piperidine (NBHP) via asymmetric bioreduction. Romain Lacheretz et al.10 used the tissue of Daucus carota as biocatalyst to reduce 1-boc-3-piperidinone, and moderate chiral purity (ee 95%) and low yield of product (73%) was obtained. Other reductases reported to reduce 1-boc-3-piperidinone more efficiently were the alcohol dehydrogenase from Thermoanaerobacter brockii,11 and KR-110 screened form a commercial ketoreductase library.12 However, expensive exogenous coenzyme has to be added to the reaction system to allow the reaction to proceed.
The expensive cofactor NAD(P)H was required in the bioreduction process, thus its regeneration attracted extensive attention in the last few decades. As early as the 1980s, researchers began to develop different cofactor regeneration systems, including biological (intracellular coenzyme regeneration, substrate coupling method and enzyme coupling method), electrochemical, photochemical and pure chemistry and so on. Currently, enzyme coupling and substrate coupling have been favoured.13,14 The reduction of the substrate and the regeneration of the coenzyme are performed by two different enzymes in the double enzyme-coupled system. At present, formate dehydrogenase (FDH) and glucose dehydrogenase (GDH) mediated coenzyme regeneration system proved to be feasible in the industrial application.15 There are many successful examples using GDH to construct a coenzyme regeneration system. For instance, Gröger et al. had constructed a ‘Designer cell’ containing GDH from Bacillus subtilis and ADH from Rhodococcus erythropolis, and it had been used for the asymmetric reduction of a variety of aryl ketones in the absence of exogenous coenzyme.16,17 However, no such report on the production of S-NBHP was found.
The alcohol dehydrogenase from Thermoanaerobacter brockii (TbADH) was an attractive ADH in the biotechnology due to its high thermostability, excellent activity and enantioselectivity for the reduction of 1-boc-3-piperidinone.11 In current research, a novel two-enzyme (ADH and GDH) co-overexpressing system was developed for efficient asymmetric reduction of 1-boc-3-piperidinone without needing exogenous coenzyme. The vector pRSFDuet-1 contains two multiple cloning sites and each of them is preceded by a T7 promoter and ribosome binding site. In present work, the gene TbADH18 and glucose dehydrogenase from Bacillus subtilis (BsGDH)19 were designed to ligate into the multiple cloning sites (MCS) of vector pRSFDuet-1 and construct a recombinant E. coli BL21 (DE3) with both TbADH and BsGDH activity. S-NBHP can be prepared via the asymmetric bioreduction of 1-boc-3-piperidine catalyzed by the TbADH and the coenzyme NADPH can be recycled by the BsGDH and cosubstrate glucose in situ (Scheme 1). The enzyme production and reaction efficiency were further improved via the optimization of microorganism cultivation and reaction condition, respectively.
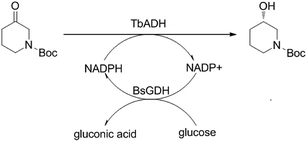 | ||
| Scheme 1 Asymmetric reduction of 1-boc-3-piperidinone to (S)-N-boc-3-hydroxy piperidine by recombinant E. coli with both TbADH and BsGDH activity. | ||
2. Experimental
2.1 Chemicals
Starch and yeast power were purchased from Aladdin Industrial Corporation (Shanghai, China). Tryptone was supplied by China National Medicines Co. Ltd. (Beijing, China). Unless otherwise stated, all other chemicals were obtained from commercial sources and were of analytical grade.2.2 Strains and culture conditions
The LB medium was used as seed medium, and the composition was (g L−1): yeast power 5; tryptone 10; NaCl 10; pH 7.0. The fermentation medium was optimized based on LB medium.Recombinant E. coli BL21 (DE3) cells was inoculated into LB medium containing 50 mg L−1 kanamycin and cultivated at 37 °C for 12 h on a rotary shaker (200 rpm). Then the seed culture was inoculated into 50 mL fermentation medium containing 50 mg L−1 kanamycin and grown at 37 °C. After 3 h, IPTG was added to the culture with final concentration of 0.025 mM, and then the recombinant E. coli were continue cultivated at 40 °C for another 10 h for protein expression.
2.3 Construction of co-expression system of TbADH and BsGDH
The TbADH and BsGDH gene were synthesized by the Generay Biotech (Shanghai) Co., Ltd. The exogenous gene was ligated into the first multi clone site (MCS) via BamH/HindIII restriction sites, and the second MCS via the NdeI/XhoI restriction sites. The construction of recombinant pRSFDuet-1 plasmid and its transformation into E. coli were performed according to standard protocols.2.4 Enzyme assay
To assay the activity of recombinant E. coli for reducing 1-boc-3-piperidinone, 10 mg of dry cell pellet was suspended in 950 μL phosphate buffer (pH 7.0, 200 mM) which contained 0.5% glucose. The mixture was incubated at 30 °C and 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm on thermo-mixer compact (Zuofei Co., China) for 15 min. Then the substrate was added into the mixture with a final concentration of 50 mM and the reaction was carried out for 15 minutes under the same conditions. The sample was extracted twice with equal volume of ethyl acetate. The combined extracts were dried over anhydrous sodium sulfate and analyzed by gas chromatography (GC).
000 rpm on thermo-mixer compact (Zuofei Co., China) for 15 min. Then the substrate was added into the mixture with a final concentration of 50 mM and the reaction was carried out for 15 minutes under the same conditions. The sample was extracted twice with equal volume of ethyl acetate. The combined extracts were dried over anhydrous sodium sulfate and analyzed by gas chromatography (GC).
2.5 Optimization of culture conditions for recombinant E. coli BL21 (DE3)
The media composition and culture condition for recombinant E. coli were optimized through the ‘one-factor-at-a-time’.20,21 The factors of carbon source, nitrogen source, inducer, pH and temperature were varied in each experiment, and the corresponding cell mass and enzyme activity after cultivation were determined as described above.2.6 Asymmetric reduction of N-boc-3-piperidone catalyzed by recombinant E. coli co-expressing alcohol dehydrogenase and glucose dehydrogenase
Whole cell of recombinant E. coli was used as biocatalyst in this study. The collected cells were resuspended into the phosphate buffer (pH 7.0, 200 mM) and were incubated at 30 °C for 15 min. Then, adequate amount of substrate, glucose, and NADP+ were added into the suspension. The reaction mixture was incubated at 30 °C and 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm on thermo-mixer compact (Zuofei Co., China). The reaction conditions including the initial pH, temperature, co-solvent, the amount of glucose and NADP+ were optimized in this research. When the substrate concentration was increased to 500 mM, Na2CO3 solution (1 M) or ammonia solution (5 M) was added to neutralize the gluconic acid formed during the reaction.
000 rpm on thermo-mixer compact (Zuofei Co., China). The reaction conditions including the initial pH, temperature, co-solvent, the amount of glucose and NADP+ were optimized in this research. When the substrate concentration was increased to 500 mM, Na2CO3 solution (1 M) or ammonia solution (5 M) was added to neutralize the gluconic acid formed during the reaction.
2.7 Analytical methods
The conversion of the sample was analyzed by GC (GC-2010, Shimadzu Co., Japan) equipped with a β-DEX™ 120 Capillary Column (30 m × 0.25 mm × 0.25 μm, Supelco, Sigma-Aldrich Co., USA). The carrier gas is nitrogen, and total flow is 20.0 mL min−1. The temperature of detector is set as 280 °C and injector's temperature is 230 °C. The oven program of the column temperature is as follows: 1 min at 100 °C, first ramp at 5 °C min−1 to 150 °C (hold for 2 min), and second ramp at 2 °C min−1 to 160 °C (hold for 2 min). The whole analysis process is 35 minutes. The retention time for 1-boc-3-piperidinone and NBHP were 28.9 min and 30.9 min, respectively.The ee value of the product was analyzed by HPLC (LC 20AT, Shimadzu Co., Japan) using a chiral OD-H column (4.6 mm × 250 mm, Daicel Co., Japan). The mobile phase was hexane/isopropanol (97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, by vol.). The extracted and dried samples were analyzed at 27 °C with the flow rate of 0.8 mL min−1, and the detection wavelength is 210 nm. The retention time for S and R-NBHP were 12.8 min and 13.9 min, respectively.
3, by vol.). The extracted and dried samples were analyzed at 27 °C with the flow rate of 0.8 mL min−1, and the detection wavelength is 210 nm. The retention time for S and R-NBHP were 12.8 min and 13.9 min, respectively.
2.8 Preparative synthesis of S-NBHP
The preparative reduction of 1-boc-3-piperidinone was conducted at the substrate concentration of 0.5 M in a 20 ml aqueous buffer system containing 1 g recombinant E. coli. Glucose of 2.7 g was added for the NADPH regeneration. The reaction was carried out at 50 °C and ammonia solution (5 M) was periodical added to the reaction system for neutralizing the gluconic acid formed during the reaction. After the substrate was completely converted into product, the reaction mixture was extracted twice with equal volumes of ethyl acetate. Then the organic phase was combined, dried over anhydrous sodium sulphate and then evaporated in vacuum. The obtained yellow liquid was validated based on the chiral HPLC and 1H NMR spectra.3. Results and discussion
3.1 Construction of the recombinant plasmids co-expressing alcohol dehydrogenase and glucose dehydrogenase
Two kinds of recombinant pRSFduet-1 with different position of TbADH and BsGDH gene, pRSFduet-1-tbadh-bsgdh and pRSFduet-1-bsgdh-tbadh were constructed (Fig. S1†) and transformed into E. coli BL21(DE3). The expression of target protein was induced by addition of 0.1 mM IPTG. The enzyme activity for asymmetric reduction of 1-boc-3-piperidinone was determined as described in 2.4 to compare the efficiencies of these two kinds recombinant E. coli and the results were shown in the Table S1†. Both of the specific activity and activity of recombinant E. coli harboring pRSFduet-1-tbadh-bsgdh was inferior to the recombinant E. coli harboring pRSFduet-1-bsgdh-tbadh. The enzymes expression of the recombinant E. coli harboring pRSFduet-1-bsgdh-tbadh was further verified by SDS-PAGE (as shown in Fig. S2†), and the results showed that both TbADH and BsGDH expressed as soluble proteins. Therefore, the recombinant E. coli harboring pRSFduet-1-bsgdh-tbadh was used in the subsequent study.3.2 Optimization of culture conditions
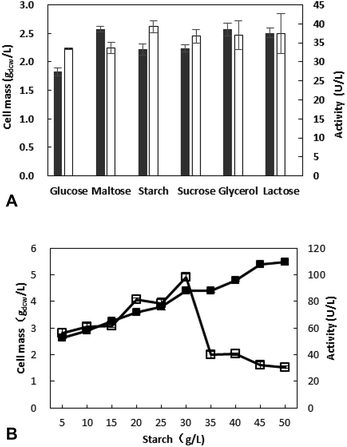
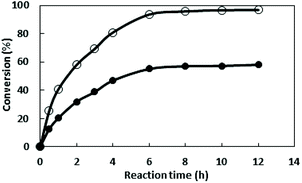
The concentration of starch was optimized in further work and the results revealed that the most appropriate concentration is 30 g L−1, and the activity is 98.4 U L−1 (Fig. 1B). With the increase of starch concentration, the cell mass is increasing, however a decrease of activity was observed when the starch concentration exceed 30 g L−1. This may be due to the fact that something from sugar metabolism affects the enzyme production at high sugar concentration.
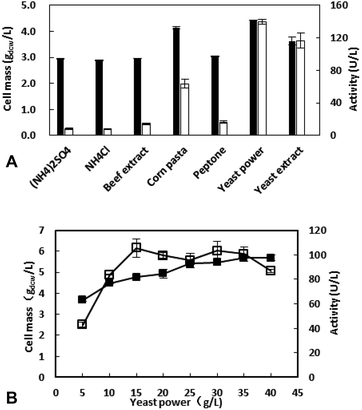
The concentration of yeast power was further investigated and the results showed that the appropriate concentration is 15 g L−1 with the highest enzyme activity of 105.4 U L−1. Although the amount of cells increased with the increasing of nitrogen source concentration, the metabolites of amino acids could inhibit the expression of the target protein when the concentration >15 g L−1.
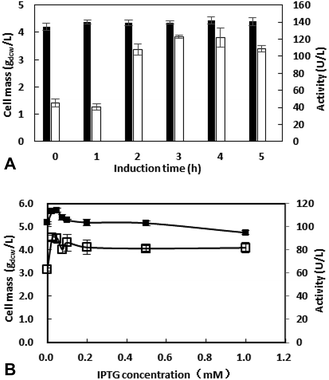
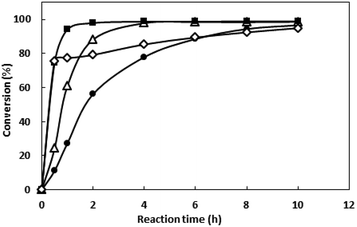
IPTG with exorbitant concentrations might affect the growth of E. coli BL21 (DE3),23 and the price of IPTG is expansive. Thus, IPTG with concentrations ranging from 0 to 1 mM were investigated and the results were shown in Fig. 3B. Between 0.1 to 1 mM, enzyme activity was similar. When IPTG concentration decreased to 0.025 mM, enzyme activity grows to 90.7 U L−1. Thus, the IPTG of 0.025 mM was added after inoculation for 3 h in the subsequent work.
3.3 Optimization of enzymatic asymmetric reduction of 1-boc-3-piperidinone
| Buffer | pH | Conversion (%) |
|---|---|---|
| a The collected cells were resuspended into the following buffers (final concentration, 200 mM): sodium phosphate (pH 6–8) and Tris–HCl (pH 8–9) and Gly-NaOH (pH 9–10). The suspension was incubated at 30 °C, 1000 rpm in thermo shaker for 15 min, and then added the substrate and glucose with a final concentration of 50 mM. The reaction was carried out at 30 °C for 10 h. | ||
| PBS | 6 | 52.5 ± 2.8 |
| 7 | 76.3 ± 1.8 | |
| 8 | 86.2 ± 0.1 | |
| Tris–HCl | 8 | 73.4 ± 0.1 |
| 9 | 74.6 ± 0.1 | |
| Gly-NaOH | 9 | 70.4 ± 0.2 |
| 10 | 56.4 ± 0.2 | |
| 5% | 10% | 15% | 20% | |
|---|---|---|---|---|
| a The collected cells were resuspended into PBS buffer (pH 8.0, 200 mM). The suspension was incubated at 30 °C, 1000 rpm in thermo shaker for 15 min, and then added the substrate and glucose with a final concentration of 50 mM. Various organic solvent were added into the reaction mixture with different final concentration (5%, 10%, 15% and 20%). The reaction was carried out at 30 °C for 10 h. | ||||
| Methanol | 80.1 ± 0.1 | 75.2 ± 0.2 | 68.7 ± 1.9 | 67.0 ± 2.4 |
| Isopropyl alcohol | 47.8 ± 2.5 | 21.4 ± 0.8 | 10.9 ± 0.3 | 6.3 ± 1.0 |
| Acetonitrile | 83.1 ± 0.5 | 46.4 ± 1.1 | 45.5 ± 0.8 | 2.5 ± 1.4 |
| Dimethyl sulfoxide | 9.7 ± 0.5 | 8.1 ± 0.4 | 7.4 ± 0.4 | 7.4 ± 0.4 |
| Glucose (mM) | Glucose/substrate (n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n) n) |
Conversion (%) |
|---|---|---|
| a Reaction condition: the collected cells were resuspended into PBS buffer (pH 8.0, 200 mM). The suspension was incubated at 30 °C, 1000 rpm in thermo shaker for 15 min, and then added the substrate (final concentration 50 mM) and glucose (final concentration of 25, 50, 75, 100 and 125 mM). Methanol of 5% (v/v) was used as the co-solvent. The reaction was carried out at 30 °C for 10 h. | ||
| 25 | 0.5 | 55.7 ± 4.3 |
| 50 | 1.0 | 86.4 ± 0.4 |
| 75 | 1.5 | 91.1 ± 2.3 |
| 100 | 2.0 | 93.1 ± 0.9 |
| 125 | 2.5 | 94.4 ± 1.8 |
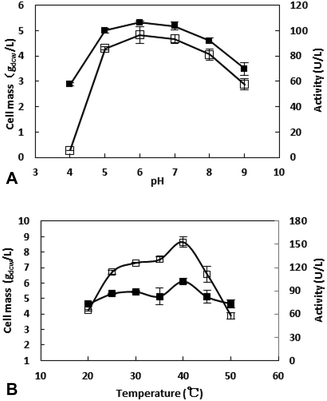
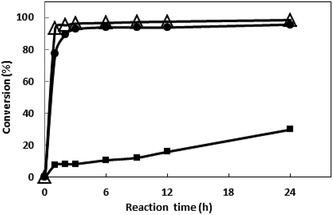
The bioreduction catalyzed by the recombinant E. coli was conducted at temperatures from 30 to 70 °C to investigate the effect of temperature. As shown in the Fig. 6, the conversion reached 96% after 10 h when the temperature was 30 °C. As the temperature increase, the reaction velocity increased gradually. At 50 °C, the conversion reached to 97.8% after 2 h, but as the temperature continues to rising, the reaction speed is reduced, which may be due to the enzyme inactivation. At 70 °C, the substrate was not converted (data no shown), and the enzyme may have been completely inactivated. The reaction was determined to be conducted at 50 °C in the subsequent work.
3.4 Asymmetric reduction of 1-boc-3-piperidinone in preparative scale
Finally, the asymmetric reduction of N-boc-3-piperidone was conducted at preparative-scale (see Section 2.8 for details). Optically pure S-NBHP of 1.67 g was recovered in a total yield of 83% and >99% ee (Fig. S3†). 1H NMR (500 MHz, CDCl3) δ 3.78–3.81 (m, 1H), 3.71 (s, 1H), 3.58 (s, 1H), 3.01–3.06 (m, 1H), 2.99 (dd, J = 12.7, 8.0 Hz, 1H), 2.74 (s, 1H), 1.90 (s, 1H), 1.75–1.77 (m, 1H), 1.45 (s, 11H).4. Conclusions
A recombinant E. coli BL21 (DE3) was constructed for co-expressing alcohol dehydrogenase and glucose dehydrogenase using the plasmid pRSFduet-1, which contains two multiple clone sites. The application potential of the constructed recombinant E. coli harbouring pRSFduet-1-bsgdh-tbadh was further improved through the optimization of the enzyme production condition and reaction condition. Finally, the bioreduction catalyzed by the recombinant E. coli was conducted at the substrate concentration of 500 mM without external addition of cofactors, giving satisfactory conversion (96.2%) and enantiomeric excesses for S-NBHP (ee value > 99%). The STY reported in this study (774 g L−1 d−1) was the highest reported for S-NBHP preparation so far. S-NBHP was obtained in a preparative experiment in 83% isolated yield and 99% ee value. Thus, the recombinant E. coli harbouring pRSFduet-1-bsgdh-tbadh was competitive and promising compared to other biocatalysts for asymmetrically reducing 1-boc-3-piperidinone. Further work about the immobilization of this recombinant E. coli and optimization of the reaction for large scale preparation is in the progress.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by Shanghai Collaborative Innovation Center of Fragrance, Flavor and Cosmetics (1021ZK170004002), the Shanghai Institute of Technology (YJ2015-36) and Shanghai Municipal Education Commission (ZZyyx15011).References
- Y. Chen, Y. Xu and J. B. Chen, MATEC Web Conf., 2015, 22, 1–5 CrossRef.
- I. Dragutan, V. Dragutan and A. Demonceau, RSC Adv., 2012, 2, 719–736 RSC.
- T. J. Harrison, B. O. Patrick and G. R. Dake, Org. Lett., 2007, 9, 367–370 CrossRef CAS PubMed.
- W. Y. Shen, Z. C. Shen, Z. Hu, Y. L. Lin and J. M. Wang, Chin. J. Pharm., 2013, 44, 436–437 CAS.
- M. S. Reddy, M. Narender and K. R. Rao, Tetrahedron, 2007, 63, 331–336 CrossRef CAS.
- G. W. Zheng, H. L. Yu, J. D. Zhang and J. H. Xu, Adv. Synth. Catal., 2009, 351, 405–414 CrossRef CAS.
- Y. Ni, J. Y. Zhou and Z. H. Sun, Process Biochem., 2012, 47, 1042–1048 CrossRef CAS.
- X. T. Zhou, R. Z. Zhang, Y. Xu, H. B. Liang, J. W. Jiang and R. Xiao, Process Biochem., 2015, 50, 1807–1813 CrossRef CAS.
- Q. Y. Wang, T. T. Ye, Z. Z. Ma, R. Chen, T. Xie and X. P. Yin, J. Ind. Microbiol. Biotechnol., 2014, 41, 1609–1616 CrossRef CAS PubMed.
- R. Lacheretz, D. G. Pardo and J. Cossy, Org. Lett., 2009, 11, 1245–1248 CrossRef CAS PubMed.
- Z. T. Sun, R. Lonsdale, A. Ilie, G. Li, J. Zhou and M. T. Reetz, ACS Catal., 2016, 6, 1598–1605 CrossRef CAS.
- X. Ju, Y. Y. Tang, X. L. Liang, M. Q. Hou, Z. H. Wan and J. H. Tao, Org. Process Res. Dev., 2014, 18, 827–830 CrossRef CAS.
- H. M. Zhao and V. D. D. WA, Curr. Opin. Biotechnol., 2003, 14, 583–589 CrossRef CAS PubMed.
- F. Hollmann and A. Schmid, Biocatal. Biotransform., 2004, 22, 63–88 CrossRef CAS.
- G. W. Huisman, J. Liang and A. Krebber, Curr. Opin. Chem. Biol., 2010, 14, 122–129 CrossRef CAS PubMed.
- M. Kataoka, L. P. S. Rohani, M. Wada, K. Kita, H. Yanase, I. Urabe and S. Shimizu, Biosci., Biotechnol., Biochem., 1998, 62, 167–169 CrossRef CAS PubMed.
- H. Gröger, F. Chamouleau, N. Orologas, C. Rollmann, K. Drauz, W. Hummel, A. Weckbecker and O. May, Angew. Chem., Int. Ed., 2006, 45, 5677–5681 CrossRef PubMed.
- M. Peretz, O. Bogin, S. Tel-Or, A. Cohen, G. Li, J. S. Chen and Y. Burstein, Anaerobe, 1997, 3, 259–270 CrossRef CAS PubMed.
- K. Yamane, M. Kumano and K. Kurita, Microbiology, 1996, 142, 3047–3056 CrossRef CAS PubMed.
- A. Banerjee, P. Kaul and U. C. Banerjee, Appl. Microbiol. Biotechnol., 2006, 72, 77–87 CrossRef CAS PubMed.
- A. K. Khandelwal, V. K. Nigam, M. K. Mohan, P. Ghoshand and B. Choudhury, J. Chem. Technol. Biotechnol., 2007, 82, 645–651 CrossRef.
- J. F. Liu, Z. J. Zhang, A. T. Li, J. Pan and J. H. Xu, Appl. Microbiol. Biotechnol., 2011, 89, 665–672 CrossRef CAS PubMed.
- P. Dvorak, L. Chrast, P. I. Nikel, R. Fedr, K. Soucek, M. Sedlackova, R. Chaloupkova, V. Lorenzo, Z. Prokop and J. D. Dvorak, Microb. Cell Fact., 2015, 14, 201–215 CrossRef PubMed.
- X. Wu, X. D. Gou and X. J. Chen, Process Biochem., 2015, 50, 104–110 CrossRef CAS.
- W. Zhu, B. Wang, Q. P. Zhang, H. Wu and B. H. Li, CN Patent 104059952 A, Syncozymes (Shanghai) Co., Ltd., 2014.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra08100j |
| This journal is © The Royal Society of Chemistry 2019 |